Abstract
The Africanization process of the honeybee Apis mellifera in America is considered one of the most spectacular biological invasions documented so far. It started in São Paulo (Brazil) in 1956 and the process has extended to almost all the continent (from the United States, to Argentina and Uruguay). The aim of this study was to evaluate the present status of Africanization in Uruguay after 40 years of its entrance, as well as to determine the circulating honeybee haplotypes through genetic (mitochondrial DNA) and morphometric approaches. Results showed that Africanized bees are completely established in Uruguay, representing 80 % of the analyzed colonies, distributed throughout the whole country. A great genetic diversity was found, detecting eight different haplotypes. Some of them have been previously reported in the country, while others are new. The persistence of the non-African haplotypes in western provinces (Paysandú, Río Negro, Soriano and Colonia) may be due to a strong importation of European queens. This fact is probably avoiding the complete Africanization of the Uruguayan colonies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The Africanization process of the honeybee Apis mellifera in America is considered one of the most spectacular biological invasions so far documented. This process started with the unplanned dispersion of African queens from Rio Claro (São Paulo, Brazil) in 1956. African bees were introduced in Brazil as an attempt to obtain an organism well adapted to the Brazilian environmental conditions, in order to increase production rates (Kerr 1957). Since then, the A. mellifera scutellata queens have been hybridizing with local drones of European origin (Smith 1991) and the process has extended to almost the whole continent (from California, United States, to the Argentinian and Uruguayan provinces located between the 35° and 40° south latitude) (Abrahamovich et al. 2007; Kim and Oguro 1999; Sheppard et al. 1999).
In Uruguay, a country with 3,200 beekeepers and 500,000 colonies (MGAP 2012), local honeybees are hybrids generated by European honeybees (A. m. mellifera, brought to the country in the 1800s; Cordara 2005) and Africanized bees from Brazil (derived from Apis mellifera scutellata; Burgett et al. 1995; Diniz et al. 2003; Collet et al. 2006).
In previous studies, a latitudinal Africanization gradient was established according to the percentage of Africanized colonies and to the percentage of African genes present in the bee genome (Burgett et al. 1995; Diniz et al. 2003). Taking into account the intensive trading between beekeepers, it is possible to hypothesize that Africanized colonies have spread around the country.
The aim of this work was to evaluate the present status of Africanization in Uruguay as well as to identify circulating mitochondrial COI-COII haplotypes.
2 Material and methods
2.1 Samples
Samples from all over the country were collected during March 2011. A total of 103 non-migratory apiaries were sampled, being the number of selected apiaries representative of the number of colonies registered in the National Register of Honeybee Colonies (DIGEGRA-MGAP). The number of colonies sampled per province was representative of the density of colonies per province.
One colony was randomly selected from each apiary. Adult bees were collected from peripheral combs, and stored in ethanol (70 %).
2.2 Genetic characterization
Taking into account that all the honeybees of each colony have inherited the same mitochondrial DNA, only one bee per colony was randomly selected and genetically characterized. The mitochondrial DNA was extracted using the protocol described by Walsh et al. (1991) with some modifications. One posterior leg was incubated at 37 ºC for 30 min to evaporate the residual ethanol. After that, 100 μl of Chelex 5 % (Sigma-Aldrich, SL, USA) and 5 μl proteinase K solution (20 mg ml−1; Promega, Madison, WI, USA) were added and the sample was incubated for 1 h at 55 ºC, 15 min at 99 ºC, 1 min at 37 ºC, and 15 min at 99 ºC (T1 Thermocycler; Biometra, Goettingen,Germany). Two microlitres of the DNA solution was used for the PCR amplification of the intergenic COI-COII region using the primers E2 (5′-GGCAGAATAAGTGCATTG-3′) and H2 (5′-CAATATCATTGAT-GACC-3′) (Garnery et al. 1992). The PCR mixture of 20 μl contained 1× reaction buffer, 1 mM MgCl2, 0.15 μM of each primer, 0.5 mM of each dNTP and 0.5 U/μl Taq polymerase (Invitrogen, Carlsbad, CA, USA). A negative control without DNA was included. The PCR cycling program included an initial denaturation step for 5 min at 94 ºC, followed by 35 cycles of 45 s at 92 ºC, 45 s at 48 ºC, 2 min at 62 ºC, and a final extension of 20 min at 65 ºC (T1 Thermocycler; Biometra) (Garnery et al. 1993). The size of the amplified DNA product was determined by electrophoresis in a 1 % agarose gel, stained with GelRed (Biotum, Hayward, CA, USA) and visualized by UV light (T13; Biometra). Then, the product was digested with FastDigest DraI (Fermentas, São Paulo, Brazil) according to manufacturer’s recommendations, analyzed by electrophoresis in a 12 % polyacrylamide gel under native conditions for 15–16 h at 80 V, and stained with GelRed (Biotium). Mitochondrial COI-COII haplotypes were determined according to Franck et al. (1998). In those cases in which the haplotypes could not be clearly distinguished due to small differences in their restriction fragments size, PCR products were sequenced (Macrogen, Seoul, Korea). The identity of these haplotypes was confirmed through comparison with published sequences of the NCBI (National Center for Biotechnology Information), and DraI in silico restriction using BioEdit version 7.0.0 software (Ibis Biosciences, Carlsbad, CA, USA).
