Abstract
Background
Lactate is an important metabolite derived from glycolysis under physiological and pathological conditions. The Warburg effect reveals the vital role of lactate in cancer progression. Numerous studies have reported crucial roles for lactate in cancer progression and cell fate determination. Lactylation, a novel posttranslational modification (PTM), has provided a new opportunity to investigate metabolic epigenetic regulation, and studies of this process have been initiated in a wide range of cancer cells, cancer-associated immune cells, and embryonic stem cells.
Conclusion
Lactylation is a novel and interesting mechanism of lactate metabolism linked to metabolic rewiring and epigenetic remodeling. It is a potential and hopeful target for cancer therapy. Here, we summarize the discovery of lactylation, the mechanisms of site modification, and progress in research on nonhistone lactylation. We focus on the potential roles of lactylation in cancer progression and cell fate determination and the possible therapeutic strategies for targeting lysine lactylation. Finally, we suggest some future research topics on lactylation to inspire some interesting ideas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lactate, an end product of glycolysis, has long been neglected because it is considered a metabolic waste product of glucose metabolism under hypoxic conditions. However, the discovery of the Warburg effect in 1956 first revealed the vital position of lactate as a waste product and a valuable organic compound produced due to a high glycolysis rate in cancer metabolism [1]. The Warburg effect results in the progression of a cancer-promoting microenvironment characterized by lactate accumulation, low O2, low nutrient and a low pH. Subsequent research on lactate has revealed its various effects on the tumor energy supply [2], signal transduction [3] and other cellular functions. Therefore, lactate pathways are gradually emerging as a potential tumor therapeutic target.
Stem cells are responsible for the development of a whole organism. Activated metabolism and proliferation are both vital features of stem cells, and thus stem cells and cancer cells share some similarities. Lactate, an important metabolic product, might have some common functions relevant to proliferation in stem cells and cancer cells. The microenvironment of lactate (such as the stem cell niche) is bound to become the next research focus. Recent studies support the aforementioned points[4,5,6]. Proteins are vital biological macromolecules performing the specific functions encoded by genes; their precise regulation is relevant to the orderliness of life processes. Genetically determined amino acid sequences ensure the structural and functional stability of a protein. Moreover, posttranslational modifications (PTMs) of proteins are a more refined, faster and more energy-efficient mechanism for regulating protein function than alterations in gene sequences [7]. Acetylation, the first discovered form of protein modification [8], initiated the field of protein PTMs. With advances in mass spectrometry (MS), various acylation reactions have been discovered and linked to the regulation of metabolic states and protein functions.
Metabolic rewiring and epigenetic remodeling are well-known cancer hallmarks. Benefitting from the high heterogeneity and complicated metabolic microenvironment of cancer, many metabolic compounds are modified by acylation, and these phenomena indicate the relevance between metabolic rewiring and epigenetic remodeling[9]. Numerous researchers also aim to detect these modifications as a method to identify novel metabolism-dominant or metabolism-associated biological mechanisms. In 2019, Zhang et al. discovered a novel modification, histone lysine acylation, called lactylation, a potential and important metabolic feedback mechanism related to glycolysis [10]. Many therapies potentially target this modification. Nevertheless, our current understanding of this novel lysine acylation modification is limited and must be expanded through further research.
Lactate is universally acknowledged as a critical regulator of cancer development, maintenance, and metastasis [11]. We postulate that the mechanisms of lactate in cancer cells and stem cells must differ. Therefore, investigations of the mechanisms by which lactate influences the progression of cancer and the proliferation and differentiation of stem cells that aim to discover novel mechanisms at different levels and describe an increasingly complete map of the function of lactate are important. The discovery of lactylation is a milestone in the study of lactate. Below, we summarize the discovery of and recent research on lactylation, and we focus on its potential effects on cancer progression and cell fate determination and the therapeutic strategies for targeting lysine lactylation. Moreover, we suggest some areas for future lactylation research to focus on to inspire some interesting ideas.
