Abstract
The tumor microenvironment contributes significantly to tumor initiation, progression, and resistance to chemotherapy. Much of our understanding of the tumor and its microenvironment is developed using various methods of cell culture. Throughout the last two decades, research has increasingly shown that 3D cell culture systems can remarkably recapitulate the complexity of tumor architecture and physiology compared to traditional 2D models. Unlike the flat culture system, these novel models enabled more cell–cell and cell–extracellular matrix interactions. By mimicking in vivo microenvironment, 3D culture systems promise to become accurate tools ready to be used in diagnosis, drug screening, and personalized medicine. In this review, we discussed the importance of 3D culture in simulating the tumor microenvironment and focused on the effects of cancer cell–microenvironment interactions on cancer behavior, resistance, proliferation, and metastasis. Finally, we assessed the role of 3D cell culture systems in the contexts of drug screening.
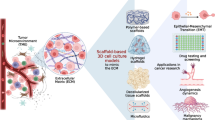
Graphical abstract
2D culture system is used to study cancer cell growth, progression, behavior, and drug response. It provides contact between cells and supports paracrine crosstalk between host cells and cancer cells. However, this system fails to simulate the architecture and the physiological aspects of in vivo tumor microenvironment due to the absence of cell-cell/ cell-ECM interactions as well as unlimited access to O2 and nutrients, and the absence of tumor heterogeneity. Recently advanced research has led researchers to generate 3D culture system that can better recapitulate the in vivo environment by providing hypoxic medium, facilitating cell-cell and cell-ECM, interactions, and recapitulating heterogeneity of the tumor. Several approaches are used to maintain and expand cancer cells in 3D culture systems such as tumor spheroids (cell aggregate that mimics the in vivo growth of tumor cells), scaffold-based approaches, bioreactors, microfluidic derives, and organoids. 3D systems are currently used for disease modeling and pre-clinical drug testing.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Throughout decades, 2D culture system has been the core for cell research and the most common in vitro model used to study cancer cell proliferation, behavior, hallmarks and drug response. This 2D culture system helped researchers to understand the biology of cancer cells, identify various carcinogenic markers, and discover new therapies. Despite its contribution to the advancement of knowledge in cancer research, 2D culture system fails to adequately simulate the complexity and heterogeneity of in vivo tumors. Therefore, it is a physiologically insufficient predictive and relevant tool for valid drug discovery. The unlimited access to O2, nutrients, and metabolites, as well as the need for consecutive passaging, can induce an accumulation of genetic mutations in cells. 2D attachment of cells leads to undesirable changes in cell morphology [1], including cell flattening, cytoskeletal alterations, and changes in nuclear morphology, which consequently modify the expression of genes and proteins [2,3,4]. Additionally, the absence of cell–cell and cell–extracellular matrix (ECM) interactions affects cell viability, proliferation, and differentiation. Thus, the in vitro 2D culture system does not adequately mimic the in vivo microenvironment.
To better understand the in vivo microenvironment, patient-derived xenografts (PDXs) were introduced as an alternative to the 2D culture system [5]. Using PDXs as pre-clinical tumor models, primary cancer cells were transplanted into an immunodeficient animal to study the metastatic capacity and test anti-cancer drugs. Despite the importance of PDXs in conserving the histological, genetic, and molecular signature of the original tumor, the use of immunodeficient animals does not simulate the real scenario taking place in the human body [5]. Moreover, the PDXs model is uncontrollable, expensive, and time- and resource-consuming. Besides, the morphological changes and cell behavior could not be monitored in this model [5]. Therefore, it was necessary to have an intermediate system that can mirror the in vivo tumor environment without altering cell features. “Goodbye flat biology,” an article published in 2003 by Alison Abbot, announced the beginning of a revolutionary era in cell biology [6]. A proper intermediary ex vivo system is the 3D culture system in which the cultured cells can maintain their morphology and polarity and interact with each other and with or without the ECM [7]. The 3D culture system can adequately recapitulate the in vivo tumor architecture, behavior, histopathological features, genetic signatures, molecular profile, and drug responsiveness [7]. In addition, cancer cells can form spheroids with a hypoxic core that recapitulates the gradient of O2 and nutrients and the drug penetration [8,9,10,11]. Thus, the proliferation rate is location-dependent and higher in the spheroids’ periphery. Hence, the 3D culture model can be a valuable tool mimicking the in vivo tumor microenvironment [7].
In this narrative review, we highlighted the 3D culture system value in mimicking the in vivo microenvironment, investigating how the interaction between tumor and microenvironment can affect cancer cell behavior, migration, invasion, and drug responsiveness.
Tumor microenvironment
The tumor microenvironment comprises stromal cells, ECM, and soluble factors surrounding the tumor cells. This microenvironment provides biophysical and biochemical support and affects cancer cell morphology, genes and proteins expression, migration, invasion, and angiogenesis. The 3D culture system aims to mimic the in vivo model by permitting cell–ECM and cell–cell interactions [7].
Stromal cells
The tumor microenvironment contains different types of stromal cells known as non-cancer cells, such as fibroblasts, stellate cells, endothelial cells (ECs), and immune cells. Fibroblasts are the predominant cell population in the stroma, secreting ECM and basement membrane (BM) proteins. Various mechanisms lead to the activation of these cells in the presence of cancer cells [12, 13]. These cancer activated fibroblasts (CAF) are characterized by their spindle-like morphology, contractile phenotype, and expression of vimentin, α-smooth muscle actin (α-SMA), and fibroblast activating protein (FAP) [12]. Activation of fibroblasts can be triggered by different factors such as inflammation, physiological stress, and ECM stiffness [12, 13]. ECs are another form of cell found in the tumor microenvironment. During tumorigenesis, the angiogenic process starts when ECs branch from pre-existing small vessels and form sprouts of capillaries. These new capillaries supply cancer cells with O2 and nutrients, making them more aggressive and resistant. They also facilitate their metastasis, rendering the tumor increasingly malignant [14, 15]. Tumor cells and CAFs secrete a wide range of chemokines and cytokines that recruit immune cells to the tumor microenvironment. The immune cell populations include tumor-associated macrophages (TAMs), T cells, B cells, natural killer (NK) cells, dendritic cells (DCs), tumor-associated neutrophils (TANs), and myeloid-derived suppressor cells (MDSCs). Among immune cells, TAM and regulatory T (Treg) cells are the major cell types that contribute to the formation of an immunosuppressive microenvironment. These cells enhance the growth and progression of cancer cells and attenuate or suppress the anti-tumor immunity. Treg and TAM release anti-inflammatory cytokines such as IL-10 and TGF-beta, that reduce the activity of DC [16], T cell and NK cells [17, 18].In addition, the direct interaction between TAM, Tregs, DCs and cytotoxic T cells via CTLA-4 /CD80/CD86 pathway, LAG-3/MHCII, and PDL1/2 /PD-pathway prevents the maturation of DC, and reduces the activation and proliferation of T cells [19,20,21,22,23]. Nowadays, researchers aim to reactivate the immune system by triggering the pro inflammatory and phagocytosis properties of macrophages, and targeting immune checkpoint using checkpoint inhibitor such as CTLA-4, LAG-3 and PDL1/2, to rebut T cell response against cancer cells and depleting Treg cells.
Extracellular matrix
The ECM is defined as an interconnected network of macromolecules distributed into interstitial and pericellular matrices. The pericellular matrix is in close contact with tumoral cells and is surrounded by the interstitial matrix [24, 25]. The BM forms a sheet layer between the epithelial cells and underlying connective tissue, consisting of laminin, collagen IV, nidogen type 1 and 2, and proteoglycan (PG) family [24, 25]. The ECM is a highly specialized framework that its characteristics depends on each tissue or organ's physical, physiological, metabolic, and other functions. For instance, while hyaluronic acid (HA) is a significant component of the brain ECM, other elements such as collagen and laminin are absent in the brain parenchyma [26]. On the other hand, the ECM of the breast cancer tissue is mainly formed of fibrous collagen [27].
