Abstract
Temozolomide (TMZ) is one of the most common drugs selected for glioma chemotherapy, but the therapeutic effect of glioma treatment is usually limited due to its resistance. Long non-coding RNA (lncRNA) is gradually found to be a vital regulator in numerous physiological and pathological processes. Lately, it was revealed that LINC00174 could promote CRC cell growth. However, the function and potential regulatory manner of LINC00174 in glioma remain unclear. Our results demonstrated that the expression level of LINC00174 was higher in glioma tissues, and LINC00174 down-regulation could remarkably prevent cell proliferation and promote cell apoptosis in both glioma cells and TMZ-resistant glioma cells. Mechanistic studies revealed that LINC00174 can sponge microRNA-138-5p (miR-138-5p) and down-regulate its expression, thereby up-regulating the protein level of miR-138-5p’s target, sex-determining region Y (SRY)-box9 protein (SOX9). Additionally, in vivo experiments revealed that LINC00174 shRNA can serve as a tumor suppressor through down-regulating SOX9 in glioma. In this study, a novel established regulatory way of LINC00174/miR-138-5p/SOX9 axis was systematically studied, which may provide a new manner for glioma therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With an annual incidence of 5 cases per 100,000 people, glioma is the most general and fatal malignant brain cancer of adult central nervous system [1]. According to their histopathological characteristics, gliomas can be classified into WHO grade I and II (low-grade glioma) and WHO grade III and IV (high-grade glioma) [2]. Although combined with surgical resection and postoperative chemoradiotherapy, the median survival time of glioma patients is below expectations [3]. As a deoxyribonucleic acid alkylation anti-tumor drug, temozolomide (TMZ) can easily cross the blood–brain barrier [4]. TMZ can effectively inhibit proliferation and induce apoptosis of glioma cells, which is a first-line chemotherapy drug for glioma treatment [5, 6]. However, a major obstacle for effective treatment is the acquisition of resistance to TMZ. Therefore, it is necessary to better understand the molecular mechanism underlying glioma development, so as to find novel intervention targets and improve the therapeutic effect of chemotherapy drugs.
With a length of over 200 nt, long non-coding RNA (lncRNA) exerts a key function in cell proliferation, apoptosis and other biological processes [7]. LncRNA has been proven to be a key molecule in the progression of various human tumors [8, 9]. Some lncRNAs, such as lncRNA CCND2-AS1, lncRNA PVT1 and lncRNA CCAT1 have been implicated in glioma [10,11,12]. Located in chromosome 7q11.21 (nr_026873.1), long intergenic non-protein coding RNA 174, namely LINC00174, is a non-coding RNA with 4426 nt. Recent studies have found that colorectal cancer patients with high LINC00174 expression have poor prognosis. Silence of LINC00174 can inhibit CRC cell proliferation in vitro and tumor growth in vivo by regulating miR-1910-3p/TAZ signal pathway [13]. However, the clinical value and biological function of LINC00174 in glioma are not clear.
As small non coding RNAs, microRNAs (miRNAs), 18-22 nt, regulate their target genes’ expression, which mainly depends on their interaction with gene’s 3′UTR. miRNAs can serve as oncogenes and tumor suppressors by regulating tumor related mRNAs [14]. Abundant evidences have certified that miRNAs, such as miR-155 [15], miR-200c [16] and miR-675 [17], play key roles in glioma progression. As an important miRNA, miR-138-5p has been found to be dysregulated and exert vital roles in many cancers, which included non-small cell lung cancer [18] and bladder cancer [19] et al. A decrease in miR-138-5p level was demonstrated in tumor tissues and cell lines of glioma. Glioma patients with lower miR-138-5p level have a shorter overall survival and poor prognosis. miR-138-5p effectively inhibits cell growth in vitro and tumor development in vivo through inhibiting the signal loop of EZH2-CDK4/6-pRb-E2F1 [20]. Down-regulated miR-138-5p could promote angiogenesis in glioma [21]. Besides, miR-138 mimic transfection increased glioma cell proliferation and TMZ resistance [22]. However, it is not clear whether miR-138-5p down-regulation could improve the TMZ resistance in glioma.
As a high-mobility group box-containing transcription factor, sex-determining region Y (SRY)-box9 protein (SOX9) regulates cell development and differentiation, and is involved in multiple tumor malignancies [23, 24]. It was indicated that SOX9 was overexpressed in glioma tissues and cell lines [25]. Glioma patients with higher SOX9 level have a poorer prognosis [26]. In addition, SOX9 over-expression could stimulate glioma cell migration and invasion, and EMT process through activating Wnt/β-catenin signaling [27]. Previous studies showed that SOX9 down-regulation could decrease multiple stem cell markers, inhibit the formation of glioma cell colonies and spheres, and increase the sensitivity of glioma cells to TMZ [28]. However, it is not clear whether miR-138-5p can regulate SOX9 and affect temozolomide sensitivity in glioma.
In this research, we figured out the up-regulation of LINC00174 in tissues from glioma patients. The influences of LINC00174 down-regulation on the proliferation and apoptosis of glioma cells and TMZ-resistant cells were studied. miR-138-5p that may be a potential target of LINC00174 was predicted and confirmed. Furthermore, the regulation relationship between miR-138-5p and SOX9 was studied. The functional role and molecular mechanism of LINC00174/miR-138-5p/SOX9 in glioma TMZ resistance were systematically researched, which highlights their potential as novel candidates for glioma therapy.
Materials and methods
Clinical samples
Human glioma tissues (tumor) and adjacent control brain tissues (normal) were gathered from patients diagnosed with glioma. These patients underwent surgery at the Department of Neurosurgery of Second Affiliated Hospital of Xi’an Medical University from 2015 to 2018. Before section, no chemotherapy or radiotherapy has been used for glioma patients. After collected from glioma patients, fresh OS tissues were immediately frozen in liquid nitrogen and stored at − 80 °C. Informed consent was obtained from each patient. The research method was under the permission of the Ethics Committee of Second Affiliated Hospital of Xi’an Medical University. After routine neuropathological evaluation, liquid nitrogen was used to freeze tumor samples immediately and preserve these samples before use. According to the 2007 WHO classification, glioma patients were classified as three groups, which included grade I–II, grade III and grade IV. Clinical information of glioma patient was listed in Table 1.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Tumor/normal tissues and cells total RNA was extracted by the TRIzol reagent (Thermo Fisher Scientific). Using a cDNA Reverse Transcription Kit (Applied Biosystems), cDNA was synthesized from total RNA. SYBR Premix Ex Taq (Takara) was used to perform qRT-PCR assays for LINC00174 and SOX9 mRNA expressions. miR-138-5p levels were examined by a SYBR PrimeScript miRNA RT-PCR Kit (Takara). U6 was used as endogenous control for miR-138-5p and GAPDH was selected forLINC00174/SOX9 expressions. Primer sequences for qRT-PCR analysis were listed in Table S1.
