Abstract
Prostate cancer (PCA) is an epithelial malignant tumor occurring in the prostate gland. It is the second most common male cancer in the world and one of the top five cancer deaths in men. To combat this disease, it is needed to identify important tumor suppressor genes and elucidate the molecular mechanisms. S100 calcium-binding protein A14 (S100A14), a member of the S100 family, is located on chromosome 1q21.3 and contains an EF-hand motif that binds calcium. S100A14 is involved in a variety of tumor biological processes in several types of cancers. Its expression level and related biological functions are tissue or tumor specific. However, its possible effects on prostate cancer are still unclear. Herein, we found the low expression of S100A14 in human prostate cancer tissues and cell lines. S100A14 suppressed the proliferation of prostate cancer cells and promoted cell apoptosis. Additionally, S100A14 suppressed the motility and EMT processes of prostate cancer cells. We further found S100A14 promoted the expression of FAT1 and activated the Hippo pathway, which, therefore, suppressed the prostate cancer progression. The in vivo assays confirmed that S100A14 suppressed tumor growth of prostate cancer cells through FAT1-mediated Hippo pathway in mice. In conclusion, we clarified the mechanism underlying S100A14 suppressing prostate cancer progression and, therefore, we thought S100A14 could serve as a tumor suppressor protein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCA) is an epithelial malignant tumor occurring in the prostate gland [1]. It is the second most common male cancer in the world and one of the top five cancer deaths in men [2]. The treatment of prostate cancer should be considered comprehensively according to the age and severity of the patient, mainly including surgical treatment, radiotherapy and chemotherapy, targeted therapy and immunotherapy, etc. [3]. To prevent and control advanced and metastatic tumors, it is necessary to find markers that predict clinical progression and treatment strategies [4]. The inactivation or overactivation of tumor suppressor genes or proto-oncogenes in many types of cancer, including prostate cancer, is often an important factor in tumor development [5]. To combat this disease, it is needed to identify important tumor suppressor genes and elucidated the molecular mechanisms.
S100 calcium-binding protein A14 (S100A14), a member of the S100 family, is located on chromosome 1q21.3 and contains an EF-hand motif that binds calcium [6]. S100A14 mediated multiple cellular processes and have several physiological functions [7, 8]. S100A14 protein is involved in cell invasion by affecting the expression and function of MMP2 via transcriptional regulation [9]. Additionally, it also stimulates cell proliferation or apoptosis at different concentrations via the receptor for advanced glycation end products (RAGE) [10].
Recently, the wide effects of S100A14 have been well revealed [11, 12]. It is involved in a variety of tumor biological processes, including proliferation, apoptosis, cell motility, and signal transduction [9, 11]. However, its expression level and related biological functions are tissue or tumor specific [13]. Studies have shown that S100A14 can activate the mitogen-activated protein kinase (MAPK) pathway, and then induce lung adenocarcinoma cell proliferation, cervical cancer cell EMT and breast cancer cell metastasis [14, 15]. S100A14 also served as a tumor suppressor gene; it suppressed FAK pathway and MMPs by blocking stored Ca2+ influx [11]. However, its possible effects on prostate cancer are still unclear.
In this study, we assessed the expression levels of S100A14 in human prostate cancer tissues and cell lines, investigated its effects on prostate cancer cell proliferation, apoptosis, motility, and EMT processes, and explore the possible mechanisms. Our data revealed that S100A14 suppressed prostate cancer progression through FAT1-mediated Hippo pathway, and therefore we thought S100A14 could serve as a tumor suppressor protein.
Materials and methods
Bioinformatic analysis
Gene Expression Profiling Interactive Analysis (GEPIA) was used to analyze S100A14 or FAT1 expression in prostate cancer tissues from The Cancer Genome Atlas (TCGA) database.
Antibodies and plasmids
Anti-S100A14 antibody (1:100 dilution for Immunohistochemical (IHC), 1:500 dilution for Immunoblot, ab251779, abcam), anti-Ki67 antibody (1:200 dilution for IHC, 1:2000 dilution for Immunoblot, ab15580, abcam), anti-E-cadherin antibody (1:200 dilution for IHC, 1:1000 dilution for Immunoblot, ab40772, abcam), anti-p-YAP1 antibody (1:200 dilution for IHC, 1:500 dilution for Immunoblot, ab62751, abcam), anti-YAP1 antibody (1:500 dilution, ab52771, abcam), anti-FAT1 antibody (1:100 dilution for IHC, 1:500 dilution for Immunoblot, ab190242, abcam) anti-N-cadherin antibody (1:500 dilution, ab245117, abcam), anti-LATS1 antibody (1:500 dilution, ab234820, abcam), anti-p-LATS1 antibody (1:500 dilution, ab111344, abcam), anti-MST-1 antibody (1:500 dilution, ab231138, abcam), anti-p-MST-1 antibody (1:500 dilution, ab247466, abcam), anti-Bax antibody (1:500 dilution, ab32503, abcam), anti-Bcl-2 antibody (1:500 dilution, ab182858, abcam), anti-PCNA antibody (1:2000 dilution, ab92552, abcam), anti-p21 antibody (1:1000 dilution, ab109520, abcam), anti-β-actin antibody (1:3000 dilution, ab8226, abcam). The plasmids of pcDNA3.1-S100A14, sh1-S100A14, sh2-S100A14, sh1-FAT1, and sh2-FAT1 plasmids were constructed by ourselves.
