Abstract
Validation of marker-QTL association for genes grain size 3 (GS3), grain weight 2 (GW2), seed width 5 (qSW5) and a QTL qgrl7.1 for grain length was undertaken in a set of 242 diverse rice germplasm. Further, the study was extended to an F2 mapping population derived from cross of Sonasal, a short grain aromatic rice landrace with Pusa Basmati 1121, a variety with extra long slender grains. Seven gene specific markers, namely, SF28, SR17, RGS1and RGS2 based on GS3, W004 for GW2, MS40671 for qSW5 and RM505 for qgrl7.1, were used for validation. Single marker analysis revealed significant association of these markers to grain size and shape. The marker SF28 explained highest phenotypic variance (37 %) while the marker W004 explained lowest variance (2.6 %) for grain length in the germplasm set at the significance level P < 0.05. Three markers namely, SF28, MS40671 and RM505 were polymorphic between the parents Sonasal and Pusa Basmati 1121. In the F2 population, the marker SF28 linked to gene GS3 explained highest phenotypic variance (32.5 %), while RM505 linked to qgrl7.1 explained 5.4 % of phenotypic variance for grain length. The marker SF28 was found to be most robust in the validation studies both in germplasm and F2 population. The validated gene specific markers can be utilised in marker assisted selection for improving grain size and shape as these traits have significant contribution towards grain quality and grain yield. This is the first study on validation of gene based markers for grain dimension traits in Indian rice germplasm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice grain size and shape before and after cooking are determined by a combination of traits that include grain length, grain breadth, length/breadth ratio and elongation ratio. These complex traits not only determine grain shape and size but also contribute to grain yield (Fan et al. 2008). Grain physical appearance is imperative for consumers because preference for rice varies with consumer from region to region and also across countries. Short to medium bold grain rice with low amylose and stickiness is preferred in countries like China, Japan, Taiwan, Thailand and two Koreas whereas in countries like India, Pakistan, Malaysia, Myanmar and Brazil long slender grain is preferred (Unnevehr et al. 1992). India has a wide range of varietal difference for grain size ranging from short, medium, long to extra long grain (Khush et al. 1979). Continuous efforts are being made by rice researchers towards developing new varieties with desirable physical grain characteristics based on the understanding gained through inheritance studies in past (Chau 1928; Bollich 1957; Ramaiah and Parthasarthy 1933), mapping of genes/QTLs (Aluko et al. 2004; Huang et al. 1997; Li et al. 2004; Tan et al. 2000; Wan et al. 2005 and 2006) and cloning of genes determining grain dimension traits (Takano–Kai et al. 2009; Song et al. 2007; Shomura et al. 2008; Wang et al. 2008).
Studies on grain dimension traits have led to significant progress in finding chromosomal regions determining these traits. Although QTLs responsible for grain dimension traits have been identified on each of the 12 chromosomes (www.gramene.org), only few genes affecting grain size namely GS3, GW2 and qSW5 have been fine mapped, cloned and functionally validated. A genomic region identified in pericentromeric region of chromosome 3 has been consistently detected explaining the high proportion phenotypic variance for grain length in different genetic backgrounds (Huang et al. 1997; Redona and Mackill 1998; Tan et al. 2000; Kubo et al. 2001; Xing et al. 2002; Thomson et al. 2003; Aluko et al. 2004). Eventually, GS3 gene underlying the QTLs from the same region of chromosome was cloned using a BC3F2 population from the cross between Minghui63/Chuan7 (Fan et al. 2006). A functional SNP in exon-2 of the gene was identified resulting into pre-mature stop codon causing truncation of the protein leading to long grain size. A CAPS marker, SF28 was developed to differentiate long grain genotypes carrying the SNP from the short grain genotypes. The function of GS3 and its genetic pathway for determination of seed size was demonstrated through RT-PCR and GUS expression in transgenic plants (Takano–Kai et al. 2009). Three functional loci in GS3 were identified namely SR17, RGS1 and RGS2 in the second intron, the last intron and the final exon of GS3, respectively (Wang et al. 2010).