In situ hybridization (ISH) and immunohistochemistry (IHC) assays
Tissues were first fixed and paraffin-embedded. Then, 5 μm sections were cut. For ISH, LINC00174 probe was labeled with peroxidase (Thermo Fisher Scientific). An ISH kit (RiboBio, Guangzhou, China) was used to detect LINC00174. By scanning 10 nonoverlapping fields in each section through Image-ProPlus 6.0, the staining intensity was quantified. After that, the mean staining intensity was calculated. Therefore, tissue was identified as “Low expression” if the LINC00174 intensity was below the mean value. It was identified as “High expression” if LINC00174 intensity was above the average value. An overall survival curve was determined using the Kaplan–Meier method based on the follow-up data from each patient.
For IHC assay, antigen was repaired after tissue sections were rehydrated. After that, antibodies for SOX9, Ki-67 and cleaved caspase-3 were used to incubate tissue sections overnight at 4 °C. Then corresponding secondary antibodies and 3,3′-diaminobenzidine (DAB) solution were used to visualize the sections.
Cell culture
Normal human astrocytes (NHA) were obtained from ScienCell (San Diego, CA), and served as a normal counterpart of glioblastoma. Glioma cell lines (LN229, SHG-44, U118, U251 and U87) were all purchased from American Type Culture Collection (ATCC, Rockville, MD). Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) was used to culture cells. These cells were cultured at 37 °C in a humidified environment containing 5% CO2.
Cell transfection
U251 or U87 cells were transfected with shRNA for LINC00174 (sh-LINC00174#1, sh-LINC00174#2), LINC00174 expressing plasmid (pcDNA LINC00174), miR-138-5p mimic, miR-138-5p inhibitor or their corresponding controls (sh-NC, pcDNA3.1, NC mimic or NC inhibitor). Sequences of LINC00174 shRNA were listed in Table S2. And the more effective one (sh-LINC00174#2) were selected for the following assays. When cell confluence is at 50–70% confluence, Lipofectamine 3000 reagent (Thermo Fisher Scientific) was used to perform cell transfection.
Cell proliferation assay
For cell vitality curve assay, U251 or U87 cells (6 × 103 cells/well) were, respectively, transfected with sh-NC or sh-LINC00174, and then seeded onto 96-well plates. A Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan) was used to detect the viable cell numbers at day 1, 2, 3, 4. To measure the absorbance (450 nm), a microplate spectrometer (Thermo Fisher Scientific) was used.
For colony formation assay, U251 or U87 cells (1 × 103) were first mixed into top agar (1.5 ml), and then added onto base agar. 3 weeks later, colonies were stained by Crystal Violet. And a dissection microscope (TE2000-U, Nikon, Japan) was used to count the colonies.
For 5-ethynyl-2′-deoxyuridine (EDU) assay, the thymidine analog EDU was incorporated into DNA when cells undergoing DNA replication. Briefly, after fixation and permeabilization, the cells were incubated with EDU (50 µM, 3 h). DAPI (1 µg/ml, 10 min, Sigma-Aldrich) was used to stain the cell nuclei. The EDU positive cells were determined using fluorescence microscopy.
Cell apoptosis and cell cycle assay
At 48 h post transfection, U251 or U87 cells were collected for cell apoptosis and cell cycle assay. After fixed with 70% ethanol overnight, the cells were incubated with propidium iodide (PI, BD Biosciences, San Jose, CA) or PI plus Annexin V-FITC (BD Biosciences). Then, a FACSCalibur system (BD Biosciences) was used for cell apoptosis or cell cycle analysis. The results were analyzed using the ModFit_LT software.
Generation of TMZ-resistant glioma cell lines
First, the absorbance at 450 nm was determined after cells were incubated with different concentrations of temozolomide (TMZ) for 72 h. Using the sigmoidal dose–response function of GraphPad Prism, the 50% inhibitory concentration (IC50) was calculated. As described previously [29], U251 or U87 cells were exposed to of TMZ, whose concentrations gradually increased from 5 μM to 100 μM for 6 months. Thus, the TMZ-resistant glioma cell lines were established.
Plasmid constructs
A human genomic DNA of U251 cells was extracted to amplify the sequence of LINC00174 using PCR. To insert the sequence of LINC00174 into the pmiR-Glo dual-Luciferase reporter plasmid (Promega, Madison, WI, USA), a One Step Cloning Kit (Vazyme Biotech, Nanjing) was used. According to the above methods, LINC00174 expressing plasmid (pcDNA LINC00174) was constructed by inserting its sequence into the pcDNA3.1 plasmid. Additionally, pmiR-Glo plasmid containing the 3′ UTR of SOX9 was constructed. The binding-site (ACACCAGC) was mutated by (UGUGGUCG) and used as a control. For The primers for plasmid construct, see Table S3.
Dual luciferase reporter assay
Luciferase reporter assays was performed as described reported [30]. When the cell confluence is at 70–80%, the firefly luciferase reporter plasmid (1 μg) and miR-138-5p mimic/NC mimic were co-transfected into U251 or U87 cells. The protein was extracted 24 h after transfection. A Luciferase Reporter Assay System (Promega) was used to test the luciferase activities.
RNA binding protein immunoprecipitation (RIP) assay
RIP assay was conducted as previously described [31]. Briefly, antibodies for Ago2 or IgG were used to incubate the cell lysates from U251 or U87 cells. After that, the pull down complexes were performed by western blot analysis of Ago2 or qRT-PCR analysis of LINC00174 and miR-138-5p. Antibody for SNRNP70 was also used for RIP assay.
Protein extraction and western blotting
RIPA lysis buffer (Beyotime Institute of Biotechnology) was used to extract total proteins from the cells. For total protein content quantification, a BCA protein assay kit (Thermo Fisher Scientific) was used. Proteins were performed by SDS-PAGE and electrophoretically transferred to PVDF membranes. The primary antibodies were used to incubate the membranes overnight at 4 °C, which were then incubated with a horseradish peroxidase-conjugated anti-mouse/anti-rabbit IgG (Thermo Fisher Scientific). Antibodies included SOX9, PI3 K, p-PI3 K, Akt, p-Akt, GAPDH (an internal control) and corresponding secondary antibodies. Antibody information in this study is listed in Table S4.