Immunohistochemical (IHC) assays
This study is concerned with animal and human experimental compliance Guide for the Care and Use of Laboratory Animals and World Medical Association Declaration of Helsinki. The operations in the assays were all approved by the Institutional Animal Care and Use Committee (IACUC) of Fujian Medical University Union Hospital. All procedures performed in studies involving human participants were in accordance with the standards upheld by the Ethics Committee of Fujian Medical University Union Hospital. The expression levels of proteins in tumor tissues were detected through IHC assays. The sections (5 µm thick) were fixed by the use of 4% paraformaldehyde (PFA) at room temperature for 30 min. The sections were subsequently blocked using 2% BSA for another 30 min. The blocked sections were then incubated through the use of antibodies for 2 h and subsequently incubated with biotinylated secondary antibodies for another 1.5 h. At last, the chromogen substrate was used.
Cell culture and transfection
The human prostate cells (RWPE-1), and 4 types of prostate cancer cells, including DU-145, PC-3, VCaP, and LNCaP cells, were all bought from ATCC and maintained in DMEM supplemented with 10% of fetal bovine serum (FBS) and incubated at 37 °C in a 5% CO2 incubator.
The plasmids were transfected into endometrial cancer cells by the use of Lipofectamine® 2000 (Invitrogen; Thermo Fisher).
Quantitative PCR assays
Total RNA was extracted from prostate cancer cells using Trizol reagent (15596-018, Invitrogen). Then total RNA was reverse-transcribed by the use of M-MLV reverse transcriptase kit (M1701, Promega). Quantitative PCR was then performed through the use of the SYBR mixture (RR420A, Takara). The expression levels of S100A14 were normalized to the expression of GAPDH. The sequences of S100A14 quantitative PCR primers were: forward, 5′-GAGACGCTGACCCCTTCTG-3′ and reverse, 5′-CTTGGCCGCTTCTCCAATCA-3′; the sequences of FAT1 quantitative PCR primers were: forward, 5′-CATCCTGTCAAGATGGGTGTTT-3′ and reverse, 5′-ACTCTTCCGCTTTGAACAGGT-3′; the sequences of GAPDH were: 5′-CGACCACTTTGTCAAGCTCA-3′ and reverse, 5′-GGTTGAGCACAGGGTACTTTATT-3′.
Immunoblot assay
Cells or tumor tissues were lysed by the use of RIPA buffer (9800; Cell Signaling). All the cell and tissue samples were isolated to extract the proteins and separated through SDS-PAGE, then transferred onto PVDF membranes, followed by blocking with 5% fat-free milk in TBST. All membranes were subsequently treated with primary antibodies that targeted a series of proteins for 1.5 h. Subsequently, the membranes were incubated with secondary antibodies at room temperature for 1 h. Signals were then detected.
CCK-8 assay
Cells were plated into the 96-well plates with a density of 1000 cells and subsequently maintained for 48 h upon the indicated treatment. Cells were then treated with CCK-8 for 4 h and the OD value was measured at 490 nm wavelength.
Edu and TUNEL assay
Cells were plated into the 24-well plates with a density of 10,000 cells and subsequently maintained for 48 h upon the indicated treatment. Cells were then treated with the Edu (abcam) or TUNEL kit (abcam). Then Immunofluorescence assays were performed to detect Edu-positive or TUNEL-positive cells.
Scratch wound assay
Cells transfected with indicated plasmids were grown to confluency. Then scratches were made with a 20 μL pipette tip, followed by washing with PBS buffer. Subsequently, culture medium was added to induce wound closure. Images were taken to analyze cell migration ability.
Transwell assays
Cells transfected with indicated plasmids were plated into the upper chamber of Transwell chambers in serum-free culture medium. Then complete culture medium containing 10% FBS was added into the bottom chambers to stimulate cell migration. After maintaining for 24 h, cells in the upper chamber were removed, and the migration cells were fixed, stained with 0.1% crystal violet and quantified.
Immunofluorescence
Cells were washed by PBS buffer, fixed and permeabilized with PBST (0.1% Triton X-100 in PBS). After blocking in 2% BSA in PBST for 30 min, cells were incubated with the antibodies at room temperature. Cells were rinsed with PBS, followed by Alexa 488 or Alexa 555-conjugated secondary antibodies (Abcam). After being counterstained with DAPI, all the images were captured using a fluorescence microscope (Carl-Zeiss).