Other major gene, GW2 contributing to grain width and weight was identified in segregating population developed from a cross between WY3, large grain genotype and Fengaizhan-1 (FAZ1), small grain genotypes (Song et al. 2007). GW2 carrying eight exons and seven introns encodes for a new RING-type protein with intrinsic E3 ubiquitin ligase activity. A single base pair deletion resulting in a premature stop codon in GW2 gene leads to increase in seed size and number of spikelet hull cells, which increases hull size and enlarges the endosperm cell size in mature grain. Expression studies revealed that reduced expression of this gene increases grain width and results in enhanced grain weight.
Another locus contributing to grain width, qSW5 identified using F2 mapping population from the cross Nipponbare (wide grain) and Kasalath (slender grain), explained 38.5 % of phenotypic variance in F2 population (Shomura et al. 2008). Several haplogroups were identified in the qSW5 region and the deletion in the Nipponbare allele of qSW5 was found associated with an increase in grain width and associated with domestication.
A QTL, qgrl7.1 was mapped using RIL populations from a cross Pusa Basmati 1121, an extra long slender grain Basmati rice variety with exceptionally high cooked kernel length with Pusa 1342, a non-aromatic new plant type breeding line with medium grain length. This QTL was located on chromosome 7 in the marker interval RM11–RM505 with LOD score of 4.0 explaining 7.4 % of the phenotypic variation (Amarawathi et al. 2008). This QTL was also found to influence grain breadth and grain length breadth ratio contributing to 10.1 % and 10.0 % of the phenotypic variation, respectively. The same region was earlier reported to contribute for grain dimension traits (Redona and Mackill 1998; Bai et al. 2010).
The aforesaid gene based markers of grain dimension traits reported earlier need to be validated across a diverse germplam set for their effective use in marker assisted selection for improving respective traits. Therefore, the present study was undertaken to validate the marker-QTL association with respect to above said loci in a set of diverse germplasm and an F2 mapping population, and to assess the relative contribution of the loci towards trait variance and find their utility in MAS for improvement of these traits.
Materials and methods
Markers and traits
A total of four QTL/genes were investigated in present study using gene based/QTL linked markers. List of markers and traits studied is given in Table 1.
Plant material
A set of 242 diverse set of germplasm consisting of traditional landraces and improved high-yielding varieties including aromatic and non–aromatic genotypes grown in different agro ecologies of India, was used for validation study. In addition, an F2 population of 300 plants derived from cross Sonasal, a short grain aromatic landrace and Pusa Basmati 1121, having extra long sender grain Basmati rice variety with cooked kernel length of more than 20 mm and elongation ratio of 2.50 was used for validation of the markers in a segregating population. All the germplasm and F2 population were genotyped using seven gene based markers for grain dimension traits to validate their utility for MAS.
Phenotypic evaluation for grain dimension traits
Mature harvested grains of the germplasm set and F2 population along with parental lines were used for taking rough rice length and breadth. The seeds were then dried at 37 °C for 15 days, dehulled in a mini hulling machine and polished with a mini polisher (Kett Electronics, Japan). Ten representative unbroken polished grains with two replications were spread on a graph paper and photographed using CCD camera (Alpha Innotech FluorChem TM 5500) for the measurement of grain dimensions. The grain length/breadth ratio was calculated by dividing mean grain length with mean grain breadth of the ten grains in each replication. Similarly, cooked kernel length and cooked kernel elongation ratio was estimated by dividing mean grain length after cooking with mean grain length before cooking of each genotype.