2 Discovery of lactylation
With the recent development of MS and chromatography, the modification of proteins, especially lysine residues in histones, by intermediate metabolites has attracted the attention of scholars. Protein modifications are collectively referred to as PTMs of proteins. With the discovery of PTMs, the complexity of human proteomics has increased exponentially [12]. Furthermore, studies on PTMs revealed that metabolic reprogramming is closely related to epigenetic regulation [13] [14]. Reports of propionylation, butylation [15] [16], succinylation [17] [18], crotonylation [19] [20], 2-hydroxyisobutyrylation [21] [22] and benzoylation [23] of histone lysine residues indicated that, as an important metabolite produced under physiological and pathological conditions, lactate is a substrate of protein modification and thus affects the physiological and pathological functions of proteins specifically by changing protein structures. In 2019, Zhang et al. detected lysine lactylation (Kla) through a high-performance liquid chromatography–tandem mass spectrometry analysis of the core histone in human MCF-7 cells digested with trypsin. They successfully identified 26 Kla sites in core histones in human HeLa cells and 16 sites in mouse bone marrow-derived macrophages (BMDMs) [10]. These researchers further showed that lactate is a vital substrate for Kla in cultures treated with sodium L-lactate (13C3, 98%) and D-glucose (U-13C6, 99%) isotopes. They identified arginase 1 (Arg1) as a specific Kla-modified gene and the time-dependent change in Arg1 expression is mediated by an endogenous ‘lactate clock’ in bacterially challenged M1 macrophages [10]. Moreover, this group revealed that the recruitment of P300 mediated by p53-induced Kla, which is similar to acetylation, effectively promoted Arg1 transcription and enabled macrophages to acquire an M2-like phenotype [24]. The findings reported by Zhang et al. suggest a new approach to reexamine the role of glycolysis and subsequent lactate overproduction in normal cells and even in tumor tissues. More importantly, is a previously undiscovered mechanism of carcinogenesis mediated by lactylation?
3 The lysine lactylation mechanism
Lactylation is a novel PTM [10], but our understanding of its substrates, modification reactions (enzymatic or nonenzymatic), and reaction dynamics is still limited [25] [26]. Many questions remain unanswered, such as the identification of a specific acylase (writer), deacylase (eraser) and recognition enzyme (reader) involved in lactylation, as well as the quantity, distribution and relationship between lactyl-coenzyme A (lactyl-CoA) and lactate. Given the commonness of the lysine acylation modification, a biochemical analysis of lactylation may lead to new research ideas and the discovery of novel modification mechanisms for other lysine acylation reactions [10].
3.1 The accessible e-amino group of the lysine side chain is a modification substrate
Lysine, arginine, and histidine are the only amino acid residues with positive side chains at physiological pH. Moreover, lysine and arginine are mostly located on the hydrophilic surface of proteins. The hydrophobicity of the lysine and arginine side chains leads to the exposure of ε-amino groups to solvents, increasing their potential to participate in various physiochemical interactions [27]. According to recent studies, more lysine PTMs exist than arginine PTMs [28] [29], which may be due to the difference between the arginine and lysine side chains. The three-dimensional ion interaction formed by the guanidino group of arginine mainly maintains the structure and stability of proteins and drives protein folding. In addition to maintaining the protein structure, the ε-amino group of lysine also forms a single ion interaction, and thus the side chain of lysine residues is prone to modification [30]. This accessibility and reactivity of this lysine residue facilitates PTMs and affects protein function.
3.2 Enzymatic pathway of the lactylation modification
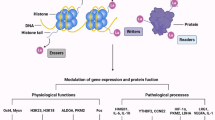
Zhang et al. showed that the substrate of Kla was lactate, the lactate level in tissues or cells was positively correlated with the histone Kla level, and lactylation was based on enzymatic action. P300, a proven acetyltransferase recruited by p53, is an important writer of histone Kla [10]. In addition to acetyl-CoA and lactyl-CoA, P300 has been shown to react with a variety of acyl-CoA species, such as butyl-CoA and crotonyl-CoA, and this reactivity decreases with increasing acyl chain length, showing robust cofactor activity [31]. As a conventional acetyl-lysine reader, the bromodomain recognizes nonacetylated lysine residues, such as butyl-lysine and crotonyl-lysine [32]. In 2022, Zhang et al. identified Class I histone deacetylases (HDAC1–3) as histone lysine delactylases, filling a research gap in delactylation and thus integrating the novel lactylation PTM into a complete biochemical process [33]. The results suggest that lactylation may be closely related to acetylation or that these PTMs may have similar writers, erasers, and readers (Fig. 1).
The metabolism, mechanism (blue box) and effects (purple and green boxes) of lactylation. Lactate is a biomarker of changes in cells (glycolysis) and the microenvironment (pO2, acidity, nutrient level and intermediate products). The metabolism and mechanism of lactylation are partially and differentially dependent on the microenvironment. Lactylation of different proteins (histones are shown in purple and nonhistone proteins are shown in green) leads to different effects, including carcinogenesis, pluripotency, transformation and stem cell activation
Abbreviations: MCT1&4, monocarboxylate transporter 1&4; GLUT, glucose transporter; LDHA, lactate dehydrogenase A; GLO cycle, glyoxalase cycle; LGSH, lactyl-glutathione; Arg1, arginase 1; TAMs, tumor-associated macrophages; YTHDF2, YTH N6-methyladenosine RNA-binding protein 2; HMGB1, high mobility group box 1
3.3 Nonenzymatic lactylation modification pathway
In 2020, a liquid chromatography–high-resolution mass spectrometry analysis with synthetic lactyl-CoA was performed to quantify lactyl-CoA levels in HepG2 cells in vitro. Researchers identified that the concentration of lactyl-CoA in cultured HepG2 cells was 1.14 × 10− 8 pmol/cell, which was 1/350 to 1/20 the concentration of acetyl-CoA, propionyl-CoA or other typical acyl-CoAs [34]. Considering the relatively low stoichiometric levels of lactyl-CoA and lactate deposition, enzymatic regulation of lactylation in most situations is relatively improbable or rare, and some other mechanisms related to lactylation might exist. An important research direction is to enrich and complement the various mechanisms of lactylation, and the contributions of multifunctional (varied acylation reactions) enzymatic reactions and nonenzymatic reactions to lactylation abundance should be generally considered [35].