Tumor growth in vivo assay
The operations in animal assays were all approved by the Institutional Animal Care and Use Committee (IACUC) of Fujian Medical University Union Hospital. The BALB/c nude mice (18–22 g, male, 8 weeks old) were purchased from Beijing Vital River Company. All mice in this study were fed with enough food and water, and kept at a SPF level. 12 nude mice were used in this study (6 for each group). To measure tumor growth capacity in vivo, PC-3 cells were stably transfected with S100A14 overexpression plasmids and injected into the right flank of male nude mice. The volume of tumors was measured every 5 days and calculated. After 30 days, the tumor growth curves were calculated and the tumor weight and volume was compared between different groups.
Statistics
GraphPad 6.0 was used for statistical analysis. Data were represented as mean ± SD. Student’s t test was used for the comparisons, and p < 0.05 was considered as significant. * or # indicates p < 0.05, **or ## p < 0.01 and ***or ### p < 0.001, respectively.
Results
S100A14 was low expression in human prostate cancer tissues and cell lines.
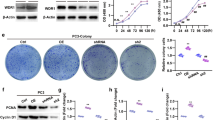
To clarify the possible role of S100A14 in the progression of prostate cancer, we first detect its expression levels in prostate cancer tissues and normal tissues according to the TCGA database. Importantly, we found the expression of S100A14 in 492 tumor tissues was obviously lower than that in 152 normal tissues (Fig. 1a). In addition, it was shown that the transcript per million of S100A14 in 497 primary tumor tissues was lower than 52 normal tissues (Fig. 1b). Through quantitative PCR assays, we detected the mRNA levels of S100A14 in prostate cancer tissues and normal tissues collected in the hospital, and found low expression of S100A14 in human prostate cancer tissues, compared to normal tissues (Fig. 1c). Similarly, we found the protein levels of S100A14 in 4 representative prostate cancer tissues were significantly low, compared to the 4 paired normal tissues, through Immunoblot assays (Fig. 1d). Performing IHC assays, we also noticed the low expression of S100A14 in prostate cancer tissues, compared to normal tissues (Fig. 1e).
S100A14 was less expressed in human prostate cancer tissues and cell lines. a TCGA database showed the mRNA levels of S100A14 in 492 prostate cancer tissues and 152 normal tissues. b TCGA database showed the transcript per million of S100A14 in 497 primary tumor tissues and 52 normal tissues. c qPCR assays showed the mRNA levels of S100A14 in prostate cancer tissues and adjacent tissues from our hospital. d Immunoblot assays showed the expression of S100A14 in 8 prostate cancer primary tissues and adjacent tissues. e IHC assays showed the expression of S100A14 in 3 representative prostate cancer primary tissues and adjacent tissues (100 and 200 magnification, respectively). f Immunoblot assays showed the expression of S100A14 in RWPE-1, DU145, PC-3, VCaP, and LNCaP cells. All experiments were repeated three times. Data are presented as mean ± SD
We further detected the expression of S100A14 in human prostate cancer cell lines DU-145, PC-3, VCaP, and LNCaP, and normal prostate cell line RWPE-1. Through Immunoblot assays, we noticed the low expression of S100A14 in prostate cancer cell lines, compared to that in normal cell line (Fig. 1f). Collectively, we thought S100A14 was low expression in human prostate cancer tissues and cell lines.
S100A14 mediates prostate cancer cell proliferation and apoptosis
To further confirm the effects of S100A14 on the progression of prostate cancer, we then performed series of in vitro assays. Two prostate cancer cell lines, PC-3 and LNCaP, were used as cell model of prostate cancer. The overexpression plasmid, pcDNA3.1-S100A14, and two types of shRNA plasmids, sh1-S100A14 and sh2-S100A14, were transfected into PC-3 and LNCaP respectively, to change the expression of S100A14 in prostate cancer cells. Through Immunoblot assays, we confirmed the increased expression of S100A14 after the transfection of pcDNA3.1-S100A14 in PC-3 and LNCaP cell, respectively (Fig. 2a). Additionally, the transfection of sh1-S100A14 and sh2-S100A14 in PC-3 and LNCaP cells successfully decreased the expression of S100A14 (Fig. 2a).
S100A14 mediates prostate cancer cell proliferation and apoptosis. a Immunoblot showed the expression of S100A14 in PC-3 and LNCaP cells transfected with the indicated plasmids. b CCK-8 assays showed the proliferation capacity PC-3 and LNCaP cells transfected with the indicated plasmids. c Colony formation assays were performed and showed the colony number of PC-3 and LNCaP cells transfected with the indicated plasmids. d Edu assays showed the percentage of Edu-positive PC-3 and LNCaP cells transfected with the indicated plasmids. e TUNEL assays showed the apoptosis degree of PC-3 and LNCaP cells transfected with the indicated plasmids. Data are presented as mean ± SD, pcDNA3.1-S100A14 vs pcDNA3.1,*p < 0.05, **p < 0.01, sh-S100A14 vs sh-NC, #p < 0.05, ##p < 0.01. NC negative control. All experiments were repeated three times
Performing CCK-8 assays, we found the overexpression of S100A14 dramatically decreased the OD value at 490 nm wavelength in PC-3 and LNCaP cells, whereas its depletion obviously increased OD value, suggesting S100A14 suppressed the proliferation of prostate cancer cells (Fig. 2b). Similarly, colony formation assays also confirmed that S100A14 overexpression suppressed the colony formation of PC-3 and LNCaP cells, reversely, its depletion increase colony number remarkably (Fig. 2c). Furthermore, performing Edu proliferation assays, we noticed the decrease percentage of Edu-positive PC-3 and LNCaP cells after S100A14 overexpression, whereas knockdown of S100A14 increased Edu-positive cell number (Fig. 2d). These results confirmed S100A14 suppressed the proliferation of prostate cancer cells.
