Abstract
Osteoporosis is a highly prevalent disease characterized by bone mass loss and structural deterioration. There are evidences that altered differentiation of human bone marrow mesenchymal stromal/stem cells (hBMSCs) is a major cause for osteoporosis. Recent studies suggest that circular RNAs (circRNAs) are dysregulated in osteoporosis patients and involved in the pathogenesis of osteoporosis. In the present study, we are aimed to analyze the circRNA expression profiles in osteoporosis patients and identify potential circRNAs that involved in the differentiation of hBMSCs during osteoporosis. Transcriptome RNA-sequencing was conducted to search for differentially expressed circRNAs. Transwell assay, ARS and ALP staining, and ectopic bone formation model were performed to evaluate osteogenic differentiation of hBMSCs. RNA pull-down assay, RNA immunoprecipitation, western blot, and in vitro binding assay were conducted to evaluate the interaction of circRNAs and RNA-binding protein HuR. We found that hsa_circ_0008842 (designated as circZNF367) was upregulated in osteoporosis patients and decreased in hBMSCs during osteogenic differentiation. CircZNF367 overexpression suppressed migration, invasion and osteogenic differentiation of hBMSCs in vitro and in vivo. In comparison, knockdown of circZNF367 promoted migration, invasion and osteogenic differentiation of hBMSCs. CircZNF367 could interact with the RNA-binding protein HuR, thus reduced the mRNA stability of LRP5. Furthermore, HuR overexpression or LRP5 restoration abrogated the effects of circZNF367 overexpression on osteogenic differentiation of hBMSCs. Our results indicated that circZNF367 played a role in osteogenic differentiation of hBMSCs via reducing HuR-mediated mRNA stability of LRP5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoporosis is characterized by decreasing of bone mass, impairment of bone microarchitectural and increase risk of fragility fractures [1]. It is defined as bone mineral density (BMD) T score of – 2.5 or less. Osteoporosis is a highly prevalent disease. It is estimated that one in three women and one in five men that aged over 50 will experience osteoporotic fractures during their lifetime [2]. There are more than 8.9 million fractures caused by osteoporosis every year worldwide [3]. Osteoporotic fractures can dramatically reduce the quality of life and cause a two–eightfold increase risk of mortality [3]. Indeed, the mortality rate of osteoporotic fractures of the hip and spine is as high as 20% [4]. The main risk factors for osteoporosis are old age and postmenopausal period, and other risk factors include inflammatory arthropathy, immobilization, medications and endocrine disorders [5]. Osteoporosis is caused by an imbalance between bone resorption and formation. Bones are constantly resorbed by osteoclasts and regenerated by osteoblasts, and thus remain a dynamic balance throughout life [6]. The bone remodeling process is critical for maintaining bone density and mineral homeostasis. Osteoblasts are derived from mesenchymal stromal/stem cells that reside in the bone marrow (BMSCs). During intramembranous bone formation, BMSCs proliferate and differentiate into osteoblasts, and thus promote extracellular matrix proteins production and mineral deposition. Therefore, elucidating the molecular mechanism of osteoblasts differentiation might help to develop new treatments for osteoporosis.
BMSCs, a non-hematopoietic stem cell population first found in bone marrow, can differentiate into osteoblasts or adipocytes, and thus act an important role in maintaining bone stability. There are evidences that altered differentiation of BMSCs is a major cause for osteoporosis [7]. Age-related osteoporosis is correlated with marrow fat accumulation and decrease of bone formation [8]. BMSCs of aging person exhibit lower capacity to differentiate into osteoblasts and higher capacity to differentiate into adipocytes [9]. Actually, bone marrow fat accumulation has been found in the majority of bone loss diseases [10]. The differentiation of BMSCs to osteoblasts or adipocytes is delicately regulated by many factors, including various signaling pathways and transcriptional factors. Wnt/β-catenin signaling pathway acts a critical role in cell fate determination and differentiation. It is also critical for bone mass maintaining [11]. Previous studies demonstrate that Wnt/β-catenin signaling promotes osteogenic differentiation and suppresses adipogenic differentiation of BMSCs [12]. Low-density lipoprotein receptor-related protein 5 (LRP5), a coreceptor of β-catenin, can activate Wnt/β-catenin signaling and prevent phosphorylation and degradation of β-catenin [13]. LRP5 variants have been associated with reduced bone marrow density, increased fracture risk and osteoporosis [14].
Circular RNAs (circRNAs) are one kind of non-coding RNAs with covalently closed-loop structure. Most of circRNAs are generated by back-splicing of exons from precursor mRNAs. CircRNAs have various biological functions due to their diversity, stability, evolutionary conservation and tissue-specific patterns [15]. CircRNAs can interact with proteins and function as protein decoys, scaffolds and recruiters [16]. RNA-binding proteins (RBP) are important regulators for post-transcriptional events, including RNA splicing, stability, translation and degradation. Accumulated studies indicate that circRNAs can interact with RNA-binding proteins and modulate their functions [17]. For example, circular RNA circDLC1 interacts with HuR to decrease the stability of MMP1 mRNA, and thus inhibits liver cancer progression [18]. There are growing evidence that circRNAs are involved in the development of many diseases, including osteoporosis [19]. Moreover, circRNAs are dysregulated in osteoporosis patients [20]. Dysregulated circRNAs are involved in bone metabolism and differentiation of bone marrow BMSCs, too [20]. In the present study, circRNA expression profiles of bone tissues from osteoporosis patients were determined by transcriptome RNA-sequencing. CircZNF367 (hsa_circ_0008842), one of the top ten upregulated circRNAs, was found to suppress migration, invasion and osteogenic differentiation of hBMSCs in vitro and in vivo. Furthermore, CircZNF367/HuR interaction reduces the mRNA stability of LRP5, thus reduced LRP5 protein levels and inactivated Wnt/β-catenin signaling. Our study provided a novel role of circZNF367 in osteoporosis and suggested it as a potential therapeutic target for osteoporosis treatment.
Materials and methods
Patient samples and cell culture
All participants enrolled in our study signed the written informed consents and all procedures involved human tissues were approved by the Ethics Committee of The Third Xiangya Hospital (Approve no. 2017-R17021). The protocol of our study for human was followed with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Bone tissues of osteoporosis patients (n = 23) and healthy controls (n = 18) were collected at The Third Xiangya Hospital from March 2017 to December 2018. Some pieces of bone tissue were collected when the participants were suffering from external traumatic fracture. The bone tissue samples were stored in liquid nitrogen for further use. The osteoporosis patients did not take any medicine that could affect bone metabolism, nor did they suffered from any other disease that might affect their skeletal tissue before surgery. The healthy controls were known to have not suffered from any chronic condition or disease that may have affected their skeletal tissue. Human bone marrow mesenchymal stromal/stem cells (hBMSCs) were isolated as previously described via immunomagnetic isolation of STRO-1 positive cells from a whole population of bone marrow mononuclear cells [21]. The isolated hBMSCs were maintained with Alpha Modified Eagle’s Medium (α-MEM) (Thermo Fisher, USA) supplemented 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin and 100 mg/mL streptomycin (Thermo Fisher, USA) at 37 °C in a humidified atmosphere containing 5% CO2. For osteogenic differentiation, hBMSCs were cultured with osteogenic-induced medium (OIM) (Sigma-Aldrich #SCM121) for 7–14 days. For adipogenic differentiation, hBMSCs were cultured with adipogenic-induced medium (AIM) (Sigma-Aldrich #SCM122) for 14 to 21 days.
Transcriptome RNA-sequencing
TRIzol reagent (Invitrogen, USA) was used to extract total RNAs from bone tissues. Ribo-off rRNA Depletion Kit (Human/ Mouse/ Rat) (Vazyme #N406) was used to purify the total RNA. Complementary DNA library was built using the VAHTS Universal V8 RNA-seq Library Prep Kit for lllumina (Vazyme #NR605). Next, the cDNA libraries were sequenced by the Illumina HiSeq2500 platform (Illumina, USA). Gene expression levels was calculated by RPKM values. Dysregulated genes were defined as |log2 Fold Change|≥ 1 and adjusted p < 0.05. Each sample had three repeats Table 1.