Biochemical studies have shown that the chemical processes of nonenzymatic lysine acylation in mitochondria are related to the deprotonation of the lysine ε-amino group in the alkaline mitochondrial environment, and the nucleophilic effect of the deprotonated ε-amino group on the thioester bond of acyl-CoA greatly promotes the chemical mechanism of acylation [36]. Carboxyl groups in acyl donors exert a nucleophilic attack on the thioester bond, resulting in the formation of a cyclic anhydride that is more active than the original acyl-CoA to achieve nonenzymatic chemical processes [37]. These biochemical analyses provide us some insights. This nonenzymatic reaction might occur during the process of lactylation.
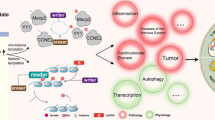
In 2020, Dominique et al. reported a unique nonenzymatic lactylation pathway [38] that differed from the pathway discovered by Zhang in 2019 [10], whose substrate is lactyl-glutathione (LGSH), not lactyl-CoA, as in conventional studies. LGSH is derived from methylglyoxal (MGO) and is a glycolytic byproduct in the glyoxalase (GLO) cycle. MGO, a byproduct of glycolysis, has important signaling functions at low concentrations but is cytotoxic at higher concentrations, which was partially shown in the nonenzymatic process known as glycation [39, 40]. The formation and accumulation of MGO have been implicated in the pathogenesis of type 2 diabetes and its vascular complications, cardiovascular diseases, and cancers [41,42,43]. The Warburg effect promotes MGO biosynthesis, but an increase in the MGO level due to the upregulated GLO system has not been observed [44, 45]. Glutathione (GSH) is the reductant of MGO detoxification in the GLO cycle mediated by glyoxalase I and II (GLO1 and GLO2, respectively). LGSH, the detoxification product, is a vital cancer biomarker for lung cancer [46], and a highly relevant finding is that LGSH is the substrate of lactylation in the tumor metabolic environment [38]. With the activation of the GLO cycle, the efficient synthesis of LGSH provides the substrate required for nonenzymatic lactylation. The study by Dominiqu revealed another unique lactylation pathway, and the authors tried to identified some links between oxidative stress and epigenetics [38] (Fig. 2).
Research on lactylation sites and related interactions. (A) General strategy for studying lactylation. The identification of a lactylation site is the basis for research on the interaction between lactylated proteins (cyan) and the corresponding writers (yellow), erasers (orange) and readers (dark blue). (B) Three types of interactions (in different color blocks) between lactylation (green residues with La) and other PTMs (orange residues with R) are shown. These relationships are typically observed among interactions of proteins after lactylation and/or other PTMs (cyan), corresponding PTM readers (dark blue), and other proteins in the pathway (blue)
Abbreviations: PTM, posttranslational modification
4 Research methods for identifying lactylation sites
After studying the mechanism of lactylation, identifying lactylation sites is vital for analyzing the function and structure of lactylated proteins. Various methods have been developed for studying protein PTM sites [47], but more general, accurate, and efficient methods are needed to identify complicated PTMs (Fig. 2).
4.1 Limitations of conventional methods for studying lactylation sites
First, research on PTM sites was performed with individual proteins and isotope-labeled anti-acyl-CoA or anti-acyl-lysine antibodies [7]. In MS analyses, the target protein is digested into peptides containing several amino acids with trypsin and then analyzed to obtain exact information on the modified sites [48]. In 2006, Zhao et al. successfully identified hundreds of acetylation sites through a combination of acetylated peptide immunoaffinity enrichment and high-resolution MS [49].
Considering the universality and complexity of PTM sites, large-scale screening is indispensable. Large-scale screening with MS potentially ensures the accuracy of the identification of lactylation sites, but this process is neither economical nor efficient. Therefore, a method that combines accuracy, scalability, and efficiency is urgently needed. Some researchers have tried to perform statistical analyses based on the identified lactylation sites to identify conserved amino acid sequences or structural features close to lactylation sites and thus determine the processes governing their distribution [50].