Molecular Analysis
A total of seven markers namely SF28, SR17, RGS1, RGS2, W004, MS40671 and RM505 were used for genotyping germplasm set and F2 population. All PCR primer sequences were taken from respective publications (Fan et al. 2008; Song et al. 2007; Shomura et al. 2008; Amarawathi et al. 2008; Wang et al. 2010). These markers were also used for survey of polymorphism in parents, Sonasal and PB1121. Markers RM505, MS40671 and SF28 were found to be polymorphic and used to genotype 300 F2 plants. Total DNA was extracted from each of 300 F2 plants and 242 germplasm lines following the procedure of Murray and Thompson (1980). The quantification of DNA was carried out with 0.8 % agarose gel using uncut Lamda DNA as standard and diluted to concentration of approximately 25 ng/μl for PCR analysis. PCR reactions were performed in thermalcycler (G–storm, KAPA Biosystems, UK). PCR reactions were carried out in a volume of 10 μl reaction mixture consisting of 10x PCR assay buffer (Bangalore Genei Pvt. Ltd., India), 200 μM of each dNTP (MBI Fermentas, Lithuania, USA), 12 ng (1.8 picomole) each of forward and reverse primers (Sigma), 0.5 units of Taq DNA polymerase (Bangalore Genei Pvt. Ltd., India). Template DNA was initially denatured at 94 °C for 5 min followed by 35 cycles of PCR amplification with the following parameters: 1 min denaturation at 94 °C, 1 min annealing at 55 °C and 2 min of primer extension at 72 °C followed by final extension of 72 °C for 7 min. The PCR products were separated on 3.5 % Agarose gel (Bangalore Genei Pvt. Ltd., India) using 1X TAE buffer. The size of the amplified fragments was determined using 50 bp DNA ladder (Fermentas) and gel images were documented in Biorad gel–doc system.
Markers–traits association analysis
Chi–square analysis was carried out to test goodness of fit of observed segregation ratio at individual marker locus using the formula,
where, O is an observed value and E is the expected value. Statistical analyses were performed using single marker analysis using SAS software (SAS Institute Inc 2008). Analysis of variance were analysed at a P ≤ 0.05 level of significance.
Results
Variation for grain dimension traits
Phenotypic data for eight grain dimension traits namely, rough rice length, rough rice breadth, rough rice length to breadth ratio, milled rice length, milled rice breadth, milled rice length breadth ratio, cooked kernel length and elongation ratio were recorded in both the germplasm and F2 population. In the germplasm, rough rice length ranged from 5.50 to 12.50 mm with mean of 8.80 mm while rough rice breadth ranged from 1.70 to 3.30 mm with mean value 2.50 mm. The traits, viz., cooked kernel length and elongation ratio ranged from 5.50 to 17.30 mm and 1.20–2.10 with mean values of 9.10 mm and 1.50, respectively. The rough rice length of parental lines, Sonasal and Pusa Basmati 1121 were 5.60 mm and 11.60 mm, respectively. In the F2 mapping population rough rice length ranged from 5.10 to 11.40 mm while rough rice breadth ranged from 1.80 to 2.90 mm with mean of 2.40 mm. The cooked kernel length and elongation ratio in F2 population ranged from 5.76 to 15.23 mm and 1.00–2.30 with mean values of 8.86 mm and 1.67, respectively. The details of the variability parameters for grain dimension traits as observed in the germplasm set and F2 population are presented in Table 2. Frequency distributions in the germplasm for eight grain dimension traits are shown in Fig. 1.
Genotyping of germplasm set and F2 population with gene specific markers
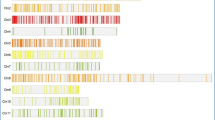
Number of different alleles amplified along with their fragment sizes with respect to each of the seven markers in germplasm set is given in Supplementary Table 1. The CAPS marker SF28 amplified a fragment of 136 bp in germplasm set, parents and F2 population, which on restriction digestion with PstI produced two fragments of sizes 110 bp + 26 bp in majority of short grain genotypes (Fig. 2). While fragment of 1,438 bp and 1,100 bp were amplified with SR17, 196 bp and 180 bp with RGS1, 269 bp and 260 bp with RGS2, 1,100 bp and 700 bp with W004, 320 bp and 300 bp with MS40671 and 220 bp and 180 bp with RM505.