Plasmid constructs
The full length of circZNF367 was cloned into the pLC5-ciR vector (Geenseed, China) to build the circZNF367 overexpression lentivirus vector. The empty pLC5-ciR vector was regarded as control. The AU-rich sequence mutant circZNF367-mut was generated using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies #210,518, USA) according to the manufacturers’ instruction. HuR or LRP5 expression lentivirus vector was constructed by cloning the coding sequence for HuR or LRP5 into the pCDH-Puro (System Biosciences #CD510B-1) lentivirus vector. To knock down HuR, LRP5 or circZNF367, short hairpin RNAs (sh-RNAs) specifically targeting HuR (sh-HuR-1 and sh-HuR-2), LRP5 (sh-LRP5-1 and sh-LRP5-2) or the junction sites of circZNF367 (sh-circ-1 and sh-circ-2) were introduced into the pLKO.1 plasmid. The empty pLKO.1 plasmid introduced with a non-targeting sequence was used as sh-NC control. GST or Flag-tagged HuR truncates were constructed as previously reported [22]. The sequences for sh-RNAs were listed in supplementary Table 1.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNAs from tissue samples and cell lines were extracted by TRIzol reagent (Invitrogen, USA). For subcellular RNA fractionation assays, cells were lysed by lysis buffer (50 mM Tris–HCl pH7.5, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40) for 90 s, then centrifuged at 9000 rpm for 3 min. The supernatant and the precipitant were used to isolate RNA from cytoplasm and nucleus, respectively. PrimeScript RT reagent Kit (Takara, Japan) was used to synthesize complementary DNA. Real-time PCR was performed by SYBR Premix Ex Taq kit (Takara, Japan). The gene expression level was calculated by the 2−ΔΔCq method and normalized to U6 or GAPDH. The sequences for primers are listed in supplementary Table 2. All samples were done in triplicates.
RNase R treatment
A total of 3 μg RNAs were incubated with RNase R (5 U/μg, Epicentre Technologies, USA) for 30 min at 37 °C. The treatment was stopped by heating at 70 °C for 10 min. The products was used for qRT-PCR analysis. All samples were done in triplicates.
Actinomycin D assay
Cells (2 × 105) were seeded in 6-well plates for 48 h. When the confluency reached 80% -90%, cells were treated with 2 μg/ml Actinomycin reagent (Sigma, USA) for 0, 4, 8 and 12 h. Collected total RNAs by TRIzol reagents at the indicated time points and evaluated by qRT-PCR. All samples were done in triplicates.
RNA fluorescence in situ hybridization (FISH)
CircZNF367 probes specifically targeting the junction sites were labeled with Cy3 and synthesized by Sangon Biotech (Shanghai, China). The hBMSCs were seeded on coverslip for 48 h, then fixed by 4% paraformaldehyde for 10 min at room temperature and permeabilized by 1% Triton X-100 for 15 min. The cells were incubated with Cy3-labeled circZNF367 probe at 37 °C overnight. DAPI was used to stain the nucleus. Images were taken by a fluorescence microscope (Eclipse NI, Nikon, Japan). The sequences of circZNF367 probe were: 5’-GGCTG GCGAG GATGG AATCC-3’.
Cell Counting Kit-8 (CCK-8) assay
The hBMSCs (2500/well) were seeded in 96-well plate and treated as indicated. At each time points, cell viability was determined by Cell Counting Kit-8 (Beyotime, China) as protocol instructed. Briefly, 10 μL CCK-8 solution was added in each well and incubated at 37 °C for 1 h. Then, optical density at 450 nm was recorded by a microplate reader. Each sample was done in triplicates.
Colony formation assay
Seeded the cells (4000/well) in 6-well plates and cultured for 14 days without disturbance. The colonies were then fixed by 4% paraformaldehyde for 15 min and stained with crystal violet for 60 min at room temperature. Images were obtained by a scanner. Each sample was done in triplicates.
Flow cytometry
Digested the cells into single cells suspensions. Then, 1 × 106 cells were incubated with Annexin-V-FITC (Sigma, USA) and propidium iodide (PI) (Sigma, USA) for 20 min avoiding light. Signal at 488/530 nm were evaluated by FACS LSR Fortessa (BD Biosciences, USA). Each sample was done in triplicates.
Transwell migration and invasion assay
The transwell chamber (Millipore, USA) was used for transwell migration and invasion assay. Cells (1 × 105) were seeded in the upper chamber without serum. The lower chamber was filled with medium with 10% fetal bovine serum. Cells were allowed to migrate towards the lower chamber for 48 h. To evaluate cell invasion, the chamber was pre-coated with 80 μL Matrigel (Corning, USA). The migration or invasion cells were fixed by 4% PFA for 15 min at room temperature and stained with 0.5% crystal violet for 30 min at room temperature. Images were obtained by a microscope. Each sample was done in triplicates.
Quantification of alkaline phosphatase (ALP) activity
ALP activity was detected by ALP detection kit (Beyotime #P0321S, China) according to the manufacturers’ instructions. Briefly, samples were mixed with the detection reagent, then incubated at 37 °C for 10 min. The optical density at 405 nm by a microplate reader. Each sample was done in triplicates.
Alkaline phosphatase staining
Human BMSCs were seeded in 6-well plates and cultured with osteogenic-induced medium for 14 days, then cells were washed with PBS and fixed by 95% cold ethanol. Next, cells were stained by the ALP staining kit (CWBIO, China) according to the manufacturers’ instructions. The images was taken by a microscope.
Alizarin red staining (ARS) and quantification
Human BMSCs were seeded in 6-well plates and cultured with osteogenic-induced medium for 14 days, then cells were washed with PBS and fixed by 4% paraformaldehyde for 15 min at room temperature. Cells were stained with 0.1% Alizarin red (Solarbio, China) according to the manufacturers’ instructions. Images were taken by a fluorescence microscope (Eclipse NI, Nikon, Japan). To quantify mineralization, the bound dye was dissolved in 10% cetylpyridinium chlorid and absorbance at 570 nm was detected by a microplate reader (BioTek Synergy 2, BioTek, USA). Each sample was done in triplicates.
Ectopic bone formation assay
To evaluate the role of circZNF367 in regulating new bone formation in nude mice, hBMSCs (2 × 106) were transduced with circZNF367, EV, sh-circ-1, sh-circ-2 or sh-NC expression lentivirius vector and resuspended with tricalcium phosphate (TCP) carrier (Bicon, USA) scaffolds at 37 °C for 1 h. Then, the cells were implanted into the dorsal subcutaneous space of nude mice (n = 5 for each group). After 8 weeks of ectopic implantation, the mice were anaesthetized by inhalation with 3% isoflourane and killed by broking the neck. The samples were harvested for hematoxylin and eosin (HE) and Masson's staining.
RNA immunoprecipitation (RIP) assay
RIP assay was performed via the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instruction. Briefly, 2 × 107 cells were lysed with RIP lysis buffer (20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.5% NP-40, 0.5% Triton X-100) supplemented with proteinase inhibitors and RNase inhibitors (Invitrogen, USA), then incubated with magnetic beads bounded with antibody against IgG (Cell signaling #3900, 1: 50) or HUR (Cell signaling #12,582, 1: 50). The RNAs pulled down by these antibodies were evaluate by qRT-PCR.
Western blot
RIPA buffer (Beyotime, China) was used to extract total protein from culture cells. BCA kit (Thermo Fisher, USA) was used to determine protein concentration. A total of 20 μg protein lysates was separated by 10% or 12% SDS-PAGE and transferred to a PVDF membrane (Bio-Rad, USA). Next, membrane was blocked with 5% non-fat milk for 1 h at room temperature, and incubated with primary antibodies at 4 °C overnight and secondary antibodies for 1 h at room temperature. The protein band was revealed by ECL plus kit (ThermoFisher, USA) using the ChemiDoc Touch Imaging System (Bio-Rad, USA). The specific antibodies were: Lamin A/C Mouse mAb (Cell signaling #4477, 1: 1000), GAPDH Rabbit mAb (Cell signaling #5174, 1: 1000), Anti-Osteocalcin antibody (Abcam #ab133612, 1: 1000), Anti-RUNX2 antibody (Abcam #ab192256, 1: 500), HUR (Cell signaling #12,582, 1: 1000), FMRP Antibody (Cell signaling #4317, 1: 1000), IGF2BP1 Rabbit mAb (Cell signaling #8482, 1: 1000), TAF15 Rabbit mAb (Cell signaling #28,409, 1: 1000), LRP5 Rabbit mAb (Cell signaling #5731, 1: 1000), β-Catenin Rabbit mAb (Cell signaling #8480, 1: 1000), active β-Catenin (Cell signaling #4270, 1: 1000), CEBPα Rabbit mAb (Cell signaling #8178, 1: 1000) and PPARγ Rabbit mAb (Cell signaling #2443, 1: 1000).