2.3 Morphometric characterization
Bees from colonies which showed a non-African origin after mitochondrial DNA analysis were morphometrically characterized using the FABIS method, a simple technique that allows the discrimination between African and European honeybees (Rinderer et al. 1987). In order to determine the Africanization probability, the average wing and femur lengths of ten bees were used.
The percentage of Africanized colonies was defined as the number of colonies which showed African origin (by genetic and morphometric analysis) in relation with the total colony numbers.
3 Results
In order to determine the percentage of Africanized colonies in Uruguay as well as the circulating honeybee mitochondrial haplotypes, a genetic and morphometric approach was performed including honeybee samples from all over the country.
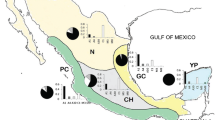
The intergenic COI-COII region was amplified and 600 or 850-bp products were visualized in all the samples (Supplementary material). After digestions of these products, different patterns were obtained.
Most of the patterns were associated with the African origin of the samples, with haplotype A4 being the most frequently detected (52.4 %) as present in almost every province (Supplementary material) (Figure 1). Haplotypes A30 (16.5 %), A1 (9.7 %) and A26 (1 %) were also detected. The non-African haplotypes found were C1 (8.7 %), C2 (6.8 %), M4 (3.8 %) and M7 (0.97 %) (Supplementary material) (Figure 1). Samples in which the haplotypes could not be clearly distinguished due to small differences in their restriction fragments size were sequenced and identified (Accession numbers KJ661741, KJ661742 and KJ661743).
Genetic analyses indicated that 79.6 % (82 out of 103) of the analyzed colonies were Africanized. Since it is possible that the 21 remaining colonies inherited African characteristics by a paternal line (not represented in the mitochondrial DNA), the African origin probability of these samples was calculated by a morphometric approach. All those samples presented 0 % of an African origin probability, except one, which showed 65 % of African origin probability taking into account the wing length, but a 3 % of African origin probability considering together the wing and leg lengths.
4 Discussion
In this study we determined that Africanized bees are completely established in Uruguay and widely distributed within the country, representing 80 % of the colonies. A high genetic diversity was detected, including haplotypes from African and non African origins. Haplotypes A1, A4, A26, A30, C1 and M4 had already been described in our country a few years ago (Collet et al. 2006), while haplotypes C2 and M7 are described for the first time in Uruguay.
Haplotypes C1, A4 and A1 have been identified in close countries. However, their prevalence and distribution were quite different, which could reflect different colony management, and adaptation to the different environmental conditions. For example, in Buenos Aires (Argentina) most of the haplotypes corresponded to C1 (92.31 %) and the African ones were in low proportion (5.02 %) (Abrahamovich et al. 2007). Haplotype A4 was the most common in Uruguay, Centre and South of Brazil, and Venezuela (Clarke et al. 2001), while it has been reported that A1 is one of the most common haplotypes in Mexico (Clarke et al. 2001). Collet et al. (2006) proposed the existence of a negative correlation between the latitude and the presence of A1 haplotype, which is in agreement with the low prevalence of this haplotype in Uruguay.
Geographic distribution of the mitochondrial COI-COII haplotypes in Uruguay was not uniform, as most of them (A4, A1, A30, C1, C2, M4 and M7) were concentrated in Paysandú, Río Negro, Soriano and Colonia. These provinces are located in the West region of the country, which is the main beekeeping area. Here, experienced beekeepers frequently import European honeybee queens (mainly A. mellifera ligustica) looking for behavioral and productive features, which could explain our results.
The loss of the latitudinal Africanization gradient from north to south observed by Sheppard et al. (1999) and Diniz et al. (2003), and the high Africanization rate detected in this study, can be explained by the commercialization of honeybee queens from Northern provinces (close to the border with Brazil) to the rest of the country.
For all these reasons, we suggest that the complete Africanization of the country is limited mainly by the artificial introduction of European honeybees. This is in accordance with Abrahamovich et al. (2007), who attributed the low level of Africanization in Buenos Aires province, Argentina, to the artificial introduction of Italian queens and selection programs in that country.