We then detected the effects of S100A14 on the apoptosis of prostate cancer cells through TUNEL assays. We found that S100A14 overexpression stimulated the apoptosis of PC-3 and LNCaP cells, with the decrease of TUNEL-positive cell number, and the ablation of S100A14 inhibited prostate cancer cell apoptosis (Fig. 2e). Therefore, we thought S100A14 could mediate prostate cancer cell proliferation and apoptosis.
S100A14 restrained the motility and EMT of prostate cancer cells
Previous data suggest that S100A14 might suppress the progression of prostate cancer via mediating cell proliferation and apoptosis, and we then investigated the effects of S100A14 on the motility and EMT of prostate cancer cells.
Through wound closure assay, we found S100A14 overexpression suppressed the migration of PC-3 and LNCaP cells, and its ablation promoted cell migration (Fig. 3a). In addition, performing transwell assays, we also found the inhibition of cell invasion caused by S100A14 overexpression, whereas its depletion promoted the invasion of PC-3 and LNCaP cells (Fig. 3b). In addition, we further found S100A14 overexpression suppressed the EMT process of PC-3 and LNCaP cells, reversely, its depletion promoted the EMT of prostate cancer cells (Fig. 3c). Therefore, we thought S100A14 restrained the motility and EMT of prostate cancer cells.
S100A14 restrained the motility and EMT of prostate cancer cells. a Wound closure assays showed the wound healing capacity PC-3 and LNCaP cells transfected with the indicated plasmids. b Transwell assays showed the invasion capacity PC-3 and LNCaP cells transfected with the indicated plasmids. c Immunofluorescence assays were performed, and the expression levels of N-cadherin and E-cadherin were detected in PC-3 and LNCaP cells. Data are presented as mean ± SD, pcDNA3.1-S100A14 vs pcDNA3.1,*p < 0.05, **p < 0.01, sh-S100A14 vs sh-NC, #p < 0.05, ##p < 0.01. NC negative control. All experiments were repeated three times
S100A14 promotes FAT1 expression and activates the Hippo pathway
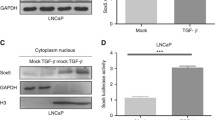
Since the previous study showed S100A14 affected cancer progression via promoting FAT1 expression [15]. We then investigated whether S100A14 suppressed prostate cancer progression via activating FAT1 expression. We first detected the expression of FAT1 in tumor tissues and normal tissues through qPCR assays and found the obvious decrease of FAT1 mRNA levels in tumor tissues compared to normal tissues (Fig. 4a). The results of IHC assays also confirmed the decrease of FAT1 protein expression in tumor tissues (Fig. 4b).
S100A14 promotes FAT1 expression and activates the Hippo pathway. a. qPCR assays showed the mRNA levels of FAT1 in prostate cancer tissues and adjacent tissues from our hospital. b. IHC assays showed the expression of FAT1 in 3 representative prostate cancer primary tissues and adjacent tissues (100 and 200 magnification, respectively). c The correlation analysis between the mRNA levels of FAT1 and S100A14 in prostate cancer tissues. d Immunoblot showed the expression of indicated proteins in PC-3 and LNCaP cells transfected with the plasmids of control, S100A14, and shRNA plasmids of control and S100A14. Data are presented as mean ± SD, pcDNA3.1-S100A14 vs pcDNA3.1, **p < 0.01, ***p < 0.001, shS100A14 vs shNC, ##p < 0.01, ###p < 0.001. NC negative control. e Immunoblot showed the expression of indicated proteins in PC-3 and LNCaP cells transfected with the plasmids of control, S100A14, and two types of FAT shRNA plasmids. Data are presented as mean ± SEM, pcDNA3.1-S100A14 vs pcDNA3.1,*p < 0.05, **p < 0.01, S100A14 + sh-FAT1 vs S100A14 + sh-NC, #p < 0.05, ##p < 0.01, ###p < 0.001. NC negative control. All experiments were repeated three times. Data are presented as mean ± SD
Then we investigated the correlation between the mRNA levels of S100A14 and FAT1 in prostate cancer tissues. Interestingly, we found S100A14 mRNA levels were positively correlated with FAT1 mRNA levels (p < 0.001), suggesting the correlation between S100A14 and FAT1 in prostate cancer progression (Fig. 4c).