Moreover, the ECM biomechanical signals might be modulated by variations in the composition and structure of the ECM along with the binding of soluble factors secreted by the tumor cells. The cell–ECM interaction interprets internal and external mechanical forces and translates them into biochemical signals regulating cell migration, proliferation, and differentiation [28,29,30,31]. ‘Internal forces’ refer to contractile forces that are generated internally by the cell cytoskeleton, whereas ‘external forces’ refer to forces that are generated from outside of the cell.
3D culture modeling in tumor microenvironment
To reproduce the tumor–ECM interaction in vitro, cancer cells are encapsulated or embedded in a natural or synthetic biocompatible and biodegradable material that recapitulates the biomechanical and biophysical properties of the in vivo microenvironment. This approach is mainly used to study the cancer cells’ migration and invasion behaviors [9].
Biomaterials are extensively used to establish a 3D culture model that reliably mimics the native ECM. Natural biomaterials can be derived from mammalian ECM such as Matrigel or proteins and polysaccharides such as collagen, alginate, chitosan, hyaluronic acid, and gelatin [32]. The majority of these materials are biocompatible, have tunable mechanical properties as well as cell adhesive features. However, these properties may vary from batch- to- batch and are challenging to control [33]. Polymers are used to make synthetic biomaterials including polyethylene glycol (PEG), polylactic acid (PLA), polyglycolic acid (PGA), poly (lactic-co-glycolic acid) (PLGA), and poly-e-caprolactone (PCL) [32]. The structure and chemical composition of these materials are well defined but they still require further modification to increase their biocompatibility and bio-adhesion properties [33]. Researchers developed these biomaterials to recapitulate the complex biochemical and biophysical structure of the native ECM using biodegradable material [34], adding protease-sensitive sequences such as MMP sensitive sequence [35], and incorporating bioactive molecules such as adhesive ligands integrin-binding domains (Arginine -Glycine- Aspartic Acid (RGD) [36, 37] and peptides that facilitate the interaction between cancer cells and the synthetic ECM. To obtain a matrix that resembles the natural ECM, researchers constructed “hybrid biomaterials” that combine the features of both natural and synthetic biomaterials [33]. It is important to note that the physical properties of these biomaterials, such as stiffness, porosity, elasticity, and adhesivity, can all be rigorously controlled. Compared to the natural or fully synthetic matrices, the hybrid matrix represents a potential tool for assessing the role of the ECM in tumor progression, metastasis and invasion.
Decellularized matrices are obtained from the decellularization and handling of living malignant or healthy tissues, or through in vitro ECM production by 2D-cultured cells [38, 39]. It's a promising tissue engineering tool because it conserves molecular composition, complex 3D structure, and biochemical and biophysical properties of the native ECM. Decellularized matrices still have a long way to go to be part of the go-to repertoire of scaffolds for 3D in vitro tumor model assembly, due to challenges in their production in high throughput compatible platforms, as well as limited control over structure and architecture [39].
Tumor Cells-ECM interactions and the effect of ECM composition and physical properties on cancer cell migration and invasion in 3D culture models
The exact composition of the ECM surrounding the primary tumor site can vary according to the tissue type. Hence, the in vitro 3D culture model offers a wide range of customized biomaterials that bio-mimic the dynamic and reciprocal interaction between cancer cells and ECM allowing the researchers to explore cancer cells’ behavior.
Composition of ECM in 3D culture models
Nguyen-Ngoc et al. used a 3D ex vivo system to mimic the tumor microenvironment and compared cancer cells’ behavior in Matrigel and collagen I ECM. Fragments from the same primary human mammary carcinoma were embedded in the Matrigel and collagen I ECM. In Matrigel, a solubilized BM was extracted from the Engelbreth-Holm-Swarm (EHS) mouse sarcoma and was rich in ECM proteins such as laminin (a major component), collagen IV, heparan sulfate proteoglycans, and entactin/nidogen [40]. Cells embedded in Matrigel preserved their epithelial-like appearance without protrusions and demonstrated a collective migration behavior. However, cells embedded in collagen I ECM showed protrusion migration and local dissemination. To investigate the effect of the ECM microenvironment, researchers tested various combinations of Matrigel and collagen I ECM. Accordingly, the tumor fragments, which were primarily cultured in Matrigel, were transferred to collagen I ECM and vice versa. The results revealed that cells with collective migration in Matrigel started to form protrusions once transferred to collagen I ECM. In compliance with these findings, retraction of protrusions and reversal of collective migration were detected when collagen I embedded cells were transferred to Matrigel [41]. In consistency with these results, glioblastoma cell lines grown as spheroids on HA-RGD hydrogel under 3D conditions display a migration and invasion pattern close to that seen in human brain slices which is completely distinct from 2D culture and a 3D collagen I-based scaffold [26]. To better appreciate the behavior of cancer cells, Guzman et al. tried to replicate the in vivo environment by creating “shelled” spheroids, which were surrounded by a BM layer and assembled into a scaffold. The analysis of these shelled spheroids embedded in the collagen I matrix revealed laminin and collagen IV in the BM layer [42]. Therefore, the heterogeneous ECM can imitate the in vivo microenvironment and permit the evaluation of cancer cell migration and invasion more efficiently than a homogenous ECM (including collagen I, fibronectin, laminin). The researchers investigated two distinct breast cancer cell lines; MCF 10 H Ras, characterized by a mesenchymal-like morphology, and MD-MB468, described by a grape-like appearance. They reported that the shelled spheroids embedded within the collagen I matrix displayed a combined invasion phenotype. Inside the BM layer, the cells exhibited a multicellular invasion phenotype with a stream of densely packed cells moving through the BM towards the collagen I matrix. When these cells reached the collagen I matrix, the invasion pattern of an individual cell was detected. However, the BM layer-free spheroids showed an individual invasion phenotype in the collagen I matrix. Moreover, the presence of MMP inhibitors in the collagen I matrix decreased invasion frequency and the mean number of the individual invasive cells. Despite the presence of the MMPs, researchers noted the formation of multicellular streams and a successful invasion [42]. Because the behavior of cancer cells in vitro is greatly affected by the composition of the biomaterial, Koh et al. developed a decellularized patient-derived ECM to study the behavior of primary glioblastoma cells in vitro. They noticed that patient-derived glioblastoma cells exhibit a heterogeneous shape; cells with elongated shapes developed elongated protrusions and displayed a directionally movement, however, round cells showed poorly defined protrusions, and random movement. Moreover, they demonstrated that patient-derived glioblastoma cells alter their invasion pattern to achieve aggressive motility in a 3D microenvironment [43]. In conclusion, the wide range of customized biomaterials allows researchers to thoroughly study the metastatic and invasion potential of cancer cells in their original ECM and their behaviors in the invaded matrix.
Physical properties of the ECM in 3D culture models
Noticeable alterations in ECM have been detected during tumorigenesis [26, 44,45,46,47,48], including increased deposition, cross-linking, orientation, and organization of the ECM. The high deposition and cross-linking of the ECM lead to increased ECM stiffness (rigidity). ECM stiffness is found to be higher in malignant tissue than in normal tissue, and it has been linked to cancer cell migration and invasion features [49,50,51]. Hence, using 3D culture systems, researchers will be able to control the biophysical properties of the ECM, and therefore interrogate their effect on cancer cell migration and invasion.