Tumor xenografts in nude mice
First, using a recombinant lentivirus, U251 cells stably encoding shRNA for LINC00174 (sh-LINC00174) or the corresponding control cells (sh-NC) were established. To construct a mouse glioma xenograft model, six-week-old thymic BALB/c nude mice (16–18 g) and stable U251 cells were used. All experiments with nude mice were constructed in accordance with the Guidelines for the Care and Use of Laboratory Animals, which was presented by the National Institutes of Health [32]. The right flanks of mice (6 mice/group) were subcutaneously injected with stable U251 cells (1 × 106). Mice were administered orally with TMZ (10 mg/kg) 4 days post injection. Using Kaplan–Meier survival, the survival curve analysis was determined. Tumor volume was measured every week post injection, which was calculated by the formula: volume (glioma3) = length × width2/2. The mice were killed 5 weeks post injection, and the corresponding tumors were isolated, weighted and photographed. Additionally, the IHC analysis of SOX9, Ki67 and cleaved caspase-3 were performed.
Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM). Student’s t test or one-way ANOVA were used to evaluate the statistical analysis. The correlations between LINC00174 and miR-138-5p, miR-138-5p and SOX9 mRNA, LINC00174 and SOX9 mRNA were, respectively, analyzed by Pearson correlation analysis. An overall survival curve was determined using the Kaplan–Meier method. P < 0.05 was considered significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Results
LINC00174 level is up-regulated in both glioma tissues and cells
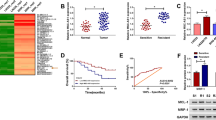
As shown in Fig. 1a, we determined the LINC00174 levels in tissue samples from glioma patients (n = 40) using qRT-PCR, the results showed that the LINC00174 level was markedly increased in tumor samples in contrast with normal samples. The LINC00174 levels in tumor samples from grade I–II, grade III and grade IV were stepwise increased. Additionally, the results of ISH also demonstrated that LINC00174 up-regulation is an usual event in glioma (Fig. 1b). In addition, through the Kaplan–Meier method, the relationship between glioma patients’ survival rate and the LINC00174 level was analyzed. As indicated in Fig. 1c, glioma patients with high LINC00174 expression (n = 16) have a lower survival rate. Additionally, LINC00174 levels in five glioma cell lines (LN229, SHG-44, U118, U251 and U87) were higher than that in normal human astrocytes, NHA (Fig. 1d). Among the glioma cell lines, U251 and U87 cells expressed higher levels of LINC00174, therefore, were subsequently used to investigate the function of LINC00174 down-regulation in glioma. Together, these data revealed that LINC00174 is up-regulated in glioma tissue samples and cell lines, which might play an oncogenic role in glioma.
LINC00174 is up-regulated in glioma tissues and cells. a The relative LINC00174 levels in glioma tissues (tumor, n = 40) and control tissues (normal, n = 40), as determined using qRT-PCR (left). The relative LINC00174 levels in glioma tissues from patients at grade I–II, grade III and grade IV (right). b The LINC00174 levels in glioma tissues (tumor, n = 40) and control tissues (normal, n = 40), as determined using ISH. Representative three sets of images were shown. c Kaplan–Meier curves of overall survival of 40 glioma patients, stratified by LINC00174 expression. d The relative LINC00174 levels in five glioma cell lines (LN229, SHG-44, U118, U251 and U87) and normal human astrocytes (NHA), as determined using qRT-PCR. Independent experiments were performed three times. The data are expressed as the mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001
LINC00174 effects the proliferation, apoptosis and cycle progression of glioma cells
To determine the influences of LINC00174 on glioma cells, two shRNAs for LINC00174 (sh-LINC00174#1, sh-LINC00174#2) were designed. The results in Fig. 2a demonstrated that LINC00174 level was markedly decreased after transfected shRNAs for LINC00174 compared to the control cells. Because sh-LINC00174#2 results in more effective interference efficiency, thus was selected for the following experiments. The results in Fig. 2b revealed that the cell proliferation was suppressed in U251 or U87 cells after LINC00174 was inhibited. Colony formation results demonstrated that LINC00174 down-expression caused a significant decrease of colony number (Fig. 2c). Moreover, Fig. 2d indicated that number of U251 or U87 cells incorporating EDU in the sh-LINC00174 transfected group markedly reduced compared to the control cells. Furthermore, the results of cell apoptosis assay in Fig. 2e showed that LINC00174 down-expression significantly increased the percentage of apoptosis cells. Additionally, as shown in Fig. 2f, LINC00174 interference in U251 or U87 cells led to an increased cell population in the G1-phase and a decreased cell population in the S-phase. These results above indicated that LINC00174 down-regulation can inhibit cell proliferation, suppress cell cycle progression and promote cell apoptosis in U251 or U87 cells.
LINC00174 effects the proliferation, cell apoptosis and cell cycle of glioma cells. a The relative expression levels of LINC00174 in U251 or U87 cells, after transfection with shRNAs for LINC00174 (sh-LINC00174#1, sh-LINC00174#2), or the corresponding control (sh-NC). b Growth curves of U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. The measurements of the cell growth rate were obtained using a CCK-8 kit. c Colony formation analysis of U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. Colony numbers were quantified and shown as histograms. d Representative profiles of EDU cell growth in U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. EDU positive cell numbers were quantified and shown as histograms. e Cell apoptosis analysis of U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. Apoptosis rates were quantified and shown as histograms. f Cell cycle analysis of U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. Cell percentage in G1, S and G2 phages were quantified and shown as histograms. Independent experiments were performed three times. The data are expressed as the mean ± SEM, *P < 0.05, **P < 0.01
LINC00174 inhibition improves the chemoresistance to TMZ of glioma cells
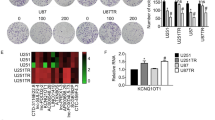
To detect the cytotoxicity of TMZ, we first used a CCK-8 assay to determine the sensitivity of U251 or U87 to the TMZ. As shown in Fig. 3a, TMZ exerted an inhibitory effect on glioma cells in a dose dependent manner. The result showed that the IC50 values of sh-LINC00174 groups were significantly lower than those of sh-NC groups. Additionally, to analyze the effects of LINC00174 inhibition on the TMZ chemoresistance of glioma cells, U251 or U87 cells resistance to TMZ were established with TMZ treatment (100 mM). Then, the proliferation and apoptosis assays were performed in TMZ-resistant glioma cells. The results in Fig. 3b revealed that in TMZ resistant U251 or U87 cells, LINC00174 down-regulation caused more significant cell growth inhibition. Colony formation results demonstrated that LINC00174 down-expression significantly reduced colony numbers in TMZ-resistant glioma cells (Fig. 3c). Moreover, Fig. 3d indicated that in TMZ-resistant U251 or U87 cells, the numbers of cells incorporating EDU in the sh-LINC00174 group were significantly reduced compared to the control cells. Furthermore, the results of cell apoptosis assay in Fig. 2e showed that LINC00174 down-expression significantly promoted the percentage of apoptosis cells in glioma cells resistance to TMZ. Because MGMT promoter methylation can decrease MGMT protein expression, thereby abrogating the DNA repair activity necessary for TMZ resistance [33]. The MGMT methylation status were then determined in the established TMZ-resistant sublines of U251 and U87 by methylation-specific polymerase chain reaction [34], which were further down-regulated than those in parent U251 and U87 cell lines (Fig. S1A). As a MGMT-expressing glioblastoma cell line, LN229’s chemoresistance to TMZ was also significantly decreased after inhibiting LINC00174 (Fig. S1B). These results above indicated that LINC00174 down-regulation plays a key function in the TMZ resistance of glioma cells.