It was reported that FAT1 affected cancer progression via activating the Hippo pathway [15]. Subsequently, we detected the effects of S100A14 on FAT1-mediated Hippo pathway in prostate cancer cells through Immunoblot assays. We noticed the increase of FAT1 expression in prostate cancer cells after S100A14 overexpression, whereas its depletion decreased FAT1 expression (Fig. 4d). Furthermore, the phosphorylation levels of LATS1, MST1, and YAP1 were obviously increased after S100A14 overexpression and dramatically decreased after S100A14 depletion (Fig. 4d). This result suggested that S100A14 activated FAT1 expression and LATS1–MST1–YAP1 axis in prostate cancer cells.
Furthermore, we detected the effects of FAT1 depletion on the Hippo pathway of prostate cancer cells after S100A14 overexpression. Through Immunoblot assays, we found S100A14 overexpression increased FAT1 expression and activated LATS1–MST1–YAP1 axis, whereas the depletion of FAT1 by two shRNAs, sh1-FAT1 and sh2-FAT1, decreased the phosphorylation levels of LATS1, MST1, and YAP1 caused by S100A14 overexpression (Fig. 4e). Therefore, we thought S100A14 promoted FAT1 expression and activated Hippo pathway.
The depletion of FAT1 reversed the suppression of cell proliferation and EMT caused by S100A14 overexpression in prostate cancer cells
Subsequently, we detected the effects of FAT1 on prostate cancer cell growth and EMT processes. We noticed S100A14 overexpression suppressed PC-3 cell proliferation, through CCK-8 and Edu assays (Fig. 5a, b). Whereas the depletion of FAT1 rescued the suppression of PC-3 cell proliferation caused by S100A14 overexpression (Fig. 5a, b). Through TUNEL assays, we found S100A14 overexpression stimulated cell apoptosis, and the knockdown of FAT1 reversed the stimulation of cell apoptosis (Fig. 5c).
The depletion of FAT1 reversed the suppression of cell proliferation and EMT caused by S100A14 overexpression in prostate cancer cells. a CCK-8 assays showed the proliferation capacity PC-3 cells transfected with the indicated plasmids. b Edu assays showed the percentage of Edu-positive PC-3 cells transfected with the indicated plasmids. c TUNEL assays showed the apoptosis degree of PC-3 cells transfected with the indicated plasmids. d Wound closure assays showed the wound healing capacity PC-3 cells transfected with the indicated plasmids. e Transwell assays showed the invasion capacity PC-3 cells transfected with the indicated plasmids. f Immunoblot assays showed the expression of the indicated proteins in PC-3 cells transfected with the plasmids of control and S100A14, and two types of FAT sh-RNA plasmids. Data are presented as mean ± SD, pcDNA3.1-S100A14 vs pcDNA3.1,*p < 0.05, **p < 0.01, ***p < 0.001, sh-S100A14 vs sh-NC, #p < 0.05, ##p < 0.01, ###p < 0.001. NC negative control. All experiments were repeated three times
Additionally, through wound closure and transwell assays, we found S100A14 overexpression suppressed PC-3 cell migration and invasion, and FAT1 depletion promoted migration and invasion of prostate cancer cells after S100A14 overexpression (Fig. 5d, e). We then performed Immunoblot assays, and found the decrease of PCNA, Bcl-2, and N-cadherin expression, and the increase of p21, Bax, and E-cadherin expression in PC-3 cells after S100A14 overexpression (Fig. 5f). However, FAT1 depletion obviously rescued the alteration of expression of these proteins caused by S100A14 overexpression (Fig. 5f). We, therefore, thought the depletion of FAT1 reversed the suppression of cell proliferation and EMT caused by S100A14 overexpression in prostate cancer cells.
S100A14 suppressed tumor growth of prostate cancer cells through FAT1-mediated Hippo pathway
We then detected the effects of S100A14 on tumor growth of prostate cancer cells in vivo. We constructed the stably S100A14 overexpressed PC-3 cells, and then a total of 105 PC-3 cells were injected subcutaneously into the abdomen of nude mice (n = 6 for each group). After 5 days, tumor began formation. The tumor volume every 5 days and until 30 days was measured. Interestingly, S100A14 overexpression obviously suppressed tumor growth of prostate cancer cells (Fig. 6a). Additionally, the tumor weight was decreased significantly in S100A14 overexpression groups (Fig. 6a).