4.2 Application of new technology represented by artificial intelligence in the study of lactylation sites
Statistical methods used to study lactylation sites are the basis of a theoretical prediction method using existing experimental data. In conjunction with the rapid development of artificial intelligence (AI), Jiang et al. began to apply AI to predict PTM sites, optimizing complicated AI experiments and statistical analyses with computer algorithms [51]. First, according to the identified lactylation sites that had been previously published, an integrated dataset was established with amino acid sequences [52], protein structures [53] and other features as model parameters. Then, these datasets were processed using machine learning to design an algorithm for the prediction of lactylation sites in unknown protein sequences. However, since only 382 sites in 191 proteins have been identified [10] [50] [54], these training datasets are too small and uneven for machine learning, which is prone to overfitting. Therefore, in 2021, Jiang et al. creatively introduced few-shot learning [55] and ensemble deep learning [56] in a joint design for use in dataset processing. They constructed 11 feature-encoding sets to cover amino acid sequences, the physicochemical environment of each amino residue, structural information, and other parameters that can be used to describe lactylation sites and these environments in general. The area under the curve of this joint algorithm was 11.7% (from 0.667 to 0.745), which was better than that of a deep neural network, random forest, logistic regression and other general algorithms, verifying its superior sensitivity and specificity [51]. When computer technology development and multiomics studies provide large-scale data and samples for our research, revolutionary changes also occur in biological and medical research. PTM site prediction by AI is a discovery that changed research thinking. When faced with a large number of PTM sites, conventional physical, chemical and biological experiments no longer must be planned immediately, and we can predict and evaluate these sites first. The trial-and-error cost can be saved, but the accuracy of the AI may not be sufficient.
5 Lactylation of nonhistone proteins
Lactylation was first discovered on histones; therefore, the role of lactylation on histones has been relatively well studied. However, recent research revealed that lactylation was ubiquitous [57, 58], and thus studies exploring lactylation of nonhistone proteins with multiple functions is important [7]. Currently, many researchers are focusing on the lactylation sites in nonhistone proteins (Table 1). In 2020, Gao et al. completed a lactylation analysis of Botrytis cinerea (a destructive necrotrophic fungal pathogen) using proteomics with LC–MS/MS to identify 273 lactylation sites in 166 proteins, and approximately 2/3 of the identified lactylated proteins were distributed in the cytoplasm (27% in mitochondria). In addition, 1/3 of these proteins were related to translational control, suggesting that lactylation may be enriched in ribosome-related protein subsets and play an important role in determining the expression of nonhistone proteins [50]. In the same year, James et al. identified 350 lactylated proteins in a study of nonenzymatic lactylation. After analysis with the Database for Annotation, Visualization, and Integrated Discovery (DAVID) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database, these researchers found that lactylated proteins were mainly enriched in glycolysis and carbon metabolism [38], which further suggests that lactylation in mammals may have a greater and broader effect on glycolysis. In 2021, Li et al. found that lactylation/acetylation of high mobility group box-1 (HMGB1) was promoted by a P300/CBP-dependent mechanism, which increased HMGB1 exosome secretion and subsequently induced endothelial dysfunction in individuals with sepsis [59] (Fig. 1). In fact, nonhistone proteins are the most direct executors of gene function. The functions of organisms are usually accompanied by changes in protein expression, modification, or stability. Nonhistone proteins have received more attention; we believe that lactylation of nonhistone proteins will be proven to be an important aspect of the mechanism by which lactate affects biological processes.
6 Lactylation is involved in cancer progression
Lactylation was initially discovered in research on inflammation and immunity, but some clues suggest its involvement in cancer progression. Since the discovery of the Warburg effect by Otto Warburg in 1956 [1], lactate, a core metabolite produced by the Warburg effect, has attracted increasing attention [60]. Among the recognized tumor characteristics [61], glycolysis is associated with lactate production and its accumulation in the tumor microenvironment (TME), which constitutes a critical tumor phenotype; this connection made us question whether lactate plays a role in cancer progression. From metabolic waste to a ‘general fuel’ and ‘information transmitter’ and then to an important substrate for metabolic reprogramming, the leading role of lactate in cancer progression has gradually been confirmed with research progress [2, 62]. The discovery of lactylation has provided us with a new perspective on the important mechanism of lactate metabolism in cancer progression.