LINC00174 effects the proliferation, cell apoptosis and cell cycle of TMZ-resistant glioma cells. a Relative proliferation of U251 or U87 cells after transfected with sh-LINC00174 or sh-NC and treated with different doses of TMZ were, respectively, determined by CCK-8. TMZ treatment significantly decreased cell proliferation in a dose-dependent manner. IC50 was calculated using the sigmoidal dose–response function of GraphPad Prism. b Growth curves of TMZ-resistant U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. The measurements of the cell growth rate were obtained using a CCK-8 kit. c Colony formation analysis of TMZ-resistant U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. Colony numbers were quantified and shown as histograms. d Representative profiles of EDU cell growth in TMZ-resistant U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. EDU positive cell numbers were quantified and shown as histograms. e Cell apoptosis analysis of TMZ-resistant U251 or U87 cells after transfection with sh-LINC00174 or sh-NC. Apoptosis rates were quantified and shown as histograms. Independent experiments were performed three times. The data are expressed as the mean ± SEM, *P < 0.05, **P < 0.01
LINC00174 directly targets and negatively regulates miR-138-5p
As indicated above, LINC00174 down-regulation inhibited cell proliferation and promote cell apoptosis in glioma cells and the TMZ-resistant glioma cells. Nevertheless, the regulatory mechanism is still unclear. Therefore, the potential target miRNA of LINC00174 was analyzed. By means of miRDB (http://mirdb.org/), we found that LINC00174 might probably interact with miR-138-5p. As Fig. 4a shown, there was potential base pairing binding sites between miR-138-5p and LINC00174. As indicated in Fig. 4b, miR-138-5p up-regulation down-regulated the luciferase activity of the LINC00174 reporter plasmids. While, after the binding sites of miR-138-5p were mutated, the luciferase activity was not significantly changed. As shown in Fig. 4c, both LINC00174 and miR-138-5p were significantly enriched in the RNA, which were immunoprecipitated by the Ago2 antibody. Because Ago2 is the important component of miRNA-mediated RISC protein complex, these results further confirmed the direct binding between miR-138-5p and LINC00174. Accordingly, as an important component of U4/U6-U5 snRNPs, SNRNP70 seems to catalyze an ATP-dependent unwinding of U4/U6 RNA duplices. As the right Bar chart in Fig. 4c showed, RNA was significantly enriched by the Ago2 antibody, compared with IgG antibody. After transfected with pcDNA LINC00174, the LINC00174 levels in U251 or U87 cells were markedly increased (Fig. 4d). Moreover, LINC00174 overexpression caused significantly decreased miR-138-5p levels in U251 or U87 cells, and sh-LINC00174 led to an opposite trend. On account of the direct binding between miR-138-5p and LINC00174, we tested the LINC00174 level after changing miR-138-5p expression. The results in Fig. 4e showed that miR-138-5p up-regulation caused a reduction of LINC00174, and miR-138-5p down-regulation led to an increase of LINC00174, suggesting that miR-138-5p can also inhibit LINC00174. Additionally, we have tested miR-138-5p levels in clinical specimens using qRT-PCR. Figure 4f revealed that miR-138-5p level was markedly decreased in glioma tissue samples. As the disease stage progresses from I to IV, the miR-138-5p level was gradually reduced. The Pearson’s correlation analysis demonstrated that there was an inverse relationship between miR-138-5p and LINC00174 levels in tissues from glioma patients (Fig. 4g). These data above demonstrated that LINC00174 can negatively regulate miR-138-5p by targeting its binding seeds in glioma.
LINC00174 directly binds to miR-138-5p and negatively regulate its expression. a starBase prediction identified seeds match for LINC00174 in the mature sequence of miR-138-5p. The predicted seed-recognition site in the miR-138-5p sequence and the corresponding LINC00174 sequence are depicted. b The relative luciferase activity of the LINC00174 reporter plasmid was assayed in U251 cells after transfection with miR-138-5p mimic or the NC mimic. The mutant LINC00174 reporter was also used as a control. c U251 or U87 cells were harvested and mixed with Ago2 or SNRNP70 antibodies to perform RNA binding protein immunoprecipitation (RIP) assay. LINC00174 or miR-138-5p enrichments were tested by qRT-PCR and compared to Anti IgG or Input. d The relative levels of LINC00174 in U251 or U87 cells, after transfection with pcDNA LINC00174 or pcDNA3.1 (left). The relative miR-138-5p levels in U251 or U87 cells after transfection with pcDNA LINC00174, sh-LINC00174 or the corresponding controls (pcDNA3.1 or sh-NC), as determined using qRT-PCR (right). e The relative levels of LINC00174 in U251 or U87 cells, after transfection with NC mimic, miR-138-5p mimic, NC inhibitor or miR-138-5p inhibitor, as determined using qRT-PCR. f The relative miR-138-5p levels in glioma tissues (tumor, n = 40) and control tissues (normal, n = 40), as determined using qRT-PCR (left). The relative miR-138-5p levels in glioma tissues from patients at grade I–II, grade III and grade IV (right). g The Pearson correlation analysis of the relative expressions between miR-138-5p and LINC00174. Independent experiments were performed three times. The data are expressed as the mean ± SEM, **P < 0.01, ***P < 0.001
miR-138-5p negatively regulates SOX9 through targeting its 3′UTR
In glioma cells, LINC00174 could directly target miR-138-5p. But, miR-138-5p’s role in glioma is still unknown. Next, TargetScan was used to predict miR-138-5p’s target genes. We predict that SOX9 are probably potential target gene of miR-138-5p. Accordingly, the seed binding sites between miR-138-5p and SOX9 were depicted in Fig. 5a. As Fig. 5b showed, miR-138-5p up-regulation inhibited the luciferase activity of the SOX9 3′UTR reporter plasmids. However, the mutation of the miR-138-5p binding sites eliminated the significant alteration of the luciferase activities. The qRT-PCR results in Fig. 5c demonstrated that miR-138-5p level was markedly up-regulated after transfected miR-138-5p mimic, and decreased after transfected miR-138-5p inhibitor. Accordingly, SOX9 mRNA level was markedly decreased after transfected miR-138-5p mimic, and increased after transfected miR-138-5p inhibitor. The results of western blot (Fig. 5d) showed that the SOX9 protein levels were markedly decreased after transfected miR-138-5p mimic, and miR-138-5p inhibitor caused an up-regulation of them. Additionally, SOX9 mRNA levels were tested by qRT-PCR in clinical specimens. The results in Fig. 5e showed that SOX9 mRNA levels were markedly increased in glioma tissue samples. As the disease stage progresses from I to IV, the SOX9 mRNA level was gradually increased. According to the Pearson’s correlation analysis, there was an inverse relationship between miR-138-5p and SOX9 mRNA level in tissues from glioma patients. And a positive relationship between LINC00174 and SOX9 mRNA level was analyzed (Fig. 5g). These results above suggested that LINC00174 might regulate SOX9 expression through sponging miR-138-5p.