S100A14 suppressed tumor growth of prostate cancer cells through FAT1-mediated Hippo pathway. a The control (NC), S100A14 overexpression plasmids stably transfected PC-3 cells were injected subcutaneously into the abdomen of nude mice (n = 6 for each group) to induce tumor growth. The tumor growth curves were conducted according to the measure of tumor volume every 5 days, and the tumor weight was measured. b IHC assays showed the expression of the indicated proteins in control or S100A14 overexpression tumor tissues. c HE staining showed the tumor tissues from control or S100A14 overexpression groups. d Immunoblot assays showed the expression of the indicated proteins tumor tissues from control or S100A14 overexpression groups. Data are presented as mean ± SD. ***p < 0.001. All experiments were repeated three times
Performing IHC assays, we found the expression levels of S100A14 and FAT1 were obviously increased in S100A14 overexpression tumor tissues (Fig. 6b). Moreover, the phosphorylation levels of YAP1 were increased in S100A14 overexpression tumor tissues, consistent with the previous data (Fig. 6b). We further found Ki67 was downregulated whereas E-cadherin expression was increased after S100A14 overexpression in tumor tissues (Fig. 6b), suggesting the inhibition of tumor growth and EMT process. Additionally, through HE staining assays, we found S100A14 overexpression in tumor tissues suppressed the tumor growth (Fig. 6c). Similarly, through Immunoblot assays, we found the increased expression of S100A14, p-YAP1, FAT1, and E-cadherin, and decrease expression of Ki67 in tumor tissues from the S100A14 overexpression group (Fig. 6d). Therefore, we thought S100A14 suppressed tumor growth of prostate cancer cells through FAT1-mediated Hippo pathway.
Discussion
Prostate cancer is often asymptomatic in its early stage, while advanced prostate cancer has a poor prognosis [16]. Conventional treatments, such as radioactive particle implantation, radical prostatectomy, radiotherapy, and chemotherapy, have limited effect on the treatment of advanced prostate cancer [17]. Although endocrine therapy, immunotherapy and targeted therapy have great potential in the treatment of advanced prostate cancer, there are few successful drug options for clinical use [18]. To improve the survival rate of patients, further elucidation of the pathogenesis of prostate cancer, identification of key tumor suppressor genes, and development of effective therapeutic targets are needed [19]. In this study, we noticed the low S100A14 expression in human prostate cancer tissues and cell lines. Our data further confirmed the suppression of prostate cancer cell proliferation, motility, and EMT process, and the stimulation of apoptosis by S100A14. We, therefore, thought S100A14 could act as a tumor suppressor protein of prostate cancer.
In the previous studies, the role of S100A14 in the progression of multiple types of tumors has been widely revealed [12, 20]. S100A14 inhibited the metastasis of nasopharyngeal carcinoma by the suppression of NF-kB pathway through degradation of IRAK1 [20]. As a comparison, we noticed S100A14 suppressed the progression of prostate cancer through FAT1-mediated Hippo pathway. Whereas in breast cancer, S100A14-CCL2/CXCL5 promoted the metastasis of tumor [9]. The loss of S100A14 at the tumor-invading front also correlated with a worse prognosis in oral squamous cell carcinoma and colorectal cancer [21, 22]. Previous studies also concluded that S100A14 was functionally linked with series of tumor regulatory molecules such as p21, p53, MMPs and RAGE [23]. Our study confirmed that FAT1-mediated Hippo signaling pathway is involved in the regulation of S100A14 in prostate cancer. The findings, therefore, enriched the understanding of the role of S100A14 in cancer progression.
In this study, we also investigated the effects of the Hippo pathway in the progression of prostate cancer. This pathway is widely involved in the regulation of cancer progression in different types of cancers [24, 25]. The critical members of the Hippo pathway, such as YAP1, LATS1, and MST1, could all affect the cancer progression and metastasis [25]. The alteration of phosphorylation levels of these proteins affected multiple types of tumors [25]. ACTN1 promoted tumor growth by inhibiting the Hippo pathway in hepatocellular carcinoma [26]. YAP could manipulate proliferation in lung adenocarcinomas [27]. We also confirmed S100A14 suppressed prostate cancer progression via the LATS1–MST1–YAP1 axis, suggesting that the Hippo pathway could serve as a potential molecular target for prostate cancer treatment.
As a transmembrane protein, FAT1 is involved in the regulation of intercellular adhesion, cell growth, migration, actin dynamics and orientation, and plays an important role in tumorigenesis and development [28]. FAT1 mutations lead to abnormal activation of the Wnt/β-catenin pathway in different tumors [29]. FAT1 loss promotes the occurrence of tumors through the Hippo pathway. In esophageal squamous cell carcinoma, FAT1 promotes epithelial–mesenchymal transformation (EMT) through the MAPK/ERK signaling pathway [30]. We, therefore, speculate that S100A14 may influence the progression of prostate cancer through FAT1. The experimental results also showed that S100A14 mainly affected the progression of prostate cancer through FAT1-mediated Hippo pathway.
In conclusion, we noticed the low expression of S100A14 in human prostate cancer tissues and cell lines. S100A14 suppressed cell proliferation, motility, EMT, and promoted apoptosis of prostate cancer cells. We further found S100A14 promoted the expression of FAT1 and activated FAT1-mediated Hippo pathway, and therefore suppressed prostate cancer progression. Our data, therefore, confirmed S100A14 could act as a tumor suppressor protein of prostate cancer.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Bland T, Wang J, Yin L, Pu T, Li J, Gao J, et al. WLS-Wnt signaling promotes neuroendocrine prostate cancer. iScience. 2021;24(1):101970. https://doi.org/10.1016/j.isci.2020.101970.