6.1 Tumor-associated proteins have the potential to undergo lactylation in the TME
The TME provides a preferred metabolic condition for lactylation modification. Because of their high proliferation rate, tumor cells require large amounts of energy and material. However, the newly formed microvasculature in solid tumor tissues is often disordered, resulting in inefficient perfusion and subsequent metabolic stress [63]. Therefore, hypoxia and a low pH with lactate overproduction are important TME characteristics [64,65,66]. Based on this information, conventional views on the physical and chemical environment suggest that the abnormal pH gradient in the TME promotes cancer progression [67, 68]. Although oxidative phosphorylation (OXPHOS) is proposed to be the main energy metabolism pathway in many tissues, including tumors (the conventional assumption is that OXPHOS is universally deactivated in cancer after the discovery of the Warburg effect) [69], under metabolic stress, tumor tissues adaptively modulate glycolysis to produce more energy and intermediate metabolites, especially lactate, to meet their requirements for cellular biosynthesis, such as gluconeogenesis [70, 71]. Lactate overproduction provides the substrate for lactylation, which represents the third contribution of lactate to cancer, in addition to serving as a ‘general fuel’ [2] and in cell signaling pathways (as a ‘signal transmitter’) [62, 72]. Previous research has shown that lactate is an important substrate for lactylation mediated by lactyl-CoA [10]. Nevertheless, the relatively low concentration of lactyl-CoA in cells led to lactylation being discovered later than acetylation and indicates that higher-precision techniques are needed to identify lactylation than those needed to detect acetylation [34]. The efficiency of chemical reactions partially depends on the substrate concentration; therefore, the lactate concentration in the environment may be the basal condition for lactylation.
Lactate is a powerful molecule that rewires the TME and drives “tumor–stroma symbiosis”. In this symbiosis, monocarboxylate transporters (MCTs), especially MCT1 (for lactate intake) and MCT4 (for lactate secretion), are the most important mediators of lactate exchange in the metabolic crosstalk of lactate between cancer cells and noncancer cells. The relative expression of MCT1 and MCT4 partially indicates the lactate demand of cells, which are divided into oxidative cells and hypoxic cells according to this metabolic demand. The two different cells collaborate with each other in this symbiotic relationship[60]. “Lactate metabolic coupling” in the TME indicates that lactate metabolism is not a single-cell event[73], and we also postulate that lactylation is not an isolated modification occurring in only a few cells. Lactate in tumor cells induces the overexpression of vascular endothelial growth factor and M2-like genes in tumor-associated macrophages (TAMs) (these genes include arginase 1 (Arg1), Fizz1, Mgl1, and Mgl2) [74]. Furthermore, Arg1 expression, which is closely related to M2 macrophage polarization, is positively correlated with histone Kla levels [10]. The TME is immunosuppressive, and lactate, its core metabolite, regulates the metabolism of innate and adaptive immune cells during immunosuppression [75]. In addition to the aforementioned TAMs, regulatory T cell (Treg) action is reinforced in the TME through moesin K72la (lactylation at Lys72 in moesin; this nomenclature for PTMs is presented hereafter). The interaction of moesin with the transforming growth factor-β receptor and Smad signaling is enhanced by lactylation. Anti-lactate treatment through programmed cell death protein 1 (PD-1) therapy generates a stronger antitumor effect than PD-1 alone [76]. Other researchers have examined tumor cells themselves and have shown that the lactate level is positively correlated with the malignancy of Lewis lung carcinoma and B16-F1 melanoma cancer cells [74], and histone Kla mediated by high lactate levels leads to the upregulated expression of YTH N6-methyladenosine RNA-binding protein 2 (YTHDF2) to promote cancer progression. The tumor-promoting mechanism is based on YTHDF2 recognition of m6A-modified PER1 and TP53 mRNAs, which promotes the degradation of these mRNAs, accelerating the progression of ocular melanoma [77]. All the aforementioned research suggests that a microenvironment with a high lactate level is an important basal condition for lactylation. Therefore, due to the relatively high lactate levels in the TME, lactylation in tumor parenchymal cells and stromal cells may be substantially accelerated (Fig. 1). These studies indicate that, as lactate is produced and lactate metabolism is coupled, lactylation is possible in the whole TME. Therefore, many unknown lactylation-associated mechanisms might promote cancer progression.
6.2 Lactylation is a potential tumorigenic mechanism
As a core molecule involved in tumor metabolism and the TME, lactate affects various aspects of cancer progression, including the activation and epigenetic reprogramming of tumors and tumor-related genes, tumor metabolism, metastasis, angiogenesis, cancer cell immune escape, and tumor resistance [78]. MCT and G-protein-coupled receptor 81 (GPR81) play important roles as the transport receptor and signaling receptor for lactate, respectively [60, 72, 79]. As a novel mechanism of lactate, lactylation may play an important role in cancer progression, and its potential effect is classified into histone-mediated pathways and nonhistone-mediated pathways, depending on the proteins primarily affected by lactylation.
The histone-mediated pathway refers to the epigenetic effect of lactylation on gene expression that alters tumor phenotypes. Epigenetic modification is universally acknowledged as a vital component of carcinogenesis, and its effects on oncogene and tumor suppressor gene expression influence cancer cells, cancer stem cells and even cancer mesenchymal cells [80,81,82]. Some of the aforementioned studies have preliminarily explained the epigenetic role of lactylation in cancer cells, such as lactylation-mediated upregulation of the oncogene YTHDF2 [77] and M2 polarization of TAMs [10].