Representative amplification profile of germplasm with the marker SF28. M: 50 bp Ladder; 1–48: germplasm S.No as per supplementary Table 1. Fragment of 136 bp is amplified in most of the long grain genotypes; while in majority of short grain genotypes fragments of 110 bp and 26 bp are produced on restriction digestion of 136 bp fragment with PstI
Out of seven markers used, three markers, viz., SF28, RM505 and MS40671 were found polymorphic between parents Sonasal and PB1121. The marker SF28 amplified 136 bp allele in both Sonasal and PB1121, which on restriction digestion with PstI produced two fragments of sizes 110 bp + 26 bp allele (Sonasal-A type) while PB1121 showed a uncut fragment of size 136 bp (digested as B), in the F1 plants all the fragments 136 bp + 110 bp + 26 bp were observed, confirming the hybridity of F1 plants. Similarly, RM505 amplified fragments of 220 bp and 180 bp in Sonasal and PB1121, respectively. MS40671 linked to qSW5, amplified 320 bp in PB1121 and 300 bp fragment in Sonasal. The data on segregation pattern of the three markers are presented along with probability of goodness to fit with the expected Mendelian segregation ratio of 1:2:1 for codominant marker is presented in Table 3. High degree of segregation distortion was observed for SF28 with homozygous individuals carrying mutant allele (B-type) and heterozygous individuals observed in very high frequency. The markers MS40671 and RM505 showed Mendelian segregation.
Analysis of marker-trait association
In the germplasm set, the marker SF28 showed a strong association with grain dimension traits such as grain length, grain breadth, L/B ratio, cooked kernel length and elongation ratio, explaining 37 %, 10 %, 32 %, 32 % and 12 % of phenotypic variance, respectively. The marker-trait association for qSW5 and qgrl7.1 were also found to be significant (Table 4).
In F2 population, the GS3 gene based marker SF28 was found to be strongly associated with grain length explaining 32.5 % of the phenotypic variance while RM505 explained 5.4 % of the phenotypic variance. SF28 also explained considerable amount of phenotypic variance for grain breadth, grain L/B ratio and cooked kernel length explaining 3 %, 14 % and 25 % of the phenotypic variance, respectively at significant P values. The marker MS40671, specific for gene qSW5 explained 2.3 % phenotypic variance for grain breadth while markers SF28, W004 and RM505 showed non significant association for this trait.
Discussion
In rice, QTL mapping studies have led to the identification of several markers linked to QTLs for grain and cooking quality traits. Many of these QTLs have been fine mapped and gene based markers have been developed for breeding applications. Grain length, grain breadth, cooked kernel length and elongation ratio are very important traits for improvement of grain and cooking quality in rice in general and Basmati rice in particular. These traits are under polygenic control and genetic gain in these traits through direct selection is rather low. Identification of molecular markers linked to QTLs/genes underlying these traits would go a long way in their genetic improvement through marker assisted selection.
In the present study, the marker-trait association was validated using seven gene based markers in a diverse set of rice germplasm and in F2 mapping population segregating for grain dimension traits. The proportion of phenotypic variance explained by the individual markers ranged from 2.5 % to 37 % and 2.7 % to 32.5 % in germplasm and F2 mapping population, respectively. The marker SF28 in GS3 clearly distinguished 242 germplasm lines into two groups amplifying 136 bp in long grain types and two fragments of 110 bp and 26 bp on restriction digestion of 136 bp fragments with PstI in short grain type. The SF28 marker in GS3 explained 37 % of phenotypic variance for grain length in the germplasm set and thus, appears to be a useful candidate for marker assisted introgression of the gene GS3 for improving grain size in rice. Similarly in F2 mapping population, GS3 locus explained substantially high proportion of phenotypic variance for grain length (32.5 %), grain length/breadth ratio (28 %) and cooked kernel length (25 %). Therefore, Pusa Basmati 1121 can be used as a potential donor for grain size in the breeding programme.