Researchers have to use ECM of different stiffnesses to recapitulate the different stages of in vivo breast cancer cells [52, 53]. Using a 3D culture system of various stiffnesses, breast cancer cell lines cultured on a stiff substrate showed a well spread and elongated shape with mature focal adhesion and broad lamellipodia at the leading edges [54, 55]. These morphological changes enhance cell migration and invasion processes. During migration, cells on a stiff substrate exhibit a more directional movement and a faster persistent migration than cells on a soft substrate. Thus, the migration and invasion potential of cancer cells correlates positively with the stiffness of the ECM [54, 55]. Riching et al. reported that the collagen fibers alignment increases the ECM stiffness and promotes cell migration by increasing directional movement and restricting protrusions along aligned fibers [56]. The pore size of the ECM is another factor that participates in cancer cell migration and invasion regulation. According to Lang, N., It is difficult to separate the study of the impact of matrix stiffness on cell migration from its impact on matrix pore size and adhesive ligand density [57]. Stiff matrices with large pores enhance the invasion of cancer cells cultured on a porous 3D gel; however, it impairs their invasion when the pore sizes decrease [57]. Collectively, to mimic the in vivo tumor microenvironment, researchers can tune the physical properties of the in vivo ECM to recapitulate the complex architecture and structure of the native ECM, and therefore examine their impact on cancer cell dissemination and invasion.
Aggressiveness of tumor cells in 3D culture models
The 3D culture system reveals cancer cells’ potential to remodel the ECM in vitro, dictating the migration and invasion patterns of cancer cells and regulating switches among invasion modes, such as EMT, amoeboid, and collective migration. Kwon et al. cultured multiple ovarian cell lines on a fibroblast-derived 3D matrix. They proved that cancer cells with high E-cadherin expression collectively migrate and secrete ECM-degrading enzymes. Lack of E-cadherin expression in cancer cells leads to looser agglomeration, and allowing them to invade the stroma through ROCK-dependent ECM modification. Besides, it has been shown that some ovarian cancer cells, such as SKOV3 and UPN 251, can migrate in a single direction without affecting or remodeling the ECM structure [58]. The use of 3D collagen type I hydrogels confirmed these findings by demonstrating the heterogenicity of primary breast cancer cells. In consistence with these results, primary breast tumors grown on collagen type 1 hydrogels, revealed the detection of a specific subset of cancer cells with a basal epithelial phenotype to lead the collective invasion [59]. Furthermore, the invasion pattern can be protease-dependent, in which cells facilitate invasion by synthesizing specific enzymes such as MMPs that degrade the ECM. When the ECM pore size is smaller than 3 μm, cells develop proteases secreting protrusions invading the ECM (invadopodia). On the other hand, cells can invade the ECM without using proteases, thanks to the ROCK or Rho signaling pathways. Recently, Wisdom et al. suggested that even when the pore size is smaller than the cell size (< 3 μm), cells can form invadopodia structures (protease-independent manner) and squeeze between ECM pores to widen the pores. These actions are ensured by applying protrusive and contractile forces [48]. In conclusion, both the physical properties of the ECM and the interactions of cancer cells with ECM components play a crucial role in the migration and invasion pattern.
3D multicellular models
To recapitulate the complexity and heterogeneity of the in vivo tumor microenvironment, researchers developed “double and triple co-culture systems” that consists of tumor cells, with one or two other types of cells present in the stroma such as fibroblasts, immune cells, endothelial and epithelial cells. This innovative system was used to evaluate the effect of stroma on cancer cells behavior and invasiveness, and to develop new therapies.
Double co-culture model
The in vitro 2D co-culture systems allows partial contact between cells and supports paracrine crosstalk between host cells and cancer cells. On the other hand, mutual cell–cell interactions and the secretion of soluble factors found in the 3D culture system enhance tumor growth and progression, remodel the protein composition of the ECM, induce cancer cells migration and promote cancer invasion [60]. The secretome of cancer and stromal cells generates a microenvironment that promotes tumor cell invasion and angiogenesis. In a 3D culture system, CAFs regulate cancer cells via secretion of soluble factors that enhance migration (active TGF ß and leptin), invasion (IL-8, IL-6, MMP2, MMP3, and MMP9), and angiogenesis (VEGF, FGF, and PDGF) [61,62,63,64,65,66]. Hernandez-Fernaud et al. introduced a novel mechanism underlying cell migration and invasion by showing how angiogenesis and cancer progression can be driven by secreted glutathione-dependent oxido-reductases via a transglutaminase 2 (TGM2)-dependent invasion. It has been described that CLCI3 secreted by CAFs promotes angiogenesis of ECs and invasion of cancer cells in a 3D culture system by activating TGM2. Activated TGM2 leads to an increase in ECM stiffness which contributes to a5ß1 integrin-dependent invasion [67].
In a 3D co-culture system, the direct contact between cancer cells and fibroblasts enhances cancer cell proliferation and survival. Moreover, the addition of fibroblasts results in the generation of uniform, compact spheroids [64] and preferentially promotes the metabolic activity of cancer cells [68]. The migration and invasion potential of cancer cells in co-culture with fibroblasts has been studied using various methods. For instance, Estrad et al. developed a co-culture model in which aggregates of MCF7 breast cancer were encapsulated in alginate either alone or with fibroblast [69]. In co-culture, polarized cancer cells were arranged around small lumina and were surrounded by fibroblasts. After 15 days, analysis of aggregate morphology showed that 40% of aggregates of the co-culture microcapsule lost its circularity and presented an altered phenotype compared to aggregates in the monoculture that maintained their morphology. In addition, a collective and unidirectional migration out of the co-culture microcapsule was detected. These changes are accompanied by a decrease in the expression of estrogen receptor and E cadherin. Importantly, the secretion of pro inflammatory, pro-invasive cytokines (IL-6, IL-8, and CXCL1), and sICAM (associated with tumor cell growth and angiogenesis) increased significantly in the co-culture microcapsules [69]. In consistency with these results, Knuchel et al. generated a 3D co-culture model in which SW620 spheroids and fibroblast were embedded in Matrigel and the fibroblasts surrounded the spheroids, establishing cell–cell contacts and stimulating cancer cell invasion. This process is mediated by fibroblast surface associated-FGF2, which activates FGFR on cancer cells. Once FGFR is triggered, it induces the phosphorylation of Src, which in turn stimulates integrin ɑvß5-dependent tumor cell adhesion and motility [70]. To better understand the effect of fibroblast on cancer cell migration and invasion, A431 vulvar squamous cell carcinoma and fibroblast were embedded in a 3D matrix formed by collagen and recombinant basement membrane. The results showed that fibroblast migrated toward cancer cells and established cell–cell contact with A431 via a heterophilic E cadherin/N cadherin junction that induces collective cancer cell migration [71]. Recently, a new invasion assay based on chimeric solid spheroid formed by cancer cells and fibroblast was developed. The chimeric spheroids were embedded in a 3D matrix that facilitates cancer cell invasion analysis. Using this assay, Miyazaki et al. demonstrated that the fibroblasts invade the collagen matrix, the cancer cells attached to fibroblasts via integrin ɑ5ß1, and migrate using the fibroblast protrusions as a track. Integrin ɑ5ß1 adhere to fibronectin on the surface of fibroblasts and facilitate migration and invasion of cancer cells. According to Miyazaki, heterophilic E cadherin/N cadherin junction is not essential for cancer cell migration, because cancer cells lacking the expression of E cadherin were still able to adhere to fibroblast. It is important to note that although integrin ɑ5ß1 is the major receptor that binds fibronectin, fibronectin receptors may vary depending on the type of cancer cells [72]. Stellate cells are quiescent fibroblasts that reside in the pancreas and liver [73]. Under 3D conditions, the crosstalk between pancreatic stellate cells (PSCs) and pancreatic ductal adenocarcinoma (PDACs) stimulates the arrangement of collagen fibers and directs cell migration through ROCK-mediated contractility [74]. In a recent study, Hwang et al. showed that the co-culture of PSCs and PANC1 human pancreatic cancer cell line remodels the ECM and enhances cell migration and invasion as well as increases the secretion, deposition and thickness of the collagen 1 and fibronectin fibers. Interestingly, collagen fibers and the protrusion of the cell membrane were growing in a same direction, suggesting that the remodeling of the ECM matrix enhances cell migration. The interaction between the PANC1 and PSCs increased the expression level of Ki67 and several factors detected in the conditioned medium, including IL-6, IL-8, CXCL1, VEGF, TIMP 1 and 2, MCSF, and GMCSF. Moreover, an EMT-like phenotype was observed in the co-culture tumor spheroids [75]. SEM and TEM analysis of the 3D structure of mixed cell spheroids formed by hepatocellular carcinoma (HCC) cell line Huh7 and stellate cells LX-2 showed extensive cell protrusion at the periphery of the spheroids. An elevated expression level of ɑ SMA, Collagen I, and TGFß1 was also detected in the mixed spheroids tumor obtained from human HCC patients as well as human xenografts. After embedding the mixed spheroids in matrix (Matrigel and collagen), both Huh 7 and LX-2 cells showed an invasive phenotype and migrated out of spheroids [76]. In summary, active interaction between cancer cells and fibroblasts is required for cancer cell migration and invasion. Thus, inclusion of fibroblasts in a 3D culture system helps to model the physiological features of the tumor microenvironment.