Biotin-labeled RNA pull-down
The cells (2 × 107) were lysed with IP lysis buffer (50 mM Tris–HCl pH 7.4, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40) supplemented with proteinase inhibitors and RNase inhibitors (Invitrogen, USA). The biotin-labeled RNA probes for circZNF367 specifically targeting the junction sites were synthesized by Sangon Biotech (Shanghai, China). The cell lysates were incubated with 400 pmol of biotin-labeled RNA probes for circZNF367 (anti-sense) or scramble probes (sense) for 2 h at 4 °C, and 30 μL of Streptavidin C1 magnetic beads (Invitrogen, USA) for 1 h at 4 °C. For in vitro cyclized circZNF367 RNA pull-down assay, biotin-labeled circZN367 and circZNF367-mut were in vitro transcribed using the Bition RNA Labeling Mix (Roche, Switzerland) with T7 polymerase according to previously reported [23]. Next, the linear transcripts were incubated with guide oligonucleotide targeting the circular RNA junction sites, and circularized by T4 RNA ligase, digested by RNase R, and purified by RNeasy Mini Kit (Qiagen, USA). Cell lysates were incubated with 2 ug of biotin-labeled circZN367, circZNF367-mut or the anti-sense control of circZNF367 for 4 h at 4 °C, then 30 μL of Streptavidin C1 magnetic beads (Invitrogen, USA) for 1 h at 4 °C. The retrieved mRNAs or proteins were evaluated by qRT-PCR or western blot. The sequences for circZNF367 probe were: 5'-UGGGC CUACC ACGUC GGAUU CCAUC CUCGC CAGCC ACUCG GCCGC GGCCU-3'; scramble probe: 5'-AGGCC GCGGC CGAGU GGCUG GCGAG GAUGG AAUCC GACGU GGUAG GCCCA-3'.
Statistical analysis
Data were analyzed using GraphPad Prism 8.0 software. Two-tailed Student’s t test and One-way ANOVA (LSD post hoc test) were used to compare difference between two or more groups. Data were shown as mean ± SD. P ≤ 0.05 was considered statistically significant.
Results
CircZNF367 is upregulated in osteoporosis patients
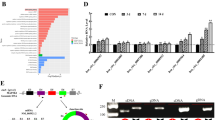
To elucidate the potential role of circRNAs in osteoporosis, bone tissues from osteoporosis patients (N = 3) and matched healthy controls (N = 3) were subjected for transcriptome RNA-sequencing. Differentially expressed circRNAs were defined as |log2 fold change|≥ 2 and P value < 0.05. In our study, 474 circRNAs were upregulated while 636 circRNAs were downregulated in bone tissues from osteoporosis patients (Supplementary Table 3). The top 20 upregulated or downregulated circRNAs were depicted in the heatmap (Fig. 1A). Previous studies indicate that differentiation of BMSCs to osteoblasts or adipocytes is involved in the development of osteoporosis [6, 7, 24]. We speculated that some of these dysregulated circRNAs might play a role in the differentiation of human bone marrow stem cells (hBMSCs). Thus, the top 10 upregulated circRNAs were introduced into hBMSCs, respectively, and the infected cells were cultured with osteogenic medium for 7 days to evaluate osteogenic differentiation. Alkaline phosphatase (ALP) is the marker of osteoblast differentiation. We found that the ALP activity of hBMSCs was significantly decreased by hsa_circ_0008842 compared with empty vector (EV) control or other circRNAs (Fig. 1B). The host gene for hsa_circ_0008842 is ZNF367, thus we denoted this circRNA as circZNF367 in our study. The upregulation of circZNF367 in bone tissues that used for transcriptome RNA-sequencing was validated by qRT-PCR (Fig. 1C). Next, circZNF367 levels were evaluated in an enlarged cohort of osteoporosis patients (n = 20) and matched healthy controls (n = 15). Our results confirmed that circZNF367 was obviously upregulated in osteoporosis patients (Fig. 1D). Besides, the expression of circZNF367 was gradually decreased in hBMSCs during osteogenic differentiation (Fig. 1E). Taken together, our results indicated that circZNF367 was upregulated in osteoporosis patients and might involve in osteogenic differentiation of hBMSCs.
CircZNF367 is upregulated in osteoporosis patients. A heatmap showed the top 20 upregulated and downregulated circRNAs in osteoporosis patients. B hBMSCs transduced with indicated circRNAs or empty vector (EV) control were cultured with osteogenic-induced medium for 7 days, then relative ALP activity was measured. C the upregulation of circZNF367 in bone tissues that used for transcriptome RNA-sequencing were validated by qRT-PCR. D circZNF367 levels in an enlarged cohort of osteoporosis patients (n = 20) and matched healthy controls (n = 15) were evaluated by qRT-PCR. E, hBMSCs were cultured with osteogenic-induced medium for indicated times, then relative circZNF367 expression was evaluated by qRT-PCR. All assays were done in triplicates. *P < 0.05, **P < 0.05
Characterization of circZNF367
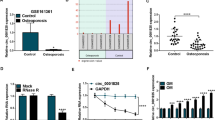
CircZNF367 is generated by back-splicing of exon 4, 5 and 6 of ZNF367 gene (Fig. 2A). The back-splicing sites of circZNF367 was confirmed by Sanger sequencing (Fig. 2A). Convergent primers were designed to detect the linear transcripts of the host gene ZNF367, and divergent primers were designed to detect the circular transcript of circZNF367. CircZNF367 could only amplify from the complementary DNA (cDNA) but not genomic DNA (gDNA), confirming the circular structure of circZNF367 (Fig. 2B). CircZNF367 was resistant to RNase R exonuclease digestion, suggesting that circZNF367 had a closed-loop structure (Fig. 2C). Furthermore, hBMSCs were treated with Actinomycin D for indicated times, then the levels of circZNF367 and linear transcripts of ZNF367 were evaluated by qPCR. CircZNF367 was proved to be more stable than the linear transcripts of ZNF367 (Fig. 2D). The cellular location of circZNF367 was evaluated by subcellular RNA fractionation assays and FISH. We found that circZNF367 was predominantly localized in the cytoplasm of hBMSCs (Fig. 2E). In FISH staining, circZNF367 was mostly accumulated in the cytoplasm of hBMSCs (Fig. 2F). Collectively, our results indicated that circZNF367 was a bona fide circRNA and mainly located in cytoplasm.