The nonhistone-mediated pathway refers to changes in general protein structure, function and localization mediated by lactylation. Because lactylation was initially discovered in histones, studies of lactylation in histones and epigenomes are ongoing [10]. However, approximately 1/3 of lactylated nonhistone proteins were found to be related to protein translation in a proteomic analysis of fungal systems [50], and lactylated nonhistone proteins are mainly enriched in glycolysis and carbon metabolism pathways in mammals [38]. This evidence indicates that lactylation might exert greater effects on protein translation and downstream pathways than on epigenetic changes and transcription. However, due to the lack of studies on individual proteins, especially the lactylation of core proteins known to play important roles in cancer progression, evidence to support the hypothesis that nonhistone proteins mediate cancer progression is lacking, and therefore further research is needed.
7 Lactylation is involved in determining the fate of stem cells
Cell fate determination is a research hotspot in the 21st century. Research on adult tissue stem cells and induced pluripotent stem cells (iPSCs) has provided suggestions for potential therapy in regenerative medicine. Metabolism guides cell fate determination, and glycolysis, OXPHOS, and other metabolic pathways influence metabolism by regulating gene expression and/or signaling pathways [83]. Metabolic reprogramming and epigenetic mechanisms in stem cells thus reveal the ‘power of metabolism’ [84]. The novel discovery of lactylation provides a new perspective for studying the mechanism of lactate metabolism in cell fate determination.
7.1 Lactate metabolism is related to the activation of stem cells
Lactate is the most important metabolite related to cell proliferation. Many of the aforementioned studies have elucidated mechanisms underlying the effect of lactate on the proliferation of certain cells, such as cancer cells, and the modulation of stem cell activation and division[4,5,6] (Table 2). First, lactate promotes stem cell activation; lactate was shown to activate hair follicle stem cells (HFSCs) [85], and another study showed that epithelial development mediated by intestinal stem cells was accelerated by microbiota-derived lactate [86]. Second, the distinct lactate metabolic flow is highly relevant to the specificity of differentiation and phenotype shifts; purification of generated cardiomyocytes for pluripotent stem cell (PSC)-dependent heart regeneration depends on the microenvironment of lactate, and lactate promotes the acquisition of a synthetic phenotype by vascular smooth muscle cells derived from PSCs [87, 88]. Lactate induces the global upregulation of genes involved in embryo cleavage, such as members of the Zscan4 gene family. Further explorations indicated that lactate stimulated increased H3K18 lactylation on germline and embryo cleavage-related genes, which in turn promoted transcript elongation [89]. The aforementioned research has described three potential effects of lactate on stem cells, which directs our attention to the roles of lactate, particularly the novel modification lactylation, in stem cells: participation in metabolism, signal transmitters or epigenetic modulators (Fig. 1).
7.2 Lactylation is related to somatic cell reprogramming
Lactylation is an important epigenetic modification regulating somatic and pluripotency genes. As the ‘fifth Yamanaka reprogramming factor’, Gli-like transcription factor 1 (Glis1) expressed in iPSCs regulates the activation of somatic genes (Atf, Batf, and Jun) and pluripotency genes (Klf4, Sox2, Oct4, Nanog, etc.), promotes glycolytic gene expression and, with substrate accumulation, accelerates the lactylation and acetylation of histones, which initiates the so-called ‘second transcriptional wave’ at the level of chromatin [54]. This study was the first to introduce a role for lactylation in somatic cell reprogramming and reveal a new mechanism by which lactylation regulates gene transcription by influencing the shift in the chromatin state (Fig. 1). However, unfortunately, the relationship between lactylation and acetylation was not addressed in this study, and it may be the next vital question to be answered. Histone acetylation plays a vital role in chromatin remodeling, which is the premise of cell reprogramming and differentiation [90]. Studies on genes related to embryo cleavage and pluripotency genes indicate that lactylation affects cell remodeling, reprogramming and differentiation at the epigenetic level, similar to acetylation [54, 89]. This valuable discovery provides a better understanding more of the mechanism by which metabolism affects cell remodeling and differentiation, the number of mediators involved and the complexity of epigenetic reprogramming.