Triple co-culture
Cancer cells, fibroblasts, and “Macrophages”
The 3D triple culture system simulates the complexity and heterogeneity of the in vivo tumor microenvironment that consists of tumor cells, stroma, and ECM. In two inventive studies, Kuen et al. and Rebelo et al. developed a novel triple culture system to study the interaction between cancer cells, CAF, and monocytes [64, 77]. In the first study, monocytes were incubated with mixed spheroids formed by fibroblasts and PDACs [64]. In the second investigation, tumor spheroids containing non-small cell lung carcinoma (NSCLC) were primarily grown in a stirred culture, and then an alginate microcapsule was created containing tumor spheroids, fibroblasts, and macrophages [77]. In both studies, CD163/CD68 macrophages were detected within the tumor spheroid which proved the migration capacity of these macrophages. Moreover, they found that tumor spheroids secret IL-4, IL-13, and CXCL1 molecules which attract macrophages to the tumor site and activate their M2-like phenotype [77]. A high levels of cytokine and growth factors such as GM-SCF and M-SCF, known to recruit monocytes, were detected in the supernatant of the mixed spheroids [64]. Additionally, a 3D co-culture system demonstrated that CXCL1 is necessary in cell–cell adhesion, and in proliferation, aggressiveness and angiogenesis of cancer cells. In the 3D triple culture system, macrophage migration, ECM protein (Collagen I, IV and FN) deposition, and increased level of pro-invasive factors (MMP1-MMP9) were detected [78]. Collectively, these findings verified that circulatory monocytes recruited to the tumor area acquire an invasive and M2-like phenotype, inhibit T lymphocytes proliferation and suppress their activity, in addition to secreting CCL22 and CCL 24, which mediate the recruitment and differentiation of regulatory T lymphocytes [78].
Cancer cells, fibroblasts, and “Endothelial cells”
Amann A et al. generated multicellular spheroids “microtissues” using the non-small cell lung cell line A549, fibroblast, and endothelial cell lines. After few days, the ECs were detected in the core of the spheroids near the fibroblast and a tubule-like structure was observed within the spheroids. Moreover, an altered morphology of A549 and an overexpression of ɑ-SMA were detected on the surface of both cancer and fibroblast cells reflecting the acquired aggressive phenotype of cancer cells under these conditions [79]. Recently, Paek et al. developed a new micro-physiological model that combined perusable microvasculature with 3D spheroids. This model was used to model human lung vascularized adenocarcinoma. Tumor spheroids formed by A549 and HUVEC were embedded in an ECM hydrogel scaffold containing ECs and lung fibroblast. After seven days, a tubule-like network was detected enwrapping the spheroids. This vascularized solid tumor model can be used to screen the efficiency and toxicity of chemotherapeutic agents [80]. Additionally, Wang et al. showed that the presence of stromal cells is required for the formation and maintenance of multicellular spheroids resembling organoids. They demonstrated that the multicellular tumor spheroid containing HCC culture with fibroblast and endothelial cell line produced a higher level of various proangiogenic (VEGFR2, VEGF, HIF-a), proinflammatory (CXCR4, CXCL12, TNF-a), and EMT (TGFb, vimentin, MMP9) factors compared to monocultured HCC spheroids [63]. The invasive capacity of cancer cells and the potential of triple co-culture to develop tubule-like structures was confirmed using layer by layer (LbL) nanofilm fibronectin-gelatin technique. Oral squamous cell carcinoma (OSCC) was able to invade a 3D culture system formed by 10 layers of normal human dermal fibroblasts (MRC-5) separated by one layer of human dermal lymph ECs (HDLECs), which built a lymphatic capillary structure within the 3D culture system [81]. In another study, Lazzaari et al. created a stable multiculture spheroid using the same technique. Their co-culture using three types of cells, adding VEGF in the medium, with the Layer-by-Layer nanofilm technology clearly revealed the presence of tubule-like web of CD31 + ECs [68]. Collectively, the triple co-culture model can recapitulate the complexity of the TME, allow interaction between cancer cells, stromal cells and the ECM, reveal their role in tumor progression and drugs screening.
3D multicellular cancer model for drug development
3D cancer models are useful tools for diagnosis, drug screening, genome editing, biobanking, and personalized medicine [7, 82]. In this section, we focus on the use of 3D systems as a cancer model for drug development.
Increased resistance to cytotoxic drugs in 3D culture models could be because of several reasons, including higher expression of cancer stem cells (CSC)-associated proteins [83,84,85], upregulation of several drug resistance genes and microRNAs [84], overexpression of multi-drug resistance (MDR) proteins in cultured cells, decreased drug penetration in the multicellular 3D models, and ECM remodeling [84, 86]. In addition, the fact that cells in 3D culture are exposed to gradient levels of the necessary compounds can also explain why cells in different models have different responses [86]. Cell–cell and cell–ECM interactions also contribute to the cellular response to drugs. For instance, Fong et al. found a negative correlation between cell–cell contact and sensitivity of Ewing sarcoma to doxorubicin using a polycaprolactone (PCL)-scaffold-based 3D model [87]. In addition to the abovementioned reasons, different cancer cells can develop dense or loose 3D spheroids thus affecting response to the drug. Cells forming dense spheroids had increased resistance to paclitaxel and doxorubicin compared to cells in 2D culture. In contrast, cells forming loose 3D spheroids displayed the same response to drugs compared to 2D-cultured cells [88]. Drug resistance in 3D culture models is also affected by matrix stiffness that is a determining factor influencing the cell response to anti-cancer agents. A study done by Ki et al. showed that immobilization of EGFR inhibitor (NYQQNC) on PANC-1 cells resulted in decreased cell viability in stiff hydrogels, but not in hydrogels with low stiffness [89]. However, MDA-MB231 breast cancer grown in spheres with stiffer matrixes were more resistant to paclitaxel compared to those grown in soft hydrogels [90].