Characterization of circZNF367. A schematic map illustrated the formation of circZNF367 via back-splicing of exon 2, exon 3 and exon 4 of ZNF367 gene. The back-splicing junction sites were verified by Sanger sequencing. B circZNF367 levels were detected by RT-PCR or PCR using convergent or divergent primers from cDNA or genomic DNA (gDNA) of hBMSCs, then analyzed by agarose gel electrophoresis analysis of PCR products. C total RNAs of hBMSCs were treated with RNase R or mock control, then circZNF367 or ZNF367 expression levels were evaluated by qRT-PCR and evaluated by agarose gel electrophoresis analysis of PCR products. D hBMSCs (1 × 106) were seeded in 6-well plates for 24 h, then treated with 2 μg/ml Actinomycin reagent at indicated time points. Relative circZNF367 and ZNF367 expression were evaluated by qRT-PCR. E The RNA expression levels of circZNF367, GAPDH and U6 in nuclear and cytoplasmic fractions of hBMSCs were evaluated by qRT-PCR. The nuclear Lamin A/C and cytoplasmic GAPDH expression were evaluated by western blot. F the subcellular location of circZNF367 was detected by RNA fluorescence in situ hybridization using Cy3-labeled circZNF367 probes. DAPI was used to stain the cell nucleus. Scale bar = 10 μm
Overexpression of circZNF367 suppresses osteogenic differentiation of hBMSCs in vitro an in vivo
As circZNF367 was upregulated in osteoporosis patients (Fig. 1D) and gradually decreased during osteogenic differentiation (Fig. 1E), we speculated that circZNF367 might involve in osteogenic differentiation of hBMSCs. Thus, circZNF367 was overexpressed in hBMSCs and evaluated for proliferation, apoptosis, migration, invasion and osteogenic differentiation. A circZNF367 expression lentivirus vector was constructed and introduced into hBMSCs. The empty vector (EV) was used as control. Successful overexpression of circZNF367 was verified by qRT-PCR using the divergent primers (Fig. 3A). In addition, the expression of host gene ZNF367 was not affected by circZNF367 overexpression (Fig. 3A). Next, the influence of circZNF367 overexpression on growth and apoptosis of hBMSCs was evaluated. Our data indicated that overexpression of circZNF367 showed no evident influence on growth, colony formation and apoptosis of hBMSCs (Supplementary Fig. 1A–E). In contrast, forced circZNF367 expression apparently restrained migration and invasion of hBMSCs (Fig. 3B and C). Besides, hBMSCs were cultured with osteogenic-inducing medium (OIM) for osteogenic differentiation. CircZNF367 overexpression apparently decreased ALP activity of hBMSCs after culturing with OIM for 7 days (Fig. 3D). ALP and ARS staining were used to evaluate the mineralization level in vitro. Compared with EV group, circZNF367 overexpression reduced ALP staining of hBMSCs after culturing with OIM for 14 days, indicating that the cells failed to differentiate into mature osteoblasts (Fig. 3E). In ARS staining, circZNF367 overexpression suppressed mineralized nodule formation of hBMSCs after osteogenic differentiation for 14 days compared with EV control (Fig. 3F and G). ALP, osteocalcin (OCN) and Runx2 are commonly used osteoblast differentiation markers. CircZNF367 overexpression evidently reduced the mRNA levels of ALP, OCN and Runx2 (Fig. 3H). Moreover, the protein levels of OCN and Runx2 were significantly decreased by circZNF367 overexpression, too (Fig. 3I). The above results indicated that circZNF367 suppressed osteogenic differentiation of hBMSCs in vitro. To evaluate the influence of circZNF367 on osteogenic differentiation in vivo, an ectopic bone formation model was constructed. The hBMSCs introduced with circZNF367 overexpression vector or EV control were encapsulated in collagen-based hydrogels and implanted into the dorsal surface of BALB/c nude mice for 8 weeks. We found that circZNF367 overexpression reduced the volume of bone formed by hBMSCs (Fig. 3J and K). In H&E staining, less bone-like tissues were formed by hBMSCs in circZNF367 overexpression group (Fig. 3L). Meanwhile, Masson’s trichrome staining showed that collagen fiber bundles were less organized and not compactly arranged in circZNF367 overexpression group (Fig. 3L). These results indicated that circZNF367 suppressed osteogenic differentiation of hBMSCs in vivo. As hBMSCs are able to differentiate into adipocytes, we also evaluated the influence of circZNF367 overexpression on adipogenic differentiation of hBMSCs by culturing with adipogenic-inducing medium (AIM). However, circZNF367 overexpression showed no influence on the mRNA and protein expression of adipogenic markers CEBPα and PPARγ after culturing with AIM for 21 days, indicating that circZNF367 could not promote adipogenic differentiation of hBMSCs (Supplementary Fig. 2A and B). Above all, our results indicated that circZNF367 suppressed osteogenic differentiation of hBMSCs in vitro and in vivo.
Overexpression of circZNF367 suppresses osteogenic differentiation of BMSCs in vitro an in vivo. A hBMSCs were transduced with circZNF367 expression lentivirus or empty vector (EV) control, then relative circZNF367 and ZNF367 expression levels were evaluated by qRT-PCR. B–C, hBMSCs transduced with circZNF367 expression lentivirus or EV control, then used for transwell migration and invasion assays (B). Relative migration and invasion cells were shown (C). Scale bar = 20 μm. D hBMSCs infected with circZNF367 expression lentivirus or EV control were cultured with osteogenic-inducing medium for 7 days, then relative ALP activity was evaluated. E–I hBMSCs transduced with circZNF367 expression lentivirus or EV control were cultured with osteogenic-inducing medium for 14 days, then cells were used for ALP (E) and ARS staining (F–G). Relative ALP, OCN or Runx2 expression were evaluated by qRT-PCR (H) and western blot (I). Scale bar = 50 μm. J–L, hBMSCs (2 × 10.6) transduced with circZNF367 expression lentivirus or EV control were implanted into the dorsal subcutaneous space of nude mice for 8 weeks, then the bone-like tissues were dissected out (J). The average volume of the bone-like tissues was measured (K), then the bone-like tissues were evaluated by H&E and Masson’s staining (L). Scale bar = 500 μm. All assays were done in triplicates. *P < 0.05
Knockdown of circZNF367 promotes osteogenic differentiation of hBMSCs in vitro and in vivo
To further explore the role of circZNF367 in osteogenic differentiation, two sh-RNAs (sh-circ-1 and sh-circ-2) specifically targeting the junction site of circZNF367 were designed and introduced into hBMSCs. Compared with the non-targeting control (sh-NC), the expression level of circZNF367 was successfully knocked down by these two sh-RNAs (Fig. 4A). In addition, the expression level of host gene ZNF367 was not affected by sh-circ-1 or sh-circ-2 (Fig. 4A). The influence of circZNF367 knockdown on hBMSCs was further evaluated. Knockdown of circZNF367 showed no effect on cell growth, colony formation and apoptosis of hBMSCs (Supplementary Fig. 3A–C). However, circZNF367 depletion evidently increased the number of migration and invasion hBMSCs in transwell assays (Fig. 4B and C). Meanwhile, hBMSCs transduced with sh-circ-1, sh-circ-2 or sh-NC were cultured with OIM for osteogenic differentiation. Knockdown of circZNF367 obviously increased ALP activity of hBMSCs, indicating improved osteogenic differentiation (Fig. 4D). In addition, ALP and ARS staining were significantly enhanced by circZNF367 depletion, indicating that the extracellular matrix mineralization was elevated (Fig. 4E–G). The mRNA and/or protein expression of osteoblast differentiation markers ALP, OCN and Runx2 were also increased by circZNF367 knockdown (Fig. 4H and I). In the ectopic bone formation model, knockdown of circZNF367 increased the volume of bone-like tissue formed by hBMSCs (Fig. 4J and K). In H&E staining, more newly constructed bone was found in circZNF367 knockdown group (Fig. 4L). In Masson’s trichrome staining, there were more compactly arranged collagen fibers in circZNF367 knockdown group (Fig. 4L). Collectively, our results demonstrated that knockdown of circZNF367 promoted osteogenic differentiation of hBMSCs in vitro and in vivo.