Besides SF28, three markers namely SR17, RGS1 and RGS2 were earlier identified in the second intron, the last intron and exon-5 of GS3, respectively (Wang et al. 2010). On validation markers SR17, RGS1 and RGS2 were also found to have significant association with grain length explaining phenotypic variance of 7.9 %, 12.0 % and 6.3 %, respectively in germplasm set. The results in the present study were in accordance with the earlier study by Wang et al. (2010), where three allelic groups were detected in Chinese landraces of rice germplasm (O. sativa), some cultivated African rice (O. glaberrima) and wild rices (O. rufipogon, O. nivara, O. barthii, and O. meridionalis). The aforesaid markers in the GS3 region were also found to be associated with other grain dimension traits such as grain breadth, grain L/B ratio, cooked kernel length and elongation ratio. The present study confirmed that GS3 locus influencing grain dimension traits was widely distributed in the Indian rice germplasm.
The marker W004 was used for validation of GW2 gene reported for enhancing grain width and yield, grouped genotypes into two allelic groups amplifying fragments of 1,100 bp and 700 bp. Although the variability in grain width in the germplasm used, ranged between 1.7 and 3.3 mm, the phenotypic variance explained by GW2 based marker W004 was not significant indicating that this locus was not so important and there may be additional genes involved in determining grain width in the germplasm studied.
The marker MS40671 accounted 2.6 % phenotypic variance for grain breadth in germplam set, which was found to be very less compared to validation results of qSW5 in a set of landraces including both japonica and indica with grain width >3.3 mm (Shomura et al. 2008). The model for japonica rice domestication proposed by them suggested that the qSW5 gene propagated to upland areas of China and Japan, is absent in wild species O. rufipogon and indica rices. Also, deletion in the gene identified through comparative sequencing clearly associated the increase in grain width due to Nipponbare allele (japonica). In our study, the marker MS40671 based on qSW5 grouped the rice germplasm into two allelic groups of 320 bp and 300 bp but its contribution to the phenotypic variance for grain width was very low indicating that qSW5 locus was not widely distributed in the germplasm studied.
The marker RM505 flanking the QTL qgrl7.1 classified the rice germplasm into two allelic groups comprising of 220 bp and 180 bp fragment size and explained 12.4 % and 12.6 % phenotypic variance for grain L/B ratio and cooked kernel length, respectively. Recently, this region on chromosome 7 has been fine mapped to a region of 4.8Kb. Candidate gene analyses found no annotated genes in this region while two open reading frames were considered to be more likely candidate for GS7 gene (Shao et al. 2012).
Since, the grain dimension traits are quantitatively inherited and have low heritability, the present study on validation of grain dimension associated markers would lead to improvment in grain dimension related traits. The present study has clearly showed the robustness of markers namely SF28, SR17, RGS1, RGS2, W004, MS40671 and RM505 in the validation study using set of diverse Indian germplasm and a biparental F2 population. These markers hold great promise in MAS for improvement of respective traits. The identified germplasm having respective positive alleles for grain dimension traits can be used as a donor parent in marker assisted backcross breeding.