As mentioned earlier, various 3D monoculture and double/triple co-culture systems differ in cancer cell response to the anti-cancer drugs according to the composition of ECM, the cellular interactions, and the soluble factors secreted by the cells [91, 92]. In a study done by Hwang et al. pancreatic ductal adenocarcinoma PANC-1 tumor spheroids were grown in monocultures and PSCs co-cultures to evaluate response to paclitaxel in terms of IC 50. It was shown that PSCs co-cultures were 100 folds more resistant to the drug treatment rather than monocultures. In a different study, cultured Huh-7 cells in 2D model were more sensitive to sorafenib compared to those in 3D monoculture. Moreover, these cells in 3D culture showed more sensitivity to sorafenib compared to Huh-7 cells co-culture with hepatic stellate cells (HSCs) [76]. On the other hand, another study suggested that the effect of co-culturing could be drug-dependent. Co-culturing increased the sensitivity of Caco-2 spheroids to 5-FU/irinotecan (FI) but did not affect the response of Caco-2 spheroids to 5-FU/oxaliplatin (FO) [93]. Tumor microenvironment is an additional player and contributor to drug response in triple culture. Non-NSCLC cells (NCI-H157) showed increased resistance to paclitaxel in triple culture with CAF and monocytes compared to monocultured cells [77]. However, the interaction with surrounding cells in the tumor microenvironment does not always increase resistance to drug. The response of NCI-H157 to cisplatin was not changed in mono and triple culture condition with CAF and monocytes [77]. Another study considered the sensitivity to doxorubicin in a triple culture of ECs, fibroblasts, and breast tumor cells (MCF-7 and MCF-7/ADR cell lines). Cancer cells in the monoculture system were more sensitive to doxorubicin than those in triple culture condition [94]. Similarly, PANC-1 cells that were cultured with fibroblasts (MRC-5) and HUVEC cells were more resistant to doxorubicin compared to monocultured PANC-1 cells [68]. Finally, Song and colleagues established a new model using primary tumor derived cells and stromal cells [95]. They produced a patient multicellular tumor spheroid (MCTS) using four different cell types: primary hepatocellular carcinoma cells (HCC), stellate cells (LX-2), fibroblasts (WI38) and endothelial cells (HUVECs). Response of primary HCC to three chemotherapeutic drugs (sorafenib, 5-fluouracil, and cisplatin) was assessed in MCT and compared to the response of HCC in monolayer cultures and in 3D spheroids. It was shown that primary HCC cells had different drug sensitivity for the different culturing methods. For instance, primary HCC in MCT culture condition were most mimicking platform for evaluation of responsiveness to sorafenib. It was thus suggested that drug screening and optimization should be done in MCT as they better mimic the in vivo tumor microenvironment in terms of 3D cellular context and cellular heterogeneity [95]. Response of patient-derived cells cultured in MCT to different drugs are thus expected to be translated in vivo, and accordingly such a model could be essential in precision medicine.
Organoids
In early 1900, Ross Harrison used the hanging drop technique to culture a fragment of embryo nerve cord on a drop of lymph on a coverslip [96]. Henry Van Peters Wilson described in vitro organism regeneration during the same time by showing the self-organization capacity of dissociated sponge cells to regenerate a whole organism [97]. These efforts led to the creation of organoids, in vitro tissue-like structures that can recapitulate organ physiology. A few decades later, researchers unveiled the critical role of growth factors and ECM in regulating tissue function and morphogenesis. Thus, the accumulation of knowledge throughout the years guided researchers to optimize the in vitro conditions to generate organoids.
We believe that Simian M and Bissell M provided one of the most accurate definitions of the word organoid, “a unit of function of a given organ that is able to reproduce in culture, a biological structure similar in architecture and function to its counterpart in vivo” [98] Organoids can be derived from normal or malignant human biopsy and obtained by enzymatical or mechanical digestion [99, 100]. These organoids conserve the in vivo, features of original tissues, and therefore offer a new model to understand the physiology of tumor cells; including cancer cell heterogeneity, the composition of original tumor microenvironment, and the interaction between cancer cells and their microenvironment [7]. Moreover, this model promotes disease modeling, molecular diagnosis, gene therapy drug screening and personalized medicine. Organoids have the potential to bridge the gap between in vitro and in vivo models [7, 99, 101]. In this section, we will focus on recent achievements in cancer modeling by organoid technology and discuss the importance of organoids. Sato et al. generated the tumor organoids, that is derived from adult cancer stem cells. The embedment of tumor derived cells in a 3D matrix with specific stem cell media induces cancer stem cell differentiation and formation of a mini structure that resembles intestinal villi and crypts [102]. Using this method, numerous patient-derived tumor organoid that mimic hepatocellular carcinoma [103], glioblastoma [104], gastrointestinal [105], pancreatic [106], colorectal [107], prostate [108], breast [109], bladder [110] and renal cancer [111] were established. This technology is being used to establish organoid line libraries and living biobanks of cancer and normal tissues that preserve heterogeneity among colorectal cancer (CRC) patients [107, 112]. Recently and based on this technique, Brandenberg et al. developed a high throughput automated technique to generate CRC organoids by culturing stem cells in hydrogel based U shape microwell arrays. This method facilitated and enabled a fast aggregation technique of cancer epithelial stem cells, increased homogeneity of organoid size, traces single-organoid from initial cell seeding to downstream analysis, facilitates drug screening, and identifies phenotypic hits [113].
It is noteworthy that patients with the same cancer type may present different histological, genetic and molecular profiles, along with various cellular behavior and chemo-resistance. Vlachogiannis et al. generated patient-derived organoids from 110 metastatic tumor samples derived from 71 patients with colorectal and gastro-esophageal cancer. They provide evidence that organoids maintain the same histological, genetic, and molecular profiles as the original tumor. Patient-derived organoids (PDOs) from different patients exhibited different mutational and transcriptional spectra, which in turn determined the responsiveness of PDO to different chemotherapeutic drugs, and indicating that PDO can be used for drug screening [105].
Moreover, drug response in PDOs was shown to recapitulate the clinical responses. This was shown by comparing the anti-cancer drug response of PDOs during in vitro studies and PDO-based orthotropic xenografts in mice compared to the patient response in clinical trials. Interestingly, in almost all cases, drugs that couldn’t cause a therapeutic response in PDOs were unable to initiate clinical response in the patients. However, drugs that were effective on PDOs were also effective in 88% to 90% of the patients. This study proposed that the utility of PDO in pre-clinical studies was useful for drug screening and for predicting the efficacy of drugs, and thus, PDO would serve as an important tool in personalized medicine [105].
Despite its importance, this organoid composed of solely epithelial cells derived from primary cancer tissues, lacks the presence of stromal, immune and endothelial cells, and requires specific growth factors and supplements. To overcome these problems, Jahnke et al. established a new protocol based on manually generating tumor micro fragments using microdissection of tissues. In this method they do dissecting tumor tissues into 400um fragments to preserve the contact between cancer cells and their microenvironments [114]. This technique was furthermore developed by Li et al. who used air liquid interference (ALI) to culture cancer organoids. They minced fragments derived from gastrointestinal tissues and maintained them in collagen gel under air liquid interference. This methodology maintains the heterogeneous composition of the TME in vitro and supports primary organoid generation, oncogenic transformation, and long-term in vitro culture of cancer organoid derived from murine GI tissue [115]. In an innovative study, Noel et al. used the same methodology to culture clinical PDO samples derived from primary and metastatic tumor cells. Interestingly, PDOs recapitulated the parental tumor histology and genetic profile, and retain fibroblasts, macrophages, Natural Killer, and different subpopulation of T cells (CD4+, CD8+, and CD3+ expressing immune check point surface receptor programmed cell death protein-1 (PD-1). In addition, this model was used to investigate the efficacy of anti-PD1 (Nivolumab). Results showed that T cells activation occurred after treatment with nivolumab in (33%) NSCLC, (25%) RCC, and (33%) melanoma PDOs, consistent with anti-PD-1/PD-L1 response rates in clinical NSCLC trials [116]. Thus, the ALI PDO method offered an advanced model to evaluate individualized patient immune response.