miR-138-5p negatively regulates SOX9 through targeting its 3′UTR. a Intersection analysis of miR-138-5p’s potential targets using prediction software of TargetScan. The predicted seed-recognition sites in the 3′UTR of SOX9 sequence and the corresponding miR-138-5p sequence were depicted. b The relative luciferase activity of the SOX9 3′UTR reporter plasmid was assayed in U251 or U87 cells after transfection with miR-138-5p mimic or NC mimic. The mutant SOX9 3′UTR reporter was also used as a control. c The relative miR-138-5p (left) and SOX9 (right) levels in U251 or U87 cells after transfection with NC mimic, miR-138-5p mimic, NC inhibitor or miR-138-5p inhibitor, as determined using qRT-PCR. d The protein levels of SOX9 in U251 or U87 cells after transfection with NC mimic, miR-138-5p mimic, NC inhibitor or miR-138-5p inhibitor, as determined using western blotting. Relatively quantitative results was determined by Image J and shown as histogram. e The relative SOX9 mRNA levels in glioma tissues (tumor, n = 40) and control tissues (normal, n = 40), as determined using qRT-PCR (left). The relative SOX9 mRNA levels in glioma tissues from patients at grade I–II, grade III and grade IV (right). f The Pearson correlation analysis of the relative expressions between miR-138-5p and SOX9 mRNA, between LINC00174 and SOX9 mRNA. Independent experiments were performed three times. The data are expressed as the mean ± SEM, *P < 0.05, **P < 0.01
LINC00174/miR-138-5p affects chemoresistance to TMZ in human glioma cells by regulating SOX9
The results above indicated that LINC00174 can influence proliferation, cycle progression and apoptosis in U251 or U87 cells. LINC00174 knockdown improved chemoresistance to TMZ in human glioma cells. And LINC00174 can sponge miR-138-5p by targeting its binding seeds in glioma cells. We next studied the influences of LINC00174/miR-138-5p axis on the chemoresistance to TMZ in human glioma cells. As Fig. 3a showed, the IC50 value of U251 or U87 cells was significantly decreased after transfected sh-LINC00174, and it was significantly increased after transfected miR-138-5p inhibitor. However, the IC50 value was close to that of inh NC+sh-NC group after transfected sh-LINC00174 and miR-138-5p inhibitor simultaneously. Additionally, the proliferation and apoptosis assays were further performed in TMZ-resistant glioma cells to analyze the effects of LINC00174/miR-138-5p axis. As Fig. 6b revealed, the cell proliferation in TMZ-resistant glioma cells was suppressed after LINC00174 was down-regulated, and was promoted after reducing miR-138-5p. Nevertheless, the effect was erased after simultaneously inhibiting LINC00174 and miR-138-5p. Colony formation results in TMZ-resistant glioma cells demonstrated that LINC00174 down-expression significantly decreased colony numbers, miR-138-5p knockdown increased colony numbers, and sh-LINC00174+miR-138-5p inh eliminated their effects on colony formation (Fig. 6c). Moreover, Fig. 6d indicated that the cell numbers incorporating EDU in the sh-LINC00174 transfected group significantly decreased, and the EDU-positive cell number increased after inhibiting miR-138-5p. However, EDU-positive cells numbers were not significantly changed when inhibiting LINC00174 and miR-138-5p meanwhile. Furthermore, the results of cell apoptosis assay in Fig. 7a showed that LINC00174 down-expression significantly promoted cell apoptosis in TMZ-resistant glioma cells. The percentage of apoptosis cells was significantly down-regulated after transfecting miR-138-5p inhibitor, compared to that of inh NC+sh-NC cells. While, after inhibiting LINC00174 and miR-138-5p simultaneously, the cell apoptosis effect induced by LINC00174 or miR-138-5p down-regulation was abolished. Additionally, the western blot results revealed that SOX9, p-PI3 K and p-Akt expression levels were decreased after LINC00174 was down-regulated, and their levels were increased after miR-138-5p was suppressed in TMZ-resistant glioma cells (Fig. 7b). While, inhibiting LINC00174 and miR-138-5p meanwhile abolished the expression changes of these proteins caused by sh-LINC00174 or miR-138-5p inhibitor. These data together indicated that LINC00174 inhibition can improve the resistance to TMZ in U251 or U87 cells, which mainly depends on the negative regulation of miR-138-5p.