Knockdown of circZNF367 promotes osteogenic differentiation of hBMSCs in vitro and in vivo. A hBMSCs were transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector, then relative circZNF367 and ZNF367 expression levels were evaluated by qRT-PCR. hBMSCs transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector, then used for transwell migration and invasion assays (B). Relative migration and invasion cells were shown (C). Scale bar = 20 μm. D hBMSCs transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector were cultured with osteogenic-inducing medium for 7 days, then relative ALP activity was evaluated. E–I, hBMSCs transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector were cultured with osteogenic-inducing medium for 14 days, then cells were used for ALP (E) and ARS staining (F–G). Relative ALP, OCN or Runx2 expression were evaluated by qRT-PCR (H) and western blot (I). Scale bar = 50 μm. J–L, hBMSCs (2 × 10.6) transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector were implanted into the dorsal subcutaneous space of nude mice for 8 weeks. The bone-like tissues were dissected out (J). Average volume of the bone-like tissues was measured (K), then the bone-like tissues were evaluated by H&E and Masson’s staining (L). Scale bar = 500 μm. All assays were done in triplicates. *P < 0.05
CircZNF367 interacts with RNA-binding protein HuR
Accumulated studies demonstrate that circRNAs can interact with RNA-binding proteins (RBP) and act as decoys or transporters for these factors [25]. In our study, the possible RNA-binding proteins that interacted with cirZNF367 were predicted by CircInteractome and ENCORI. The RNA-binding proteins HuR, FMRP, IGF2BP1 and TAF15 were predicted to interact with circZNF367 (Fig. 5A). Biotin-labeled RNA pull-down assay was conducted to evaluate these interactions. A biotin-labeled CircZNF367-specific probe was designed to target the back-splicing sites of circZNF367. Our data indicated that circZNF367 could be specifically enriched by circZNF367-specific probe compared with the scramble control (Fig. 5B). Furthermore, the RNA-binding protein HuR was enriched by the circZNF367-specific probe, but FMRP, IGF2BP1 and TAF15 could not (Fig. 5B). The interaction of circZNF367 and HuR was verified by RIP assay. Western blot analysis indicated that HuR was successfully enriched by the HuR specific antibody (Fig. 5C). Meanwhile, circZNF367 was enriched by HuR specific antibody compared with IgG control (Fig. 5C). These results indicated that circZNF367 could interact with the RNA-binding protein HuR. HuR has three RNA-binding domains, RNA recognition motif 1 (RRM1), RRM2 and RRM3. In in vitro binding assay, the RRM2 (103–189 aa), but not RRM1 (19–100 aa), RRM3 (245–326 aa), or hinge (190–244 aa) domain, of GST-tagged or Flag-tagged HuR protein was vital for its interaction with circZNF367 (Fig. 5D and E). HuR is known to bind to AU-rich elements (AREs) [26, 27]. To determine the binding sites between HuR and circZNF367, an AU-rich sequence AUUUAAUAAAU was found in circZNF367. Thus, circZNF367 and the AU-rich sequence mutant circZNF367-mut (AUUGCAUACCU) were in vitro cyclized and used for RNA pull-down assay. Compared with the anti-sense control, in vitro cyclized circZNF367 significantly enriched HuR, but the AU-rich sequence mutant circZNF367-mut failed, indicating that the AU-rich sequence AUUUAAUAAAU was vital for the interaction between HuR and circZNF367 (Fig. 5F). Next, we wondered if circZNF367/HuR interaction had any influence on mRNA or protein levels of HuR. However, circZNF367 overexpression did not change the mRNA or protein level of HuR in hBMSCs (Fig. 5G and H). To verify if circZNF367/HuR interaction was crucial for the effects of circZNF367, we knocked down HuR in hBMSCs and evaluated for osteoblast differentiation. In ALP and ARS staining, circZNF367 overexpression promoted osteogenesis of hBMSCs, but this was totally abolished by HuR knockdown, suggesting that circZNF367/HuR interaction was important for the effects of circZNF367 on osteoblast differentiation (Fig. 5I and J). Above all, our data indicated that circZNF367 could interact with RNA-binding protein HuR in hBMSCs.
CircZNF367 interacts with RNA-binding protein HuR. A, the predicted binding sites of circZNF367 and indicated RNA-binding proteins. B, biotin-labeled RNA pull-down in hBMSCs using circZNF367-specific probes and scramble probes. The enrichment of circZNF367 was evaluated by qRT-PCR. HuR, FMRP, IGF2BP1 or TAF15 pulled down by circZNF367-specific probe was evaluated by western blot. C, RIP assay was conducted in hBMSCs using the HuR antibodies. Relative enrichment of circZNF367 was evaluated by qRT-PCR. D–E, In vitro binding assay depicting the recovered circZNF367 levels from hBMSCs detected by RT-PCR (lower panel) after incubation with full length, truncation, or mutation forms of GST-tagged (D) or Flag-tagged (E) recombinant HuR protein validated by western blot (upper panel). F, biotin-labeled in vitro cyclized circZNF367, cicZNF367-mut, and anti-sense control (Control) were used for RNA pull-down assay. Enrichment of HuR was validated by western blot. G–H, hBMSCs were transduced with circZNF367 expression lentivirus or EV control, then relative mRNA and protein expression of HuR were evaluated by qRT-PCR (G) and western blot (H). I–J, hBMSCs transduced with sh-HuR-1, sh-HuR-2 or sh-NC were cultured with osteogenic-inducing medium for 14 days, then cells were used for ALP (I) and ARS (J) staining. Scale bar = 50 μm. All assays were done in triplicates. *P < 0.05. n.s. = not significant
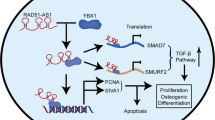
CircZNF367/HuR interaction reduces the mRNA stability of LRP5
The RNA-binding protein HuR is proved to regulate mRNA stability and translation of genes [28, 29]. Therefore, we speculated that circZNF367/HuR interaction might influence the stability of certain downstream target genes. To screen for potential genes that regulated by circZNF367/HuR interaction, hBMSCs were transduced with circZNF367 expression lentivirus or EV control for transcriptome RNA-sequencing. The dysregulated genes in circZNF367 overexpressed hBMSCs were depicted in the volcano map (Fig. 6A, supplementary Table 4). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis indicated that Wnt/β-catenin signaling was significantly repressed by circZNF367 overexpression (Supplementary Fig. 4A and B). Moreover, a series of downstream targets of Wnt/β-catenin signaling was downregulated by circZNF367 overexpression and upregulated by circZNF367 knockdown (Supplementary Fig. 4C and D). Thus, we speculated that circZNF367 might affect the activation of Wnt/β-catenin signaling in hBMSCs. LRP5, a coreceptor of β-catenin, can activate Wnt/β-catenin signaling and prevent phosphorylation and degradation of β-catenin. LRP5 is also proved to involve in regulating bone marrow density, bone mass maintaining and development of osteoporosis [14, 30]. In our study, LRP5 was among the top 5 downregulated genes in circZNF367 overexpressed hBMSCs (Fig. 6A, supplementary Table 4). This was also validated by qRT-PCR (Supplementary Fig. 4C). Therefore, we speculated that circZNF367/HuR interaction might regulate the mRNA stability of LRP5. Indeed, LRP5 is a potential target of HuR as indicated in the CLIP-seq data (https://starbase.sysu.edu.cn/index.php). In RIP assay, overexpression of circZNF367 but not the circZNF367-mut reduced the enrichment of LRP5 by HuR antibody, while circZNF367 knockdown showed opposite effects (Fig. 6B and 6C). Furthermore, knockdown of HuR decreased the mRNA and protein expression of LRP5 (Fig. 6D and E). In contrast, HuR overexpression increased the protein expression of LRP5 (Fig. 6E). To evaluate if circZNF367/HuR interaction could affect the mRNA stability of LRP5, hBMSCs were treated with Actinomycin D for indicated times, then LRP5 expression was measured by qRT-PCR. HuR knockdown evidently reduced the mRNA stability of LRP5 (Fig. 6F). Similarly, overexpression of circZNF367 but not the circZNF367-mut reduced the mRNA stability of LRP5 (Fig. 6G). In comparison, knockdown of circZNF367 increased the mRNA stability of LRP5 (Fig. 6H). As LRP5 can prevent the phosphorylation and degradation of β-catenin, we supposed that circZNF367 might affect the level of active-β-catenin in hBMSCs. As we expected, overexpression of circZNF367 but not the circZNF367-mut decreased the protein levels of LRP5 and active-β-catenin, while knockdown of circZNF367 exhibited opposite results (Fig. 6I). Next, the influence of LRP5 on osteogenic differentiation of hBMSCs was evaluated. Our data indicated that LRP5 overexpression increased ALP and ARS staining of hBMSCs, and knockdown of LRP5 decreased ALP and ARS staining of hBMSCs, indicating that LRP5 regulated osteogenic differentiation of hBMSCs (Fig. 6J–L). Taken together, our results indicated that circZNF367/HuR interaction reduced the mRNA stability of LRP5 in hBMSCs.