To facilitate organoid formation, Jacob et al. cultured glioblastoma tumor pieces in a defined media on an orbital shaker. This method led to the generation of glioblastoma organoids that can recapitulate the inter- and intra-tumoral heterogeneity and partially preserve the microvasculature and immune cells. Thus, glioblastoma organoids were proposed to be employed to evaluate conventional and new therapeutic strategies [117]. Recently, Horowitz et al. generated cuboidal-shaped micro-dissected tissues or “cuboids” using a standard McIlwain tissue chopper. In parallel, they developed a microfluidic device that enabled them to trap intact cuboids into specific wells for culture and multi-drug treatments. This new platform enabled them to produce large numbers of uniformly-sized cuboids, reduced heterogeneity between samples, facilitated their physical manipulation, and enabled drug screening [118]. Thus, organoids show promising results recapitulating the architecture, physiology and molecular signature of the original tumor. The group of Robert M. Hoffman developed 3D Sponge-matrix Gelfoam to reproduce an in vivo like microenvironment. This method, described by Joseph Leighton in the early 1950s and known as “Sponge-matrix histoculture”, conserves the architecture and physiology of tumor in vitro [119, 120]. Studies revealed that cancer cells grown on sponge matrix maintain their self-organization, proliferation and invasion capacities, heterogeneity and cell cycle phases distribution pattern [121, 122]. This approach was established to evaluate the toxicity and efficacy of chemotherapeutic drugs in vitro. Studies showed a high correlation between in vitro chemosensitivity results, the clinical response, and the survival rate of the cancer patients [123,124,125,126]. Hence, this method can be used to identify new cancer biomarkers, discover new targetable proteins, and screen chemotherapeutic drugs. In a recent study, Yamamoto et al. established patient-derived cancer cell lines using the 3D histoculture method that can be used to understand the cancer biology and to test drug sensitivity [127].
Conclusion
A great deal of evidence has been accumulated in the recent years demonstrating that 3D cell culture systems could be more promising than the traditional 2D models in modeling tumor behavior and physiology. The 3D culture model should contain specific cellular components such as fibroblasts, immune cells, and ECs to model the complex structure of the tumor microenvironment. The interaction of these components with cancer cells promotes tumor progression, remodels the ECM composition, guides cancer cell migration, and enhances cancer invasion by regulating the switches among invasion modes. Indeed, the wide range of customized and tunable biomimetic materials in the 3D models modulates the complex structure, 3D architecture, as well as biochemical and biomechanical cues of the tumor microenvironment. Besides, the 3D model can also simulate the heterogeneity using primary cancer cells instead of a cell line. Patient-derived primary cancer cells conserve the original tumor’s histological, genetic, molecular, and cellular phenotype.
In conclusion, the 3D culture system represents a more precise and comprehensive picture of the tumor microenvironment, unveils the mechanisms underlying tumor behavior, identifies novel molecular and cellular targets, and could participates in pre-clinical drug screening assays. Although these novel technologies can be useful in clinic, they still have a long way to go.
References
Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5(1):17–26.
Knight E, Przyborski S. Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J Anat. 2015;227(6):746–56.
Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2002;99(4):1972–7.
Vergani L, Grattarola M, Nicolini C. Modifications of chromatin structure and gene expression following induced alterations of cellular shape. Int J Biochem Cell Biol. 2004;36(8):1447–61.
Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73(17):5315–9.
Goodbye, flat biology? Nature. 2003;424(6951):861–861.
Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017;23(5):393–410.
Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10(4):241–53.
Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M, et al. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J. 2014;9(9):1115–28.
Weiswald L-B, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia N Y N. 2015;17(1):1–15.
Lovitt CJ, Shelper TB, Avery VM. Cancer drug discovery: recent innovative approaches to tumor modeling. Expert Opin Drug Discov. 2016;11(9):885–94.
Liao Z, Tan ZW, Zhu P, Tan NS. Cancer-associated fibroblasts in tumor microenvironment—accomplices in tumor malignancy. Cell Immunol. 2018;343:103729.
Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20(3):174–86.
Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vasc Health Risk Manag. 2006;2(3):213–9.
Hillen F, Griffioen AW. Tumour vascularization: sprouting angiogenesis and beyond. Cancer Metastasis Rev. 2007;26(3–4):489–502.
Llopiz D, Ruiz M, Infante S, Villanueva L, Silva L, Hervas-Stubbs S, et al. IL-10 expression defines an immunosuppressive dendritic cell population induced by antitumor therapeutic vaccination. Oncotarget. 2017;8(2):2659–71.
Moore KW, de Waal MR, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765.
Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790–811.
Qureshi OS, Zheng Y, Nakamura K, Attridge K, Manzotti C, Schmidt EM, et al. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science. 2011;332(6029):600–3.
Liang B, Workman C, Lee J, Chew C, Dale BM, Colonna L, et al. Regulatory T cells inhibit dendritic cells by lymphocyte activation gene-3 engagement of MHC class II. J Immunol. 2008;180(9):5916–26.
Loke P, Allison JP. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci USA. 2003;100(9):5336–41.
Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211(5):781–90.
Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumor-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416.
LeBleu VS, Macdonald B, Kalluri R. Structure and function of basement membranes. Exp Biol Med Maywood NJ. 2007;232(9):1121–9.
Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK. Extracellular matrix structure. Adv Drug Deliv Rev. 2016;1(97):4–27.
Ananthanarayanan B, Kim Y, Kumar S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials. 2011;32(31):7913–23.
Alowami S, Troup S, Al-Haddad S, Kirkpatrick I, Watson PH. Mammographic density is related to stroma and stromal proteoglycan expression. Breast Cancer Res BCR. 2003;5(5):R129-135.
Peyton SR, Ghajar CM, Khatiwala CB, Putnam AJ. The emergence of ECM mechanics and cytoskeletal tension as important regulators of cell function. Cell Biochem Biophys. 2007;47(2):300–20.
Janmey PA, McCulloch CA. Cell mechanics: integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng. 2007;9:1–34.
Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10(1):34–43.
Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol. 2010;2(12):a005066.
Kamatar A, Gunay G, Acar H. Natural and synthetic biomaterials for engineering multicellular tumor spheroids. Polymers. 2020;12(11):2506.
Ferreira LP, Gaspar VM, Mano JF. Design of spherically structured 3D in vitro tumor models—advances and prospects. Acta Biomater. 2018;15(75):11–34.
Tian H, Tang Z, Zhuang X, Chen X, Jing X. Biodegradable synthetic polymers: preparation, functionalization and biomedical application. Prog Polym Sci. 2012;37(2):237–80.
Fisher SA, Anandakumaran PN, Owen SC, Shoichet MS. Tuning the microenvironment: click-crosslinked hyaluronic acid-based hydrogels provide a platform for studying breast cancer cell invasion. Adv Funct Mater. 2015;25(46):7163–72.
Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci. 2002;115(7):1423–33.
DeLong SA, Moon JJ, West JL. Covalently immobilized gradients of bFGF on hydrogel scaffolds for directed cell migration. Biomaterials. 2005;26(16):3227–34.
Dunne LW, Huang Z, Meng W, Fan X, Zhang N, Zhang Q, et al. Human decellularized adipose tissue scaffold as a model for breast cancer cell growth and drug treatments. Biomaterials. 2014;35(18):4940–9.
Ragelle H, Naba A, Larson BL, Zhou F, Prijić M, Whittaker CA, et al. Comprehensive proteomic characterization of stem cell-derived extracellular matrices. Biomaterials. 2017;1(128):147–59.
Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15(5):378–86.
Nguyen-Ngoc K-V, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, et al. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci. 2012;109(39):E2595–604.
Guzman A, Sánchez Alemany V, Nguyen Y, Zhang CR, Kaufman LJ. A novel 3D in vitro metastasis model elucidates differential invasive strategies during and after breaching basement membrane. Biomaterials. 2017;115:19–29.
Koh I, Cha J, Park J, Choi J, Kang S-G, Kim P. The mode and dynamics of glioblastoma cell invasion into a decellularized tissue-derived extracellular matrix-based three-dimensional tumor model. Sci Rep. 2018;8(1):4608.
Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4(1):38.
Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol. 2011;178(3):1221–32.
Pedron S, Becka E, Harley BAC. Regulation of glioma cell phenotype in 3D matrices by hyaluronic acid. Biomaterials. 2013;34(30):7408–17.