Zhang Y, Wang Y, Meng L, Huang Q, Zhu Y, Cui W, et al. Targeted micelles with chemotherapeutics and gene drugs to inhibit the G1/S and G2/M mitotic cycle of prostate cancer. J Nanobiotechnol. 2021;19(1):17. https://doi.org/10.1186/s12951-020-00756-6.
Heidegger I, Necchi A, Pircher A, Tsaur I, Marra G, Kasivisvanathan V, et al. A systematic review of the emerging role of immune checkpoint inhibitors in metastatic castration-resistant prostate cancer: will combination strategies improve efficacy? Eur Urol Oncol. 2020. https://doi.org/10.1016/j.euo.2020.10.010.
Vlajnic T, Bubendorf L. Molecular pathology of prostate cancer: a practical approach. Pathology. 2021;53(1):36–43. https://doi.org/10.1016/j.pathol.2020.10.003.
Tremblay M, Viala S, Shafer ME, Graham-Paquin AL, Liu C, Bouchard M. Regulation of stem/progenitor cell maintenance by BMP5 in prostate homeostasis and cancer initiation. Elife. 2020. https://doi.org/10.7554/eLife.54542.
Pietas A, Schluns K, Marenholz I, Schafer BW, Heizmann CW, Petersen I. Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics. 2002;79(4):513–22. https://doi.org/10.1006/geno.2002.6744.
Diamantopoulou A, Mantas D, Kostakis ID, Agrogiannis G, Garoufalia Z, Kavantzas N, et al. A clinicopathological analysis of S100A14 expression in colorectal cancer. In Vivo. 2020;34(1):321–30. https://doi.org/10.21873/invivo.11777.
Sugino T, Ichikawa-Tomikawa N, Tanaka M, Shishito N, Miura T, Abe M, et al. Identification of S100A14 as a metastasis-promoting molecule in a murine organotropic metastasis model. Clin Exp Metas. 2019;36(4):411–22. https://doi.org/10.1007/s10585-019-09979-w.
Li X, Wang M, Gong T, Lei X, Hu T, Tian M, et al. A S100A14-CCL2/CXCL5 signaling axis drives breast cancer metastasis. Theranostics. 2020;10(13):5687–703. https://doi.org/10.7150/thno.42087.
Bertini I, Borsi V, Cerofolini L, Das Gupta S, Fragai M, Luchinat C. Solution structure and dynamics of human S100A14. J Biol Inorg Chem JBIC. 2013;18(2):183–94. https://doi.org/10.1007/s00775-012-0963-3.
Zhu M, Wang H, Cui J, Li W, An G, Pan Y, et al. Calcium-binding protein S100A14 induces differentiation and suppresses metastasis in gastric cancer. Cell Death Dis. 2017;8(7):e2938. https://doi.org/10.1038/cddis.2017.297.
Al-Ashkar N, Zetoune AB. S100A14 serum level and its correlation with prognostic factors in breast cancer. J Egypt Natl Cancer Inst. 2020;32(1):37. https://doi.org/10.1186/s43046-020-00048-y.
Zhao H, Guo E, Hu T, Sun Q, Wu J, Lin X, et al. KCNN4 and S100A14 act as predictors of recurrence in optimally debulked patients with serous ovarian cancer. Oncotarget. 2016;7(28):43924–38. https://doi.org/10.18632/oncotarget.9721.
Ding F, Wang D, Li XK, Yang L, Liu HY, Cui W, et al. Overexpression of S100A14 contributes to malignant progression and predicts poor prognosis of lung adenocarcinoma. Thorac Cancer. 2018;9(7):827–35. https://doi.org/10.1111/1759-7714.12654.
Wang YY, Gao YX, Gao W, Xu Y, Xu YZ, Wang YJ, et al. Design, synthesis and biological evaluation of tricyclic diterpene derivatives as novel neuroprotective agents against ischemic brain injury. Eur J Med Chem. 2015;103:396–408. https://doi.org/10.1016/j.ejmech.2015.08.057.
Freytag M, Kluth M, Bady E, Hube-Magg C, Makrypidi-Fraune G, Heinzer H, et al. Epithelial splicing regulatory protein 1 and 2 (ESRP1 and ESRP2) upregulation predicts poor prognosis in prostate cancer. BMC Cancer. 2020;20(1):1220. https://doi.org/10.1186/s12885-020-07682-8.
Gomes LF, Longhi PJH, Machado L, da Cruz IBM, Montano MAE, Martins M, et al. Lemongrass (Cymbopogon citratus (D.C.) Stapf) presents antitumoral effect and improve chemotherapy activity in prostate cancer cells. Anti-Cancer Agents Med Chem. 2021. https://doi.org/10.2174/1871520621666210112111711.