It would be interesting to evaluate if a gradual Africanization pattern (Sheppard et al. 1999; Diniz et al. 2003) persists when assessing the proportion of African alleles in the bee genome. In this aspect, a recent study based on Argentinean samples using SNPs, reported that there is still a north to south Africanization gradient (Whitfield et al. 2006).
In conclusion, the high Africanization percentage of Uruguayan honeybees reported here (80 %) suggests that these bees have adapted to the country climatic conditions and that the beekeeping industry has become used to dealing with Africanized bees.
References
Abrahamovich, A.H., Atela, O., De la Rúa, P., Galián, J. (2007) Assesment of the mitocondrial origin of honey bees from Argentina. J. Apic. Res. and Bee World. 46, 191–194
Burgett, M., Shorney, S., Cordara, J., Gardiol, G., Sheppard, W.S. (1995) The present status of Africanized honey bees in Uruguay. Am. Bee J. 135, 328–330
Clarke, K.E., Oldroyd, B.P., Javier, J., Quezada-Euan, G., Rinderer, T.E. (2001) Origin of honeybees (Apis mellifera L.) from the Yucatan peninsula inferred from mitochondrial DNA analysis. Mol. Ecol. 10, 1347–1355
Collet, T., Ferreira, K.M., Arias, M.C., Soares, A.E.E., Del Lama, M.A. (2006) Genetic structure of Africanized honeybee populations (Apis mellifera L.) from Brazil and Uruguay viewed from mitochondrial DNA COI-COII patterns. Heredity. 97, 329–335
Cordara, J.J. (2005) La Historia de la Apicultura en Uruguay. Facultad de Ciencias Agrarias. Universidad de la empresa, Montevideo, Uruguay
Diniz, N.M., Soares, A.E.E., Sheppard, W.S., Del Lama, M.A. (2003) Genetic structure of honeybee populations from southern Brazil and Uruguay. Genet. Mol. Biol. 26, 47–52
Franck, P., Garnery, L., Loiseau, A., Oldroyd, B.P., Hepburn, H.R., Solignac, M., Cornuet, J.-M. (1998) Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity. 86, 420–460
Garnery, L., Cornuet, J.M., Solignac, M. (1992) Evolutionary history of the honeybee Apis mellifera inferred from mitochondrial DNA analysis. Mol. Ecology. 1, 145–154
Garnery, L., Solignac, M., Celebrano, G., Cornuet, J.M. (1993) A simple test using restricted PCR-amplified mitochondrial DNA to study the genetic structure of Apis mellifera L. Experientia. 40, 1016–1021
Kerr, W.E. (1957) Introduction of african honey bees in Brazil. Brasil Apicola. 3, 211–213
Kim, K.T., Oguro, J. (1999) Update of the Status of the Africanized Honey Bees of the Western States. West. J. Med. 170, 220–222
MGAP. (2012). Anuario 2012. www.mgap.gub.uy/
Rinderer, T.E., Sylvester, A., Buco, S.M., Lancaster, V.A., Herbert, E.W., Collins, A.M., Hellmich, R.L. (1987) Improved simple techniques for identifying Africanized and European honey bees. Apidologie. 18, 179–196
Sheppard, W.S., Rinderer, T.E., Garnery, L., Shimanuki, H. (1999) Analysis of the Africanized honey bee mitochondrial DNA reveals further diversity origin. Genet. Mol. Biol. 22, 73–75
Smith, D.R. (1991) African bees in the Americas: Insights from Biogeography and Genetics. Tree. 6, 17–21
Walsh, P.S., Metzger, D.A., Higuchi, R. (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 10, 506–513
Whitfield, C.W., Behura, S.K., Berlocher, S.H., Clark, A.G., Johnston, J.S., Sheppard, W.S., Smith, D.R., Suarez, A.V., Weaver, D., Tsutsui, N.D. (2006) Thrice out of Africa: ancient and recent expansions of the honey bee, Apis mellifera. Science. 314, 642–645
Acknowledgements
The authors thank Ciro Invernizzi, Gustavo Ramallo, the Apiculure section of INIA La Estanzuela (Colonia, Uruguay) and the AMSUD Pasteur Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Manuscript editor: Marina Meixner
Modifications génétiques chez Apis mellifera après 40 ans d’africanisation
Abeille africaine / haplotype / ADN mitochondrial / Amérique du Sud / Uruguay
Genetische Veränderungen in Apis mellifera nach vierzig Jahren Afrikanisierung
Afrikanisierung / afrikaniserte Honigbiene / mitochondriale DNA / Haplotyp
Rights and permissions
About this article
Cite this article
Branchiccela, B., Aguirre, C., Parra, G. et al. Genetic changes in Apis mellifera after 40 years of Africanization. Apidologie 45, 752–756 (2014). https://doi.org/10.1007/s13592-014-0293-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-014-0293-2