McKenzie AJ, Hicks SR, Svec KV, Naughton H, Edmunds ZL, Howe AK. The mechanical microenvironment regulates ovarian cancer cell morphology, migration, and spheroid disaggregation. Sci Rep. 2018;8(1):1–20.
Wisdom KM, Adebowale K, Chang J, Lee JY, Nam S, Desai R, et al. Matrix mechanical plasticity regulates cancer cell migration through confining microenvironments. Nat Commun. 2018;9(1):1–13.
Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci. 2006;103(29):10889–94.
Umemoto T, Ueno E, Matsumura T, Yamakawa M, Bando H, Mitake T, et al. Ex vivo and in vivo assessment of the non-linearity of elasticity properties of breast tissues for quantitative strain elastography. Ultrasound Med Biol. 2014;40(8):1755–68.
Joyce MH, Lu C, James ER, Hegab R, Allen SC, Suggs LJ, et al. Phenotypic basis for matrix stiffness-dependent chemoresistance of breast cancer cells to doxorubicin. Front Oncol. 2018;8:337.
Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, et al. The nanomechanical signature of breast cancer. Nat Nanotechnol. 2012;7(11):757–65.
Jabbari E, Sarvestani SK, Daneshian L, Moeinzadeh S. Optimum 3D matrix stiffness for maintenance of cancer stem cells is dependent on tissue origin of cancer cells. PLoS ONE. 2015;10(7):e0132377.
Ansardamavandi A, Tafazzoli-Shadpour M, Shokrgozar MA. Behavioral remodeling of normal and cancerous epithelial cell lines with differing invasion potential induced by substrate elastic modulus. Cell Adhes Migr. 2018;12(5):472–88.
Peng Y, Chen Z, Chen Y, Li S, Jiang Y, Yang H, et al. ROCK isoforms differentially modulate cancer cell motility by mechanosensing the substrate stiffness. Acta Biomater. 2019;1(88):86–101.
Riching KM, Cox BL, Salick MR, Pehlke C, Riching AS, Ponik SM, et al. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys J. 2014;107(11):2546–58.
Lang NR, Skodzek K, Hurst S, Mainka A, Steinwachs J, Schneider J, et al. Biphasic response of cell invasion to matrix stiffness in three-dimensional biopolymer networks. Acta Biomater. 2015;1(13):61–7.
Kwon Y, Cukierman E, Godwin AK. Differential expressions of adhesive molecules and proteases define mechanisms of ovarian tumor cell matrix penetration/invasion. PLoS ONE. 2011;6(4):e18872.
Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155(7):1639–51.
Tölle RC, Gaggioli C, Dengjel J. Three-dimensional cell culture conditions affect the proteome of cancer-associated fibroblasts. J Proteome Res. 2018;17(8):2780–9.
Plou J, Juste-Lanas Y, Olivares V, Del Amo C, Borau C, García-Aznar JM. From individual to collective 3D cancer dissemination: roles of collagen concentration and TGF-β. Sci Rep. 2018;8(1):12723.
Li F, Zhao S, Guo T, Li J, Gu C. The nutritional cytokine leptin promotes NSCLC by activating the PI3K/AKT and MAPK/ERK pathways in NSCLC cells in a paracrine manner. BioMed Res Int. 2019;2019:2585743.
Wang Y, Takeishi K, Li Z, Cervantes-Alvarez E, Hortet AC, Guzman-Lepe J, et al. Microenvironment of a tumor-organoid system enhances hepatocellular carcinoma malignancy-related hallmarks. Organogenesis. 2017;13(3):83.
Kuen J, Darowski D, Kluge T, Majety M. Pancreatic cancer cell/fibroblast co-culture induces M2 like macrophages that influence therapeutic response in a 3D model. PLoS ONE. 2017;12(7):182039.
Song H-HG, Park KM, Gerecht S. Hydrogels to model 3D in vitro microenvironment of tumor vascularization. Adv Drug Deliv Rev. 2014;79–80:19–29.
Jobe NP, Rösel D, Dvořánková B, Kodet O, Lacina L, Mateu R, et al. Simultaneous blocking of IL-6 and IL-8 is sufficient to fully inhibit CAF-induced human melanoma cell invasiveness. Histochem Cell Biol. 2016;146(2):205–17.
Hernandez-Fernaud JR, Ruengeler E, Casazza A, Neilson LJ, Pulleine E, Santi A, et al. Secreted CLIC3 drives cancer progression through its glutathione-dependent oxidoreductase activity. Nat Commun. 2017;15(8):14206.
Lazzari G, Nicolas V, Matsusaki M, Akashi M, Couvreur P, Mura S. Multicellular spheroid based on a triple co-culture: a novel 3D model to mimic pancreatic tumor complexity. Acta Biomater. 2018;15(78):296–307.
Estrada MF, Rebelo SP, Davies EJ, Pinto MT, Pereira H, Santo VE, et al. Modelling the tumour microenvironment in long-term microencapsulated 3D co-cultures recapitulates phenotypic features of disease progression. Biomaterials. 2016;78:50–61.
Knuchel S, Anderle P, Werfelli P, Diamantis E, Rüegg C. Fibroblast surface-associated FGF-2 promotes contact-dependent colorectal cancer cell migration and invasion through FGFR-SRC signaling and integrin αvβ5-mediated adhesion. Oncotarget. 2015;6(16):14300–17.
Labernadie A, Kato T, Brugués A, Serra-Picamal X, Derzsi S, Arwert E, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol. 2017;19(3):224–37.
Miyazaki K, Oyanagi J, Hoshino D, Togo S, Kumagai H, Miyagi Y. Cancer cell migration on elongate protrusions of fibroblasts in collagen matrix. Sci Rep. 2019;9(1):292.
Wanless IR. Cirrhosis. In: Johnson LR, editor. Encyclopedia of gastroenterology [Internet]. New York: Elsevier; 2004 [cited 1 May 2021]. p. 356–62. https://www.sciencedirect.com/science/article/pii/B0123868602001362
Drifka CR, Loeffler AG, Esquibel CR, Weber SM, Eliceiri KW, Kao WJ. Human pancreatic stellate cells modulate 3D collagen alignment to promote the migration of pancreatic ductal adenocarcinoma cells. Biomed Microdevices. 2016;18(6):105.
Hwang HJ, Oh M-S, Lee DW, Kuh H-J. Multiplex quantitative analysis of stroma-mediated cancer cell invasion, matrix remodeling, and drug response in a 3D co-culture model of pancreatic tumor spheroids and stellate cells. J Exp Clin Cancer Res CR. 2019;38(1):258.
Khawar IA, Park JK, Jung ES, Lee MA, Chang S, Kuh H-J. Three dimensional mixed-cell spheroids mimic stroma-mediated chemoresistance and invasive migration in hepatocellular carcinoma. Neoplasia N Y N. 2018;20(8):800.
Rebelo SP, Pinto C, Martins TR, Harrer N, Estrada MF, Loza-Alvarez P, et al. 3D-3-culture: a tool to unveil macrophage plasticity in the tumour microenvironment. Biomaterials. 2018;1(163):185–97.
Miyake M, Hori S, Morizawa Y, Tatsumi Y, Nakai Y, Anai S, et al. CXCL1-mediated interaction of cancer cells with tumor-associated macrophages and cancer-associated fibroblasts promotes tumor progression in human bladder cancer. Neoplasia N Y N. 2016;18(10):636–46.
Amann A, Zwierzina M, Koeck S, Gamerith G, Pechriggl E, Huber JM, et al. Development of a 3D angiogenesis model to study tumour—endothelial cell interactions and the effects of anti-angiogenic drugs. Sci Rep. 2017;7(1):1–13.
Paek J, Park SE, Lu Q, Park K-T, Cho M, Oh JM, et al. Microphysiological engineering of self-assembled and perfusable microvascular beds for the production of vascularized three-dimensional human microtissues. ACS Nano. 2019;13(7):7627–43.