8 Lactylation is a potential target for cancer therapy and regenerative medicine
Approaches targeting lactate production and transportation to modulate are important strategies to improve tumor prognosis. Lactate dehydrogenase (LDH), especially lactate dehydrogenase A (LDHA), is the core enzyme in glycolysis and lactate metabolism, and its overexpression in various tumor cells makes it a potential target for tumor therapy [91]. In research on lactylation, the use of LDH inhibitors (such as sodium oxamate) to downregulate lactate metabolism inhibits lactylation and thus blocks the downstream lactylation pathways [10, 77]. Several effective LDH inhibitors have been identified, and some have entered phase I and II clinical trials [92]. Inhibiting the expression of genes upstream of LDH has also been an effective strategy, including the oncogene MYC (encoding c-Myc) [93], hypoxia-inducible factor [94], cAMP response element-binding protein (CREB) [95], and heat-shock factor 1 [96], MCTs, known as solute carrier family 16, are important transmembrane proteins with 4 members in the family, among which are four main transporters: MCT1 to MCT4 [79]. MCTs play an important role in lactate transport and may be important targets in lactate transportation [60, 97]. In a study on lactylation in the brain, cortical neurons were treated with the MCT2 inhibitors A-cyano-4-hydroxycinnamate and AR-C155858 in vitro, which inhibited lactylation induced by exogenous lactate treatment [98].
Direct targeting of lactylation, delactylation, and recognition of lactylation is the most accurate method for treating tumors. However, due to incomplete research on lactylation, insufficient information is insufficient to design targeted mediators. Therefore, we must thoroughly understand the writers, erasers, and readers involved in lactylation. Zhang et al. have proven that P300 mediates lactylation, which was the first discovered lactylase [10]. P300 is a well-studied lysine acetyltransferase (KAT) and transcriptional coactivator. Because P300 and CREB-binding protein (CBP) share 63% homology and contain very similar domains, these proteins are often referred to together as P300/CBP [99]. P300/CBP is an acetylase targeting more than 2/3 of acetylation sites [7], and its overexpression is strongly correlated with many diseases, especially malignant tumors [100]. The bromodomain and KAT active sites are important carcinogenic targets in hematologic malignancies [101], prostate cancer and breast cancer [102]. The discovery of lactylation expands the known carcinogenic mechanism of P300/CBP, and P300/CBP is predicted to be an important therapeutic target in the carcinogenic mechanism of lactylation. Research on micromolecules targeting P300 has progressed rapidly, especially those targeting bromodomain and KAT activity. CCS1477, developed by Cell Centric, is the only CBP/P300 inhibitor currently in phase IB/IIA clinical trials; it is a promising treatment for hematologic malignancies and advanced drug-resistant prostate cancer [100]. Although KAT and the acetylation bromodomain reportedly exert general effects on extensive lysine acylation, whether the aforementioned CBP/P300 structural target is effectively modulated through lactylation remains to be confirmed. In 2022, Zhao et al. were the first to identify sirtuins and class I histone HDAC1-3 as delactylases in vitro [33]. As a class of tumor therapeutic targets, HDAC1-3 inhibitors have been researched for 30 years and evaluated in clinical trials, and they have shown efficacy against several tumors [103, 104]. However, these results have been based on acetylation assessments. Considering the crosstalk and similarity between the writers, erasers and biochemical processes between acetylation and lactylation, lactylation may exert the same or stronger effects as acetylation, and thus lactylation-associated proteins may compete with acetylation-associated proteins for the same PTM sites.
Regenerative medicine has gained momentum in recent years [105], and the concept of precision and personalized medicine is guiding the development of promising future therapies [106]. The microenvironment (also called the niche) is a vital factor in many stem cell functions, such as their activation and proliferation, which are related to their metabolism, generalized nutrition and signaling functions [107]. Similar to cancer cells, many normal stem cells show a glycolytic phenotype [108]. Lactate is the core metabolite of glycolysis, with diverse effects on stem cells, as explained above, and some evidence has indicated an effect of lactate on the activation and maturation of stem cells. Previous studies on hair growth attracted our interest in lactate therapy in regenerative medicine because LDHA expression and accompanying lactate production were found to be essential for the activation of HFSCs [85]. LDHA is always related to cell proliferation, and inhibition of LDHA activity is a classic therapeutic strategy for cancer. We must think differently to leverage LDHA expression and promote the proliferation and regeneration of tissues for therapeutic purposes. However, we have mainly focused on the effect of lactylation reported in 2020 [54]. According to existing research, iPSCs are activated by Glis1 through the induction of glycolytic gene expression and subsequent lactate production. Lactate and its subsequent lactylation mediate metabolic reprogramming and epigenetic remodeling. It functions as a ‘switch’ that activates or inactivates pluripotency-associated genes to modulate their expression and regulate the efficiency of somatic cell reprogramming. Thus, lactate and lactylation may be keys to regenerative medicine, and we must inspire reverse thinking to link the abundant lactate-associated research on cancers to research on stem cells.
9 Perspectives
9.1 From lactylation to the proteomics of PTMs
The human genome only contains approximately 20,000 genes, but the abundance of transcripts and the so-called ‘proteoforms’ shaped by PTMs expand the proteome geometrically, making it much larger than the genome; these processes enable cells to participate in many diverse life functions [109] [110]. Other lysine acylation species are difficult to exclude when researching novel nonclassical lysine acylation (nonacetylation) modifications, such as Kla. Therefore, we should expand our horizon to include proteomics to map PTMs of target proteins identified through high-throughput MS and thus further determine the relationships and interactions between different PTMs.