LINC00174/miR-138-5p effects cell proliferation and cell apoptosis of TMZ-resistant glioma cells. a Relative proliferation of U251 or U87 cells after transfected with inh NC+sh-NC, inh NC+sh-LINC00174, miR-138-5p inh+sh-NC or sh-LINC00174+miR-138-5p inh, and treated with different doses of TMZ were, respectively, determined by CCK-8. IC50 was calculated using the sigmoidal dose–response function of GraphPad Prism. b Growth curves of TMZ-resistant U251 or U87 cells after transfection with inh NC+sh-NC, inh NC+sh-LINC00174, miR-138-5p inh+sh-NC or sh-LINC00174+miR-138-5p inh. The measurements of the cell growth rate were obtained using a CCK-8 kit. c Colony formation analysis of TMZ-resistant U251 or U87 cells after transfection with inh NC+sh-NC, inh NC+sh-LINC00174, miR-138-5p inh+sh-NC or sh-LINC00174+miR-138-5p inh. Colony numbers were quantified and shown as histograms. d Representative profiles of EDU cell growth in TMZ-resistant U251 or U87 cells after transfection with inh NC+sh-NC, inh NC+sh-LINC00174, miR-138-5p inh+sh-NC or sh-LINC00174+miR-138-5p inh. EDU positive cell numbers were quantified and shown as histograms. Independent experiments were performed three times. The data are expressed as the mean ± SEM, *P < 0.05, **P < 0.01
LINC00174/miR-138-5p effects cell apoptosis of TMZ-resistant glioma cells. a Cell apoptosis analysis of TMZ-resistant U251 or U87 cells after transfection with inh NC+sh-NC, inh NC+sh-LINC00174, miR-138-5p inh+sh-NC or sh-LINC00174+miR-138-5p inh. Apoptosis rates were quantified and shown as histograms. b The protein levels of SOX9, PI3 K, p-PI3 K, Akt and p-Akt in TMZ-resistant U251 or U87 cells after transfection with inh NC+sh-NC, inh NC+sh-LINC00174, miR-138-5p inh+sh-NC or sh-LINC00174+miR-138-5p inh, as determined using western blotting. Relatively quantitative results was determined by Image J and shown as histogram. Independent experiments were performed three times. The data are expressed as the mean ± SEM, *P < 0.05, **P < 0.01
LINC00174 down-regulation suppresses tumor growth in a mouse xenograft model
Because LINC00174 down-expression can inhibit cell proliferation, cycle progression and promote apoptosis, its function was next analyzed in vivo. To study the influence of LINC00174 on glioma growth, U251 cells stably down-expressing LINC00174 was constructed for the mouse xenograft tumor model. After two groups of nude mice were subcutaneously implanted with the stable cells, they were administered orally with TMZ (10 mg/kg). At 5 weeks after implantation, corresponding tumors were excised and Fig. 8a showed the representative photograph. As shown, the tumors in the sh-LINC00174 group exhibited smaller sizes, compared with those of the sh-NC group. As Fig. 8b showed, using the Kaplan–Meier method, mice in the sh-LINC00174 group have a higher survival rate than those in the sh-NC group. Additionally, LINC00174 inhibition led to lower SOX9 level, Ki-67 positive cell number and higher cleaved caspased-3 level in xenograft tumors from the LINC00174-shRNA group (Fig. 8c). These results indicated that LINC00174 down-expression can down-regulate the protein levels of SOX9, suppress cell proliferation, promote cell apoptosis and finally decrease the glioma growth in vivo.
LINC00174 down-regulation inhibits tumor growth in a glioma mouse xenograft model. a Representative photograph of corresponding tumors dissected from mice at 5 weeks post-implantation (up). The xenograft tumor volumes of nude mice derived from subcutaneous implantation of U251 cells stably expressing sh-LINC00174 or sh-NC (down). b Kaplan–Meier curves of overall survival of glioma xenograft mice, stratified by LINC00174 expression. c IHC staining of SOX9, Ki-67 and cleaved caspase-3 in the xenograft tumors in nude mice derived from subcutaneous implantation of U251 cells stably expressing sh-LINC00174 or sh-NC. Independent experiments were performed three times. The data are expressed as the mean ± SEM, **P < 0.01
Discussion
In the present study, we not only suggested anti-proliferation, pro-apoptosis and TMZ-sensitivity enhancement mediated by LINC00174 down-regulation, but also offered a functional relationship between LINC00174 and miR-138-5p/SOX9. Because both lncRNAs and miRNAs play a key function in glioma, targeting relationships between them in glioma progression was gradually demonstrated. For example, lncRNA HOXA11-AS could negatively regulate miR-214-3p, modulate EZH2 expression and influence cell proliferation, migration and invasion in glioma [35]. LncRNA CRNDE accelerated glioma development by targeting miR-384 and regulating PIWIL4/STAT3 axis [36]. These findings demonstrated that lncRNA/miRNA axis plays vital functional roles in glioma. Our data revealed that miR-138-5p could be sponged by LINC00174. Except for miR-138-5p, only miR-1910-3p in colorectal cancer have been found to be regulated by LINC00174. It was reported that LINC00174′s oncogenic roles are partially through negatively regulating miR-1910-3p, and thereby activating oncogenic TAZ gene [13]. So far, the expression patterns of miR-1910-3p in glioma remain unknown. Our data showed that miR-1910-3p level was down-regulated in glioma tissues. However, the average level of miR-1910-3p was significantly lower than miR-138-5p level (Fig. S2). Thus, we focused on miR-138-5p as the primary target of LINC00174 here. Whether LINC00174 also sponges miR-1910-3p needs further study. Predictably, LINC00174 may function via regulating several miRNAs and the corresponding target genes at the same time. Thus, miR-138-5p’s potential target was next studied.
By means of Targetscan prediction, it was predicted that there exists potential seed binding sites between miR-138-5p and SOX9 3′UTR. In addition, enhancer of zeste homolog 2 (EZH2) [20], Bcl-2-like protein 11 (BCL2L11) [22] and SOX13 were reported to be regulated by miR-138-5p in glioma [37]. Moreover, some other cancer-related genes can be targeted by miR-138-5p, which mainly included E-box-binding homeobox 2 (ZEB2) in lung cancer [38], surviving in bladder cancer [19], PD-L1 in colorectal cancer [39], SIRT1 in pancreatic cancer [40] et al. Based on the miRNA’s characteristics of multiple targets, whether miR-138-5p can negatively regulate these genes in glioma needs further study. On the other hand, miR-138-5p was found to be regulated by some other lncRNAs, which mainly including lncRNA TUG1 in cervical cancer [41], lncRNA RP11-476D10.1 in papillary thyroid carcinoma [42], lncRNA HOTAIR in renal cell carcinoma [43] and lncRNA DANCR in non-small cell lung cancer [44] et al. After investigating the expression characteristics of these lncRNAs in glioma, the regulatory relationships between them and miR-138-5p in glioma needs further study. Therefore, it can be speculated that miR-138-5p might act as an intermediary agent, target several genes and be regulated by multiple lncRNAs in glioma development.
SOX9 was demonstrated to be associated with the chemoresistance of several cancers. Higashihara et al. found that cells with high Sox9 expression demonstrated stronger chemoresistance to gemcitabine than cells expressing lower Sox9 level [45]. Additionally, SOX9 regulates the response of intrahepatic cholangiocarcinoma (CCA) cells to chemotherapy. CCA patients with higher SOX9 expression had shorter survival time than those with lower SOX9 [46]. Thus, SOX9 stands out as a candidate target for the chemoresistance improvement. Because of the side effects and tolerance of small molecule compounds, gene drugs based on ncRNAs show its potential to reduce resistance. Recent studies reported that some miRNAs targeting SOX9 enhanced the cisplatin sensitivity in cancers, which mainly included miR-524-5p/miR-613 in gastric cancer [47, 48] and miR-34c [49] in ovarian cancer. Our data revealed that miR-138-5p inhibitors can restored the influence of glioma cell proliferation and apoptosis, which were caused by LINC00174 inhibition. Except for miR-138-5p, miR-105, miR-145, miR-101 and miR-30c were found to regulate SOX9 at post-transcriptional level in glioma [50,51,52,53]. Because these miRNAs collectively target they all might hold promise for enhancing the TMZ sensitivity in glioma. Here, it can be further commented that these SOX9-targeting miRNAs might be influenced by LINC00174, if there exist binding sites between them and LINC00174. Thus, a combined application of chemotherapy and SOX9 inhibitors, such as LINC00174 inhibitors, miR-138-5p or other miRNAs, might improve the TMZ-resistance and therapeutic effect.