O’Dwyer E, Bodei L, Morris MJ. The role of theranostics in prostate cancer. Semin Radiat Oncol. 2021;31(1):71–82. https://doi.org/10.1016/j.semradonc.2020.07.004.
Chakravarty D, Huang L, Kahn M, Tewari AK. Immunotherapy for metastatic prostate cancer: current and emerging treatment options. Urol Clinics N Am. 2020;47(4):487–510. https://doi.org/10.1016/j.ucl.2020.07.010.
Meng DF, Sun R, Liu GY, Peng LX, Zheng LS, Xie P, et al. S100A14 suppresses metastasis of nasopharyngeal carcinoma by inhibition of NF-kB signaling through degradation of IRAK1. Oncogene. 2020;39(30):5307–22. https://doi.org/10.1038/s41388-020-1363-8.
Pandey S, Osman TA, Sharma S, Vallenari EM, Shahdadfar A, Pun CB, et al. Loss of S100A14 expression at the tumor-invading front correlates with poor differentiation and worse prognosis in oral squamous cell carcinoma. Head Neck. 2020;42(8):2088–98. https://doi.org/10.1002/hed.26140.
Wang HY, Zhang JY, Cui JT, Tan XH, Li WM, Gu J, et al. Expression status of S100A14 and S100A4 correlates with metastatic potential and clinical outcome in colorectal cancer after surgery. Oncol Rep. 2010;23(1):45–52.
Sapkota D, Costea DE, Blo M, Bruland O, Lorens JB, Vasstrand EN, et al. S100A14 inhibits proliferation of oral carcinoma derived cells through G1-arrest. Oral Oncol. 2012;48(3):219–25. https://doi.org/10.1016/j.oraloncology.2011.10.001.
Wang Y, Jia A, Cao Y, Hu X, Wang Y, Yang Q, et al. Hippo kinases MST1/2 regulate immune cell functions in cancer, infection, and autoimmune diseases. Crit Rev Eukaryot Gene Expr. 2020;30(5):427–42. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2020035775.
Wang X, Ji C, Hu J, Deng X, Zheng W, Yu Y, et al. Hsa_circ_0005273 facilitates breast cancer tumorigenesis by regulating YAP1-Hippo signaling pathway. J Exp Clin Cancer Res CR. 2021;40(1):29. https://doi.org/10.1186/s13046-021-01830-z.
Chen Q, Zhou XW, Zhang AJ, He K. ACTN1 supports tumor growth by inhibiting Hippo signaling in hepatocellular carcinoma. J Exp Clin Cancer Res CR. 2021;40(1):23. https://doi.org/10.1186/s13046-020-01821-6.
Xu W, Zhang M, Li Y, Wang Y, Wang K, Chen Q, et al. YAP manipulates proliferation via PTEN/AKT/mTOR-mediated autophagy in lung adenocarcinomas. Cancer Cell Int. 2021;21(1):30. https://doi.org/10.1186/s12935-020-01688-9.
Sadeqzadeh E, de Bock CE, Zhang XD, Shipman KL, Scott NM, Song C, et al. Dual processing of FAT1 cadherin protein by human melanoma cells generates distinct protein products. J Biol Chem. 2011;286(32):28181–91. https://doi.org/10.1074/jbc.M111.234419.
Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan S, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45(3):253–61. https://doi.org/10.1038/ng.2538.
Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang J, et al. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017;397:83–93. https://doi.org/10.1016/j.canlet.2017.03.033.
Funding
This work was supported by the Fujian Natural Sciences Foundation. (Grant no. 2017J01203), Joint Funds for the Innovation of Science and Technology, Fujian Province (Grant no. 2017Y9023) and Startup Fund for scientific research, Fujian Medical University (Grant no. 2018QH1044).
Author information
Authors and Affiliations
Contributions
SJ and YZ designed the study, supervised the data collection, SJ, ZC and ZH analyzed the data, interpreted the data, BL, YX, ZL, ZL and ML prepare the manuscript for publication and reviewed the draft of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors state that there are no conflicts of interest to disclose.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the standards upheld by the Ethics Committee of Fujian Medical University Union Hospital and with those of the 1964 Helsinki Declaration and its later amendments for ethical research involving human subjects (Approval No. 2020KY0112). All animal experiments were approved by the Ethics Committee of Fujian Medical University Union Hospital for the use of animals and conducted in accordance with the National Institutes of Health Laboratory Animal Care and Use Guidelines (Approval No. 2020-047).
Informed consent
Written informed consent was obtained from a legally authorized representative(s) for anonymized patient information to be published in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiang, S., Zhu, Y., Chen, Z. et al. S100A14 inhibits cell growth and epithelial–mesenchymal transition (EMT) in prostate cancer through FAT1-mediated Hippo signaling pathway. Human Cell 34, 1215–1226 (2021). https://doi.org/10.1007/s13577-021-00538-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00538-8