CircZNF367/HuR interaction reduces the mRNA stability of LRP5. A volcano map showed dysregulated genes between hBMSCs transduced with circZNF367 and EV control. B–C, RIP assay was conducted in hBMSCs transduced with circZNF367, circZNF367-mut, EV, sh-circ-1, sh-circ-2 or sh-NC vector using HuR or IgG antibodies. Relative LRP5 expression was evaluated by qRT-PCR. D–E, hBMSCs were transduced with sh-HuR-1, sh-HuR-2, sh-NC, EV or HuR, then relative mRNA and protein expression of HuR and LRP5 were evaluated by qRT-PCR (D) and western blot (E). F–H, hBMSCs infected with indicated genes were treated 2 μg/ml Actinomycin for 0, 4, 8 and 12 h, then relative LRP5 expression was evaluated by qRT-PCR. I, hBMSCs transduced with circZNF367, circZNF367-mut, EV, sh-circ-1, sh-circ-2 or sh-NC vector were used for western blot. J–L, hBMSCs transduced LRP5, EV, sh-LRP5-1, sh-LRP5-2 or sh-NC vector were cultured with osteogenic-inducing medium for 14 days, then cells were used for ALP (J) and ARS (K–L) staining. Scale bar = 50 μm. All assays were done in triplicates. *P < 0.05
HuR overexpression or LRP5 restoration abrogates the effects of circZNF367 overexpression on osteogenic differentiation of BMSCs
The above results suggested that circZNF367 reduced the expression of LRP5 by interacting with HuR; however, it was not certain that if this was responsible for effects of circZNF367 on osteogenic differentiation of hBMSCs. Therefore, HuR or LRP5 was overexpressed in hBMSCs that stably transduced with circZNF367 expression lentivirus or EV control. In western blot analysis, HuR was overexpressed and LRP5 was successfully restored in hBMSCs (Fig. 7A). Besides, circZNF367 overexpression reduced the level of active-β-catenin, but this was abrogated by HuR overexpression or LRP5 restoration (Fig. 7A). In transwell migration and invasion assay, circZNF367 overexpression suppressed migration and invasion of hBMSCs, but this effects was diminished by HuR overexpression or LRP5 restoration (Fig. 7B and C). CircZNF367 overexpression suppressed osteogenic differentiation of hBMSCs as indicated by reduced ALP activity, ALP and ARS staining, but HuR overexpression or LRP5 restoration completely abolished these effects (Fig. 7D–G). Collectively, our results suggested that HuR overexpression or LRP5 restoration abrogates the effects of circZNF367 overexpression on osteogenic differentiation of BMSCs.
HuR overexpression or LRP5 restoration abrogates the effects of circZNF367 overexpression on osteogenic differentiation of hBMSCs. A–C, hBMSCs were transduced with EV, circZNF367, HuR or LRP5 lentivirus vector as indicated, then collected cell lysates for western blot or used for transwell migration and invasion assay (B–C). Scale bar = 20 μm. D–G, hBMSCs were transduced with EV, circZNF367, HuR or LRP5 lentivirus vector as indicated, then cells were cultured with osteogenic-inducing medium for 7–14 days and used for ALP (D) and ARS (F–G) staining. Relative ALP activity (E) was tested. Scale bar = 50 μm. All assays were done in triplicates. *P < 0.05
Discussion
CircRNAs are highly abundant and evolutionary conserved in eukaryotes. CircRNAs have a longer half-life time and tissue-specific express pattern, which makes them as potential diagnostic biomarkers. Plenty of studies have uncovered the critical biological functions of circRNAs in human diseases, including cancer, cardiovascular disease, neurological disorder and osteoporosis [31]. In the present study, we screened for differentially expressed circRNAs in osteoporosis patients and evaluated for their potential functions. A total of 1110 dysregulated circRNAs (474 upregulated and 636 downregulated) were found in bone tissues from osteoporosis patients. CircZNF367 (hsa_circ_0008842), one of the top 10 upregulated circRNAs, was proved to suppress migration, invasion and osteogenic differentiation of hBMSCs in vitro and in vivo. Our study provided new evidences that circRNAs were enrolled in the pathogenesis of osteoporosis. Virtually, there are growing studies proving that circRNAs are dysregulated and involved in the development of osteoporosis [20]. For example, Huang et al. use circRNA microarray to identify potential cirRNAs that associate with osteoporosis and find that upregulation of circ_0002060 can be used as potential diagnostic marker for osteoporosis patients [32]. Yu et al. use transcriptome RNA-sequencing to identify differentially expressed circRNAs in six osteoporosis patients and healthy controls, and find 176 downregulated circRNAs and 211 upregulated circRNAs [33]. BMSCs are common progenitor cells for osteoblasts and adipocytes. There are increasing studies suggesting that circRNAs act an important role in osteogenic differentiation of BMSCs, too. For example, circ_0024097, a circRNA originating form YAP1, is proved to suppress osteoporosis by promoting osteogenic differentiation of BMSCs and MC3T3-E1 through sponging miR-376b-3p and increasing YAP1 expression [34]. Chia et al. report that circ-DAB1 is significantly upregulated during osteogenic differentiation of hBMSCs, whereas knockdown of circ-DAB1 suppresses proliferation and osteogenic differentiation of hBMSCs via sequestering miR-1270 and miR-944 and downregulating RBPJ expression [35]. Zhi et al. find that exosomal has_circ_0006859 is significantly overexpressed in osteoporosis patients and forced has_circ_0006859 expression inhibits osteoblastic differentiation of hBMSCs via miR-431-5p/ROCK1 axis [36]. In our study, circZNF367 overexpression reduced ALP and ARS staining, and ectopic bone-like tissue formation of human BMSCs after culturing with osteogenic-inducing medium, while circZNF367 knockdown exhibited opposite effects. Our results indicated that circZNF367 was a potential biomarker for osteoporosis and played a role in osteogenic differentiation of hBMSCs.
RNA-binding proteins play a key role in post-transcriptional regulation [37]. Accumulated studies indicate that circRNAs can bind with RNA-binding proteins and affect their interaction with downstream targets. For example, circular RNA circZKSCAN1 inhibits Wnt/β-catenin signaling activation by competing with CCARI for FMRP binding, thus suppresses stemness of hepatocellular carcinoma cells [38]. In myocardial infarction, circular RNA circFndc3b is dramatically downregulated in post myocardial infarction mouse hearts, while circFndc3b overexpression in cardiac endothelial cells elevates VEGFA expression via decreasing the protein level of FUS [39]. Human antigen R (HuR) belongs to the Hu family of RNA-binding proteins. HuR can bind with its target transcripts and control their splicing, localization, stability and translation [40]. HuR is involved in many cellular events, such as proliferation, senescence, differentiation and apoptosis [40]. Actually, previous studies have demonstrated that competitively binding with HuR might affect the mRNA stability of its downstream targets. In cancer cells, circAGO2 can interact with HuR and promote its enrichment on the 3’UTR region of target genes, and thus reduces AGO2 binding and AGO2/miRNA-mediated gene silencing [22]. In type 1 diabetes mellitus, Circular RNA circPPM1F is significantly upregulated and promotes LPS-induced M1 macrophage activation via competitively interacting with HuR and reducing mRNA stability of PPM1F, and eventually activating NF-κB signaling pathway [41]. Circular RNA circDCUN1D4 is downregulated in lung adenocarcinoma. Forced circDCUN1D4 expression suppresses invasion in vitro and metastasis in vivo through binding with HuR and enhancing its cytoplasmic transportation, and thus enhances the stability of TXNIP mRNA [42]. In the present study, we found that circZNF367 could bind with HuR. Moreover, silencing of HuR abrogated the effect of cricZNF367 on osteogenic differentiation of hBMSCs, indicating that the function of circZNF367 was possibly due to interacting with HuR. Indeed, circZNF367 was proved to compete with LRP5 for HuR binding, as circZNF367 overexpression reduced the enrichment of LRP5 by HuR antibody, while circZNF367 knockdown increased the enrichment of LRP5. Binding with HuR can enhance mRNA stability; therefore, we supposed that circZNF367/HuR interaction could reduce the mRNA stability of LRP5, and ultimately downregulated the protein level of LRP5 in hBMSCs.