In a word, our data demonstrated that LINC00174 down-expression can inhibit cell proliferation and promote cell apoptosis in both glioma cells and TMZ-resistant cells. LINC00174 could negatively regulate miR-138-5p and increase the expression level of its target, SOX9. Additionally, LINC00174 down-expression can inhibit the SOX9 expression and tumor growth in vitro and in vivo. Based on the functions of anti-proliferation, pro-apoptosis and the effect on TAZ-resistance, the signaling axis of LINC00174/miR-138-5p/SOX9 might hold promise as promising candidates for glioma treatment.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. https://doi.org/10.3322/caac.20107.
Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(9):494–503. https://doi.org/10.1038/ncpneuro0289(quiz 1 p following 16).
Milano MT, Johnson MD, Sul J, Mohile NA, Korones DN, Okunieff P, et al. Primary spinal cord glioma: a surveillance, epidemiology, and end results database study. J Neurooncol. 2010;98(1):83–92. https://doi.org/10.1007/s11060-009-0054-7.
Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11(4):448–57. https://doi.org/10.1038/sj.cdd.4401359.
Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17(2):165–72. https://doi.org/10.1016/j.cub.2006.11.033.
Stupp R, van den Bent MJ, Hegi ME. Optimal role of temozolomide in the treatment of malignant gliomas. Curr Neurol Neurosci Rep. 2005;5(3):198–206.
Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. https://doi.org/10.1093/hmg/ddl046.
Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–81. https://doi.org/10.1158/0008-5472.CAN-16-2634.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–61. https://doi.org/10.1038/nm.3981.
Cui B, Li B, Liu Q, Cui Y. lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR-181b. J Cell Biochem. 2017;118(12):4548–57. https://doi.org/10.1002/jcb.26116.
Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–57. https://doi.org/10.1007/s13311-018-0649-9.
Zhang H, Wei DL, Wan L, Yan SF, Sun YH. Highly expressed lncRNA CCND2-AS1 promotes glioma cell proliferation through Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2017;482(4):1219–25. https://doi.org/10.1016/j.bbrc.2016.12.016.
Shen Y, Gao X, Tan W, Xu T. STAT1-mediated upregulation of lncRNA LINC00174 functions a ceRNA for miR-1910-3p to facilitate colorectal carcinoma progression through regulation of TAZ. Gene. 2018;666:64–71. https://doi.org/10.1016/j.gene.2018.05.001.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. https://doi.org/10.1038/nrd.2016.246.
Yang L, Li C, Liang F, Fan Y, Zhang S. MiRNA-155 promotes proliferation by targeting caudal-type homeobox 1 (CDX1) in glioma cells. Biomed Pharmacother. 2017;95:1759–64. https://doi.org/10.1016/j.biopha.2017.08.088.
Qin Y, Chen W, Liu B, Zhou L, Deng L, Niu W, et al. MiR-200c inhibits the tumor progression of glioma via targeting moesin. Theranostics. 2017;7(6):1663–73. https://doi.org/10.7150/thno.17886.
Zheng Y, Lu X, Xu L, Chen Z, Li Q, Yuan J. MicroRNA-675 promotes glioma cell proliferation and motility by negatively regulating retinoblastoma 1. Hum Pathol. 2017;69:63–71. https://doi.org/10.1016/j.humpath.2017.09.006.
Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun. 2014;446(1):179–86. https://doi.org/10.1016/j.bbrc.2014.02.073.
Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, et al. miR-138-5p contributes to cell proliferation and invasion by targeting surviving in bladder cancer cells. Mol Cancer. 2016;15(1):82. https://doi.org/10.1186/s12943-016-0569-4.
Qiu S, Huang D, Yin D, Li F, Li X, Kung HF, et al. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim Biophys Acta. 2013;1832(10):1697–707. https://doi.org/10.1016/j.bbadis.2013.05.015.
He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu L, et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in glioma. J Exp Clin Cancer Res. 2019;38(1):65. https://doi.org/10.1186/s13046-019-1065-7.
Stojcheva N, Schechtmann G, Sass S, Roth P, Florea AM, Stefanski A, et al. MicroRNA-138 promotes acquired alkylator resistance in glioblastoma by targeting the Bcl-2-interacting mediator BIM. Oncotarget. 2016;7(11):12937–50. https://doi.org/10.18632/oncotarget.7346.
Kadaja M, Keyes BE, Lin M, Pasolli HA, Genander M, Polak L, et al. SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev. 2014;28(4):328–41. https://doi.org/10.1101/gad.233247.113.
Vidal VP, Ortonne N, Schedl A. SOX9 expression is a general marker of basal cell carcinoma and adnexal-related neoplasms. J Cutan Pathol. 2008;35(4):373–9. https://doi.org/10.1111/j.1600-0560.2007.00815.x.
Wang L, He S, Yuan J, Mao X, Cao Y, Zong J, et al. Oncogenic role of SOX9 expression in human malignant glioma. Med Oncol. 2012;29(5):3484–90. https://doi.org/10.1007/s12032-012-0267-z.
Gao J, Zhang JY, Li YH, Ren F. Decreased expression of SOX9 indicates a better prognosis and inhibits the growth of glioma cells by inducing cell cycle arrest. Int J Clin Exp Pathol. 2015;8(9):10130–8.
Liu H, Liu Z, Jiang B, Peng R, Ma Z, Lu J. SOX9 overexpression promotes glioma metastasis via Wnt/beta-catenin signaling. Cell Biochem Biophys. 2015;73(1):205–12. https://doi.org/10.1007/s12013-015-0647-z.
Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang P, et al. SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget. 2018;9(1):192–204. https://doi.org/10.18632/oncotarget.22773.
Han J, Chen Q. MiR-16 modulate temozolomide resistance by regulating BCL-2 in human glioma cells. Int J Clin Exp Pathol. 2015;8(10):12698–707.
Hu Y, Liu Q, Zhang M, Yan Y, Yu H, Ge L. MicroRNA-362-3p attenuates motor deficit following spinal cord injury via targeting paired box gene 2. J Integr Neurosci. 2019;18(1):57–64. https://doi.org/10.31083/j.jin.2019.01.12.