Abbreviations
- GS3 :
-
Grain size 3
- GW2 :
-
Grain weight 2
- qSW5 :
-
QTL for Seed width on chromosome 5
- QTL:
-
Quantitative trait locus
- SNP:
-
Single nucleotide polymorphism
- MAS:
-
Marker assisted selection
- CAPS:
-
Cleaved amplified polymorphic sequences
References
Aluko G, Martinez C, Tohme J, Castano C, Bergman CJ, Oard JH (2004) QTL mapping of grain quality traits from the interspecific cross Oryza sativa x Oryza glaberrima. Theor Appl Genet 109:630–639
Amarawathi Y, Singh R, Singh AK, Singh VP, Mohapatra T, Sharma TR, Singh NK (2008) Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol Breed 21:49–65
Bai XF, Luo LJ, Yan WH, Rao KM, Zhan W, Xing YZ (2010) Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genetic 11:16
Bollich CN (1957) Inheritance of several economic quantitative characters in rice. Diss Abstr 17:1638
Chau LF (1928) Linkage studies in rice. Genetics 13:133–169
Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Fan CC, Yu SB, Wang CR, Xing YZ (2008) A causal C–A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor Appl Genet 118:465–472
Huang N, Parco A, Mew T, Magpantay G, McCouch S, Guiderdoni E, Xu JC, Subudhi P, Angeles ER, Khush GS (1997) RFLP mapping of isozymes, RAPD, and QTLs for grain shape, brown planthopper resistance in a doubled–haploid rice population. Mol Breed 3:105–113
Khush GS, Paule CM, Cruz NM (1979) Rice grain quality evaluation and improvement at IRRI. Proc. of the workshop on Chemical Aspects of Rice Grain Quality, Int. Rice res. Inst, los Banos, Laguna, Phillippines, pp 21–31
Kubo T, Kai T, Yoshimura A (2001) RFLP mapping of genes for long kernel and awn on chromosome 3 in rice. Rice Genet Newsl 18:26–28
Li J, Xiao J, Grandillo S, Jiang L, Wan Y, Deng Q, Yuan L, McCouch SR (2004) QTL detection for rice grain quality traits using an interspecific back–cross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome 47:697–704
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Ramaiah K, Parthasarthy N (1933) Inheritance of grain length in rice (Oryza sativa L). Indian J Agric Sci 3:808–819
Redona ED, Mackill DJ (1998) Quantitative trait locus analysis for rice panicle and grain characteristics. Theor Appl Genet 96:957–963
Shao G, Wei X, Chen M, Tang S, Luo J, Jiao, Xie L, Hu P (2012) Allelic variation for a candidate gene for GS7, responsible for grain shape in rice. Theor Appl Genet. doi:10.1007/s00122-012-1914-7
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40:1023–1028
SAS Institute Inc. (2008) SAS/STAT: version 9.2 User’s Guide, Cary, North Carolina, USA, 2nd edition.
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING–type E3 ubiquitin ligase. Nat Genet 39:623–630
Takano-Kai N, Jiang H, Kubo T, Sweeney M, Matsumoto T, Kanamori H, Padhukasahasram B, Bustamante C, Yoshimura A, Doi K, McCouch S (2009) Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182(4):1323–1334
Tan YF, Xing YZ, Li JX, Yu SB, Xu CG, Zhang QF (2000) Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor Appl Genet 101:823–829
Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Unnevehr LJ, Du VB, Juliano BO (1992) Consumer demand for rice grain quality. International Rice Research Institute and International Development Research Center, Manila, Ottawa
Wan XY, Wan JM, Jiang L, Wang JK, Zhai HQ, Weng JF, Wang HL, Lei CL, Wang JL, Zhang X, Cheng ZJ, Guo XP (2006) QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor Appl Genet 112:1258–1270
Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ (2005) Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor Appl Genet 110:1334–1346
Wang E, Wang J, Zhu X, Hao W, Wang L (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40:1370–1374
Wang C, Chen S, Yu S (2010) Functional markers developed from multiple loci in GS3 for fine marker–assisted selection of grain length in rice. Theor Appl Genet. doi:10.1007/s00122-010-1497-0
Xing YZ, Tan YF, Hua JP, Sun XL, Xu CG (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet 105:248–257
Acknowledgement
This work was supported by Network Project on Transgenic in Crops (NPTC) funded by Indian Council of Agricultural Research, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
ESM 1
(DOC 441 KB)
Rights and permissions
About this article
Cite this article
Anand, D., Baunthiyal, M., Singh, A. et al. Validation of gene based marker-QTL association for grain dimension traits in rice. J. Plant Biochem. Biotechnol. 22, 467–473 (2013). https://doi.org/10.1007/s13562-012-0176-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-012-0176-4