Accumulated studies prove that LRP5 acts a vital role in bone formation and osteoporosis. Genome-wide association studies prove that LRP5 variant is connected with bone mineral density, osteoporosis, and increasing risk of fracture [14, 43, 44]. Furthermore, LRP5 is demonstrated to control bone formation locally [30, 45]. Actually, some studies also suggest that LRP5 is involved in osteogenic differentiation of BMSCs [46, 47]. As a coreceptor of β-catenin, LRP5 can activate Wnt/β-catenin signaling and prevent phosphorylation and degradation of β-catenin [13]. It is clear that Wnt/β-catenin signaling has proosteoblastic and antiadipocytic differentiation effects, and acts an important role in osteoblastic differentiation and bone mass maintaining [11, 48]. In our study, circZNF367 overexpression suppressed the protein expression of LRP5 and subsequently decreased the level of active-β-catenin in hBMSCs. In contrast, circZNF367 knockdown exhibited opposite effects. Our results indicated that circZNF367 suppressed osteogenic differentiation of hBMSCs through reducing LRP5 expression and ultimately inhibiting Wnt/β-catenin activation. As Wnt/β-catenin signaling played an important role in osteogenic differentiation of hBMSCs, this might explain the inhibitory effects of circZNF367 on osteogenic differentiation of hBMSCs.
In conclusion, we found that circZNF367 was upregulated in bone tissues of osteoporosis patients, and gradually decreased during osteogenic differentiation of hBMSCs. As a bona fide circular RNA, overexpression of circZNF367 suppressed migration, invasion and osteogenic differentiation of hBMSCs in vitro and in vivo, while knockdown of circZNF367 showed contrary effects. CircZNF367 was found to competitively bind with HuR, thus reducing the mRNA stability of LRP5. Therefore, circZNF367 overexpression decreased the protein expression of LRP5, and ultimately restrained Wnt/β-catenin activation. HuR overexpression or LRP5 restoration abrogated the effects of circZNF367 overexpression on osteogenic differentiation of BMSCs. Taken together, our results elucidated a novel role of circZNF367 in regulating osteogenic differentiation of hBMSCs, and circZNF367 might be a prognostic maker or potential target for osteoporosis treatment.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Nih Consensus Development Panel on Osteoporosis Prevention D Therapy. Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285(6):785–95. https://doi.org/10.1001/jama.285.6.785.
Sozen T, Ozisik L, Basaran NC. An overview and management of osteoporosis. Eur J Rheumatol. 2017;4(1):46–56. https://doi.org/10.5152/eurjrheum.2016.048.
Morin S, Lix LM, Azimaee M, Metge C, Caetano P, Leslie WD. Mortality rates after incident non-traumatic fractures in older men and women. Osteoporos Int. 2011;22(9):2439–48. https://doi.org/10.1007/s00198-010-1480-2.
Center JR, Nguyen TV, Schneider D, Sambrook PN, Eisman JA. Mortality after all major types of osteoporotic fracture in men and women: an observational study. Lancet. 1999;353(9156):878–82. https://doi.org/10.1016/S0140-6736(98)09075-8.
Tabatabaei-Malazy O, Salari P, Khashayar P, Larijani B. New horizons in treatment of osteoporosis. Daru. 2017;25(1):2. https://doi.org/10.1186/s40199-017-0167-z.
Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–42. https://doi.org/10.1038/nature01658.
Zhou S, Greenberger JS, Epperly MW, Goff JP, Adler C, Leboff MS, Glowacki J. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7(3):335–43. https://doi.org/10.1111/j.1474-9726.2008.00377.x.
Shen W, Chen J, Gantz M, Punyanitya M, Heymsfield SB, Gallagher D, Albu J, Engelson E, Kotler D, Pi-Sunyer X, Gilsanz V. MRI-measured pelvic bone marrow adipose tissue is inversely related to DXA-measured bone mineral in younger and older adults. Eur J Clin Nutr. 2012;66(9):983–8. https://doi.org/10.1038/ejcn.2012.35.
Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3(6):379–89. https://doi.org/10.1111/j.1474-9728.2004.00127.x.
Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, Xu C, Zhang L, Yang H, Hou J, Wang Y, Shi Y. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23(7):1128–39. https://doi.org/10.1038/cdd.2015.168.
Hartmann C. A Wnt canon orchestrating osteoblastogenesis. Trends Cell Biol. 2006;16(3):151–8. https://doi.org/10.1016/j.tcb.2006.01.001.
Yuan Z, Li Q, Luo S, Liu Z, Luo D, Zhang B, Zhang D, Rao P, Xiao J. PPARgamma and Wnt signaling in adipogenic and osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther. 2016;11(3):216–25. https://doi.org/10.2174/1574888x10666150519093429.
MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb Perspect Biol. 2012. https://doi.org/10.1101/cshperspect.a007880.
van Meurs JB, Trikalinos TA, Ralston SH, Balcells S, Brandi ML, Brixen K, Kiel DP, Langdahl BL, Lips P, Ljunggren O, Lorenc R, Obermayer-Pietsch B, Ohlsson C, Pettersson U, Reid DM, Rousseau F, Scollen S, Van Hul W, Agueda L, Akesson K, Benevolenskaya LI, Ferrari SL, Hallmans G, Hofman A, Husted LB, Kruk M, Kaptoge S, Karasik D, Karlsson MK, Lorentzon M, Masi L, McGuigan FE, Mellstrom D, Mosekilde L, Nogues X, Pols HA, Reeve J, Renner W, Rivadeneira F, van Schoor NM, Weber K, Ioannidis JP, Uitterlinden AG, Study G. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299(11):1277–90. https://doi.org/10.1001/jama.299.11.1277.
Shao T, Pan YH, Xiong XD. Circular RNA: an important player with multiple facets to regulate its parental gene expression. Mol Therapy Nucleic acids. 2021;23:369–76. https://doi.org/10.1016/j.omtn.2020.11.008.
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. https://doi.org/10.1186/s12943-020-01286-3.
Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97. https://doi.org/10.1002/jnr.24356.
Liu H, Lan T, Li H, Xu L, Chen X, Liao H, Chen X, Du J, Cai Y, Wang J, Li X, Huang J, Yuan K, Zeng Y. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics. 2021;11(3):1396–411. https://doi.org/10.7150/thno.53227.
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, Yuan W, Sun Z, Ming L. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. https://doi.org/10.1186/s12943-019-1041-z.
Chen W, Zhang B, Chang X. Emerging roles of circular RNAs in osteoporosis. J Cell Mol Med. 2021;25(19):9089–101. https://doi.org/10.1111/jcmm.16906.
Tare RS, Mitchell PD, Kanczler J, Oreffo RO. Isolation, differentiation, and characterisation of skeletal stem cells from human bone marrow in vitro and in vivo. Methods Mol Biol. 2012;816:83–99. https://doi.org/10.1007/978-1-61779-415-5_7.
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, Li D, Song H, Wang J, Hong M, Wang X, Huang K, Zheng L, Tong Q. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26(7):1346–64. https://doi.org/10.1038/s41418-018-0220-6.
Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–65. https://doi.org/10.1093/nar/gkv045.
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, Liao EY, Luo XH. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125(4):1509–22. https://doi.org/10.1172/JCI77716.
Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16): e100836. https://doi.org/10.15252/embj.2018100836.
Kim HS, Wilce MC, Yoga YM, Pendini NR, Gunzburg MJ, Cowieson NP, Wilson GM, Williams BR, Gorospe M, Wilce JA. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011;39(3):1117–30. https://doi.org/10.1093/nar/gkq837.
Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43(3):340–52. https://doi.org/10.1016/j.molcel.2011.06.008.
Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3’ untranslated regions of cytokine and angiogenic factor mRNAs. Can Res. 2001;61(5):2154–61.
Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100(14):8354–9. https://doi.org/10.1073/pnas.1432104100.
Cui Y, Niziolek PJ, MacDonald BT, Zylstra CR, Alenina N, Robinson DR, Zhong Z, Matthes S, Jacobsen CM, Conlon RA, Brommage R, Liu Q, Mseeh F, Powell DR, Yang QM, Zambrowicz B, Gerrits H, Gossen JA, He X, Bader M, Williams BO, Warman ML, Robling AG. Lrp5 functions in bone to regulate bone mass. Nat Med. 2011;17(6):684–91. https://doi.org/10.1038/nm.2388.
Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71(3):428–42. https://doi.org/10.1016/j.molcel.2018.06.034.
Huang Y, Xie J, Li E. Comprehensive circular RNA profiling reveals circ_0002060 as a potential diagnostic biomarkers for osteoporosis. J Cell Biochem. 2019;120(9):15688–94. https://doi.org/10.1002/jcb.28838.
Yu L, Liu Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun. 2019;516(2):546–50. https://doi.org/10.1016/j.bbrc.2019.06.087.
Huang Y, Xiao D, Huang S, Zhuang J, Zheng X, Chang Y, Yin D. Circular RNA YAP1 attenuates osteoporosis through up-regulation of YAP1 and activation of Wnt/beta-catenin pathway. Biomed Pharmacother. 2020;129:110365. https://doi.org/10.1016/j.biopha.2020.110365.
Chia W, Liu J, Huang YG, Zhang C. A circular RNA derived from DAB1 promotes cell proliferation and osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death Dis. 2020;11(5):372. https://doi.org/10.1038/s41419-020-2572-3.
Zhi F, Ding Y, Wang R, Yang Y, Luo K, Hua F. Exosomal hsa_circ_0006859 is a potential biomarker for postmenopausal osteoporosis and enhances adipogenic versus osteogenic differentiation in human bone marrow mesenchymal stem cells by sponging miR-431-5p. Stem Cell Res Ther. 2021;12(1):157. https://doi.org/10.1186/s13287-021-02214-y.
Pereira B, Billaud M, Almeida R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer. 2017;3(7):506–28. https://doi.org/10.1016/j.trecan.2017.05.003.
Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, Yang S, Zhao Q, Wu T, Li ZX, Liu XL, Wu R, Liu JF, Ge Y, Yang L, Wang HY, Chen L. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9(12):3526–40. https://doi.org/10.7150/thno.32796.
Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, Tang Y, Mallaredy V, Ibetti J, Grisanti L, Schumacher SM, Gao E, Rajan S, Wilusz JE, Goukassian D, Houser SR, Koch WJ, Kishore R. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10(1):4317. https://doi.org/10.1038/s41467-019-11777-7.
Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017. https://doi.org/10.1002/wrna.1372.
Zhang C, Han X, Yang L, Fu J, Sun C, Huang S, Xiao W, Gao Y, Liang Q, Wang X, Luo F, Lu W, Zhou Y. Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus. Theranostics. 2020;10(24):10908–24. https://doi.org/10.7150/thno.48264.
Liang Y, Wang H, Chen B, Mao Q, Xia W, Zhang T, Song X, Zhang Z, Xu L, Dong G, Jiang F. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Therapy Nucleic acids. 2021;23:355–68. https://doi.org/10.1016/j.omtn.2020.11.012.
Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez-Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gomez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JB, Verlaan D, Williams FM, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw KT, Lehtimaki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HA, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AW, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JP, Kiel DP, Rivadeneira F. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. https://doi.org/10.1038/ng.2249.
Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, Valdes AM, Arp P, Whittaker P, Verlaan DJ, Jhamai M, Kumanduri V, Moorhouse M, van Meurs JB, Hofman A, Pols HA, Hart D, Zhai G, Kato BS, Mullin BH, Zhang F, Deloukas P, Uitterlinden AG, Spector TD. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371(9623):1505–12. https://doi.org/10.1016/S0140-6736(08)60599-1.
Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML, Osteoporosis-Pseudoglioma Syndrome Collaborative G. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107(4):513–23. https://doi.org/10.1016/s0092-8674(01)00571-2.
Lin J, Zheng Z, Liu J, Yang G, Leng L, Wang H, Qiu G, Wu Z. LRP5-Mediated Lipid Uptake Modulates Osteogenic Differentiation of Bone Marrow Mesenchymal Stromal Cells. Front Cell Dev Biol. 2021;9: 766815. https://doi.org/10.3389/fcell.2021.766815.
Wan Y, Lu C, Cao J, Zhou R, Yao Y, Yu J, Zhang L, Zhao H, Li H, Zhao J, Zhu X, He L, Liu Y, Yao Z, Yang X, Guo X. Osteoblastic Wnts differentially regulate bone remodeling and the maintenance of bone marrow mesenchymal stem cells. Bone. 2013;55(1):258–67. https://doi.org/10.1016/j.bone.2012.12.052.
Shen L, Glowacki J, Zhou S. Inhibition of adipocytogenesis by canonical WNT signaling in human mesenchymal stem cells. Exp Cell Res. 2011;317(13):1796–803. https://doi.org/10.1016/j.yexcr.2011.05.018.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors guaranteed the integrity of the entire study. The experiments were conducted by Gengyan Liu, Jia Luo and Zhengguang Wang. Clinical studies were conducted by Yong Li, Gengyan Liu and Jia Luo. Data was analyzed by Zhou Yong. Manuscript was prepared and reviewed by Yong Li. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical standards
The protocol of our study for human were followed with the ethicaal standards of the institutional committee of The Third Xiangya Hospital (Approve no. 2017-R17021) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The use of animals in our study was approved and we strictly followed the ethical standards of the ethics committee of The Third Xiangya Hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

13577_2022_798_MOESM1_ESM.jpg
Supplementary file1 (JPG 1352 KB) Supplementary Figure 1. The effect of circZNF367 overexpression on proliferation, colony formation, apoptosis and bone formation of BMSCs. A, hBMSCs were transduced with circZNF367 expression lentivirus or EV control, then relative cell viability at indicated times was evaluated by CCK-8 assay. B–C, hBMSCs (4000/well) transduced with circZNF367 expression lentivirus or EV control were used for colony formation assay (B). Number of colony number per well was shown (C). D–E, hBMSCs were transduced with circZNF367 expression lentivirus or EV control, then stained with Annexin-V FITC and PI for flow cytometry analysis (D). The percentage of Annexin-V apoptotic cells was shown (E). All assays were done in triplicates. *P< 0.05

13577_2022_798_MOESM2_ESM.jpg
Supplementary file2 (JPG 664 KB) Supplementary Figure 2. The influence of circZNF367 overexpression on adipogenic differentiation markers of BMSCs. hBMSCs transduced with circZNF367 expression lentivirus or EV control were cultured with adipogenic-inducing medium for 14 days, then relative CEBPα and PPARγ expression were evaluated by qRT-PCR (H) and western blot (I). n.s. = not significant

13577_2022_798_MOESM3_ESM.jpg
Supplementary file3 (JPG 1573 KB) Supplementary Figure 3. The effect of circZNF367 knockdown on proliferation, colony formation, apoptosis and bone formation of BMSCs. A, hBMSCs were transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector, then relative cell viability at indicated times was evaluated by CCK-8 assay. B–C, hBMSCs (4000/well) transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector were used for colony formation assay (B). Number of colony number per well was shown (C). D–E, hBMSCs were transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector, then stained with Annexin-V FITC and PI for flow cytometry analysis (D). The percentage of Annexin-V apoptotic cells was shown (E). All assays were done in triplicates. *P< 0.05

13577_2022_798_MOESM4_ESM.jpg
Supplementary file4 (JPG 1352 KB) Supplementary Figure S4. The influence of cricZNF367 on the downstream targets of Wnt/β-catenin signaling. A–B, Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of hBMSCs transduced with circZNF367 expression lentivirus or EV control. C, hBMSCs were transduced with circZNF367 expression lentivirus or EV control, then relative LRP5 expression was evaluated by qRT-PCR. D–E, hBMSCs were transduced with circZNF367, EV, sh-circ-1, sh-circ-2 or sh-NC vector, then relative expression of indicated genes were evaluated by qRT-PCR. All assays were done in triplicates. *P< 0.05
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, G., Luo, J., Wang, Z. et al. CircZNF367 suppresses osteogenic differentiation of human bone marrow mesenchymal stromal/stem cells via reducing HuR-mediated mRNA stability of LRP5. Human Cell 36, 146–162 (2023). https://doi.org/10.1007/s13577-022-00798-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00798-y