Bierhoff H. Analysis of lncRNA-protein interactions by RNA-protein pull-down assays and RNA immunoprecipitation (RIP). Methods Mol Biol. 2018;1686:241–50. https://doi.org/10.1007/978-1-4939-7371-2_17.
Worlein JM, Baker K, Bloomsmith M, Coleman K, Koban TL. The eighth edition of the guide for the care and use of laboratory animals. Am J Primatol. 2011;73:98.
Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–4. https://doi.org/10.1056/NEJM200011093431901.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93(18):9821–6. https://doi.org/10.1073/pnas.93.18.9821.
Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504–13. https://doi.org/10.1016/j.biopha.2017.08.097.
Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24(7):1199–215. https://doi.org/10.1038/mt.2016.71.
Yan J, Xu C, Li Y, Tang B, Xie S, Hong T, et al. Long non-coding RNA LINC00526 represses glioma progression via forming a double negative feedback loop with AXL. J Cell Mol Med. 2019;23(8):5518–31. https://doi.org/10.1111/jcmm.14435.
Zhu D, Gu L, Li Z, Jin W, Lu Q, Ren T. MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol Res Pract. 2019;215(5):861–72. https://doi.org/10.1016/j.prp.2019.01.029.
Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z, et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7(29):45370–84. https://doi.org/10.18632/oncotarget.9659.
Tian S, Guo X, Yu C, Sun C, Jiang J. miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget. 2017;8(7):11071–82. https://doi.org/10.18632/oncotarget.14360.
Zhu J, Shi H, Liu H, Wang X, Li F. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017;8(39):65253–64. https://doi.org/10.18632/oncotarget.18224.
Zhao Y, Zhao L, Li J, Zhong L. Silencing of long noncoding RNA RP11-476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138-5p-dependent inhibition of LRRK2. J Cell Physiol. 2019;10:15. https://doi.org/10.1002/jcp.28702.
Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, et al. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018;37(37):5037–53. https://doi.org/10.1038/s41388-018-0175-6.
Bai Y, Zhang G, Chu H, Li P, Li J. The positive feedback loop of lncRNA DANCR/miR-138/Sox4 facilitates malignancy in non-small cell lung cancer. Am J Cancer Res. 2019;9(2):270–84.
Higashihara T, Yoshitomi H, Nakata Y, Kagawa S, Takano S, Shimizu H, et al. Sex determining region Y Box 9 Induces chemoresistance in pancreatic cancer cells by induction of putative cancer stem cell characteristics and its high expression predicts poor prognosis. Pancreas. 2017;46(10):1296–304. https://doi.org/10.1097/MPA.0000000000000945.
Yuan X, Li J, Coulouarn C, Lin T, Sulpice L, Bergeat D, et al. SOX9 expression decreases survival of patients with intrahepatic cholangiocarcinoma by conferring chemoresistance. Br J Cancer. 2018;119(11):1358–66. https://doi.org/10.1038/s41416-018-0338-9.
Xue M, Li G, Sun P, Zhang D, Fang X, Li W. MicroRNA-613 induces the sensitivity of gastric cancer cells to cisplatin through targeting SOX9 expression. Am J Transl Res. 2019;11(2):885–94.
Wang J, Xue X, Hong H, Qin M, Zhou J, Sun Q, et al. Upregulation of microRNA-524-5p enhances the cisplatin sensitivity of gastric cancer cells by modulating proliferation and metastasis via targeting SOX9. Oncotarget. 2017;8(1):574–82. https://doi.org/10.18632/oncotarget.13479.
Xiao S, Li Y, Pan Q, Ye M, He S, Tian Q, et al. MiR-34c/SOX9 axis regulates the chemoresistance of ovarian cancer cell to cisplatin-based chemotherapy. J Cell Biochem. 2019;120(3):2940–53. https://doi.org/10.1002/jcb.26865.
Liu N, Zhang L, Wang Z, Cheng Y, Zhang P, Wang X, et al. MicroRNA-101 inhibits proliferation, migration and invasion of human glioblastoma by targeting SOX9. Oncotarget. 2017;8(12):19244–54. https://doi.org/10.18632/oncotarget.13706.
Liu X, Wang H, Zhu Z, Ye Y, Mao H, Zhang S. MicroRNA-105 targets SOX9 and inhibits human glioma cell progression. FEBS Lett. 2016;590(23):4329–42. https://doi.org/10.1002/1873-3468.12458.
Liu S, Li X, Zhuang S. miR-30c impedes glioblastoma cell proliferation and migration by targeting SOX9. Oncol Res. 2019;27(2):165–71. https://doi.org/10.3727/096504018X15193506006164.
Rani SB, Rathod SS, Karthik S, Kaur N, Muzumdar D, Shiras AS. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013;15(10):1302–16. https://doi.org/10.1093/neuonc/not090.
Funding
This work was supported by The fund of Shaanxi Key Laboratory of Brain Disorders (Grant No. 18NBZD01).
Author information
Authors and Affiliations
Contributions
MSC conceived and designed the experiments, FLW and JMS analyzed and interpreted the results of the experiments, HKZ and BL performed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests, and all authors should confirm its accuracy. The authors state that there are no conflicts of interest to disclose.
Ethical approval
The animal use protocol listed below has been reviewed and approved by the Animal Ethical and Welfare Committee. Approval No. 2019010.
Informed consent
Written informed consent was obtained from a legally authorized representative(s) for anonymized patient information to be published in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13577_2019_281_MOESM1_ESM.jpg
Figure S1 (A) Relative MGMT methylation status in the established TMZ-resistant sublines of U251 and U87, and in parent U251 and U87 cell lines, as determined using methylation-specific polymerase chain reaction. (B) Relative proliferation of LN229 cells after transfected with sh-LINC00174 or sh-NC and treated with different doses of TMZ were, respectively, determined by CCK-8. TMZ treatment significantly decreased cell proliferation in a dose-dependent manner. IC50 was calculated using the sigmoidal dose–response function of GraphPad Prism. (C) Kaplan–Meier curves of overall survival of 30 glioblastoma patients, stratified by LINC00174 expression. The data are expressed as the mean ± SEM, **P < 0.01 (JPEG 744 kb)
13577_2019_281_MOESM2_ESM.jpg
Figure S2 The relative miR-1910-3p levels in glioma tissues (tumor, n = 40) and control tissues (normal, n = 40), as determined using qRT-PCR. The data are expressed as the mean ± SEM, **P < 0.01 (JPEG 462 kb)
Rights and permissions
About this article
Cite this article
Li, B., Zhao, H., Song, J. et al. LINC00174 down-regulation decreases chemoresistance to temozolomide in human glioma cells by regulating miR-138-5p/SOX9 axis. Human Cell 33, 159–174 (2020). https://doi.org/10.1007/s13577-019-00281-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-019-00281-1