Iwai S, Kishimoto S, Amano Y, Nishiguchi A, Matsusaki M, Takeshita A, et al. Three-dimensional cultured tissue constructs that imitate human living tissue organization for analysis of tumor cell invasion. J Biomed Mater Res A. 2019;107(2):292.
Li M, Izpisua Belmonte JC. Organoids—preclinical models of human disease. N Engl J Med. 2019;380(20):1982.
Reynolds DS, Tevis KM, Blessing WA, Colson YL, Zaman MH, Grinstaff MW. Breast cancer spheroids reveal a differential cancer stem cell response to chemotherapeutic treatment. Sci Rep. 2017;7(1):10382.
Ganjibakhsh M, Mehraein F, Koruji M, Aflatoonian R, Farzaneh P. Three-dimensional decellularized amnion membrane scaffold as a novel tool for cancer research; cell behavior, drug resistance and cancer stem cell content. Mater Sci Eng C. 2019;1(100):330–40.
Melissaridou S, Wiechec E, Magan M, Jain MV, Chung MK, Farnebo L, et al. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019;19(1):16.
Xu X, Farach-Carson MC, Jia X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol Adv. 2014;32(7):1256–68.
Fong ELS, Lamhamedi-Cherradi S-E, Burdett E, Ramamoorthy V, Lazar AJ, Kasper FK, et al. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc Natl Acad Sci. 2013;110(16):6500–5.
Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep. 2015;33(4):1837–43.
Ki CS, Shih H, Lin C-C. Effect of 3D matrix compositions on the efficacy of EGFR inhibition in pancreatic ductal adenocarcinoma cells. Biomacromol. 2013;14(9):3017–26.
Lam CRI, Wong HK, Nai S, Chua CK, Tan NS, Tan LP. A 3D biomimetic model of tissue stiffness interface for cancer drug testing. Mol Pharm. 2014;11(7):2016–21.
Langhans SA. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front Pharmacol. 2018;9:6.
Tepper SR, Zuo Z, Khattri A, HEß J, SEIWERT TY. Growth Factor expression mediates resistance to EGFR inhibitors in head and neck squamous cell carcinomas. Oral Oncol. 2016;1(56):62–70.
Hoffmann OI, Ilmberger C, Magosch S, Joka M, Jauch K-W, Mayer B. Impact of the spheroid model complexity on drug response. J Biotechnol. 2015;10(205):14–23.
Wang S, Mao S, Li M, Li H-F, Lin J-M. Near-physiological microenvironment simulation on chip to evaluate drug resistance of different loci in tumour mass. Talanta. 2019;1(191):67–73.
Song Y, Kim J-S, Kim S-H, Park YK, Yu E, Kim K-H, et al. Patient-derived multicellular tumor spheroids towards optimized treatment for patients with hepatocellular carcinoma. J Exp Clin Cancer Res CR [Internet]. 2018. Cited 6 Sept 2019;37. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5970513/
Harrison RG, Greenman MJ, Mall FP, Jackson CM. Observations of the living developing nerve fiber. Anat Rec. 1907;1(5):116–28.
Wilson HV. On some phenomena of coalescence and regeneration in sponges. J Exp Zool. 1907;5(2):245–58.
Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol. 2017;216(1):31–40.
Heydari Z, Moeinvaziri F, Agarwal T, Pooyan P, Shpichka A, Maiti TK, et al. Organoids: a novel modality in disease modeling. Bio Des Manuf. 2021. https://doi.org/10.1007/s42242-021-00150-7.
Zahmatkesh E, Khoshdel-Rad N, Mirzaei H, Shpichka A, Timashev P, Mahmoudi T, et al. Evolution of organoid technology: lessons learnt in Co-Culture systems from developmental biology. Dev Biol. 2021;1(475):37–53.
Aboulkheyr Es H, Montazeri L, Aref AR, Vosough M, Baharvand H. Personalized cancer medicine: an organoid approach. Trends Biotechnol. 2018;36(4):358–71.
Sato T, Stange DE, Ferrante M, Vries RGJ, van Es JH, van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and barrett’s epithelium. Gastroenterology. 2011;141(5):1762–72.
Broutier L, Mastrogiovanni G, Verstegen MMA, Francies HE, Gavarró LM, Bradshaw CR, et al. Human primary liver cancer -derived organoid cultures for disease modelling and drug screening. Nat Med. 2017;23(12):1424.
Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, et al. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465–77.
Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920.
Boj SF, Hwang C-I, Baker LA, Chio IIC, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–38.
van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45.
Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–87.
Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(12):373–38610.
Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173(2):515–52817.
Grassi L, Alfonsi R, Francescangeli F, Signore M, De Angelis ML, Addario A, et al. Organoids as a new model for improving regenerative medicine and cancer personalized therapy in renal diseases. Cell Death Dis. 2019;10(3):201.
Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18(6):827–38.
Brandenberg N, Hoehnel S, Kuttler F, Homicsko K, Ceroni C, Ringel T, et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat Biomed Eng. 2020;4(9):863–74.
Jahnke H-G, Poenick S, Maschke J, Kendler M, Simon JC, Robitzki AA. Direct chemosensitivity monitoring ex vivo on undissociated melanoma tumor tissue by impedance spectroscopy. Cancer Res. 2014;74(22):6408–18.
Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20(7):769–77.
Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175(7):1972-1988.e16.
Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180(1):188-204.e22.
Horowitz LF, Rodriguez AD, Au-Yeung A, Bishop KW, Barner LA, Mishra G, et al. Microdissected “cuboids” for microfluidic drug testing of intact tissues. Lab Chip. 2021;21(1):122–42.
Leighton J. The growth patterns of some transplantable animal tumors in sponge matrix tissue culture. J Natl Cancer Inst. 1954;15(2):275–93.
Sherwin RP, Richters A, Yellin AE, Donovan AJ. Histoculture of human breast cancers. J Surg Oncol. 1980;13(1):9–20.
Hoffman RM, Freeman AE. In vivo-like growth patterns of multiple types of tumors in Gelfoam® Histoculture. Methods Mol Biol Clifton NJ. 2018;1760:19–28.
Yano S, Miwa S, Mii S, Hiroshima Y, Uehara F, Kishimoto H, et al. Cancer cells mimic in vivo spatial-temporal cell-cycle phase distribution and chemosensitivity in 3-dimensional Gelfoam® histoculture but not 2-dimensional culture as visualized with real-time FUCCI imaging. Cell Cycle. 2015;14(6):808–19.
Vescio RA, Redfern CH, Nelson TJ, Ugoretz S, Stern PH, Hoffman RM. In vivo-like drug responses of human tumors growing in three-dimensional gel-supported primary culture. Proc Natl Acad Sci USA. 1987;84(14):5029–33.
Hoffman RM. Clinical correlation of the histoculture drug response assay in gastrointestinal cancer. Methods Mol Biol Clifton NJ. 2018;1760:61–72.
Hoffman RM, Jung P-S, Kim M-B, Nam J-H. Prospective clinical correlation of the histoculture drug response assay for ovarian cancer. Methods Mol Biol Clifton NJ. 2018;1760:73–81.
Hoffman RM. Clinical correlation of the histoculture drug response assay for head and neck cancer. Methods Mol Biol Clifton NJ. 2018;1760:83–92.
Yamamoto J, Sugisawa N, Hamada K, Nishino H, Miyake K, Matsuyama R, et al. A universal gelfoam 3-D histoculture method to establish patient-derived cancer cells (3D-PDCC) without fibroblasts from patient-derived xenografts. Anticancer Res. 2020;40(12):6765–8.
Funding
This paper was not funded.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no declarations or other relevant affiliations with any organization or entity with a financial interest in or conflict with the subject matter or materials discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Atat, O.E., Farzaneh, Z., Pourhamzeh, M. et al. 3D modeling in cancer studies. Human Cell 35, 23–36 (2022). https://doi.org/10.1007/s13577-021-00642-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00642-9