Lysine acetylation (Kac) is a classical, well-studied, multifunctional and nonnegligible lysine acylation. In some lactylation-related research on pluripotent reprogramming [54] and sepsis [59], lactylation and acetylation have been studied in parallel. However, the relationship and potential interaction between Kla and other PTMs, such as Kac, have not been thoroughly discussed. Therefore, we propose the following assumptions (Fig. 2):
-
(1)
Competitive relationship: Generally, only one acyl group is added to a lysine site, indicating that competition exists among various lysine acylation sites. This competitiveness depends on complicated factors, such as the concentration of the substrate, accessibility of lysine residues, and activity level of acylases. In the classic example of ubiquitination and SUMOylation, SUMOylation of nuclear factor κB (NF-κB) inhibits proteasome-mediated degradation mediated by ubiquitin. NF-κB is a major transcriptional activator involved in the immune response and cell survival, and it translocates into the nucleus through its nuclear import signals and the inactivation of IκBα (an inhibitor of NF-κB) to activate transcription. During this process, the ubiquitination of the K21 and K22 sites in IκBα induces its proteasomal degradation. Ubiquitination events also promote the nuclear translocation of NF-κB and transcriptional activation. Nevertheless, the K21 site is a target of SUMOylation; competitive SUMOylation blocks the ubiquitination pathway and thus stabilizes IκBα to inhibit NF-κB pathway activation [111]. We have a certain understanding of important Kac sites, such as K370ac, K372ac, K373aC, K381ac and K382ac of P53 [112], K418ac, K423ac, K1542ac, K1546ac, K1549ac, K1699ac, K1704ac, K1707ac of P300 [113], and K3016ac of ataxia telangiectasia-mutated [114]. Competitive substitution at Kla sites occurs under certain conditions. This change possibly affects the conformation and/or function of previously acetylated proteins to counteract the effect of acetylation or generate some new molecular pathological changes.
-
(2)
Synergetic relationship: Close synergistic relationships have been identified among PTMs.
The best characterized relationship involves ubiquitination and SUMOylation [115]. Both SUMOylation and ubiquitination determine circadian rhythms; the heterodimeric transcription factor CLOCK/BMAL1 controls the expression of circadian CLOCK-related genes. When K259 of BMAL1 was SUMOylated by SUMO2/SUMO3, researchers found that the ubiquitination and degradation of BMAL1 were significantly attenuated. In contrast, the transcriptional activity of circadian regulation-related genes was inhibited by the BMAL1 K259 mutant protein. Overexpression of SUMO protease reduced the SUMOylation and ubiquitination levels of BMAL1, while the ubiquitin protease affected only the ubiquitination of BMAL1. Based on these results, the SUMOylation of BMAL1 is a prerequisite for ubiquitination and that the synergy of these two PTMs mediates the degradation of BMAL1 [116]. In another example of this synergy, researchers studying pluripotent reprogramming [54] found that H3K27ac and H3K18la might exert a synergistic effect on the opening and closing of chromatin and thus affect gene transcription. Nevertheless, they did not explore the mechanism. Hence, Kla and Kac may be localized at the same protein and simultaneously or successively recognize specific molecules and activate the same downstream pathway to induce greater activation or inhibition.
-
(3)
Crosstalk relationship: PTM crosstalk refers to the complementary modulation of the structures, functions, and interactions of proteins undergoing various PTMs. PTM components positively or negatively regulate each other, and various proteins and various pathways may engage in extensive crosstalk. This signaling mechanism was first discovered in studies on phosphorylation and ubiquitination. The ubiquitination activity of the E3 ubiquitin ligase is regulated by phosphorylation [117], and phosphorylation is a potential recognition signal (phosphodegron) [118]. In contrast, the degradation of many kinases is mediated by ubiquitination [119]. In addition to the ubiquitin pathway, PTM crosstalk often involves successive protein modifications; the first modification may affect the subsequent modification. For example, P300 recruitment mediated by P53 promotes H3 acetylation to stabilize P300 itself, and then P300 stabilizes SET1C recruited by P53 to promote H3K4 trimethylation of a promoter [24]. Glycogen synthase kinase 3β promotes the phosphorylation of four amino acid residues upstream of the catalytic site, which generates a binding site for a substrate to ensure that Glycogen synthase kinase 3β phosphorylates the substrate [120]. PTM crosstalk leads to fine-tuned adjustments in a cellular response to slight changes in the environment [121]. Considering traces of Kla, we predict that Kla and other nonclassical lysine acylation species may fine-tune downstream signaling by changing the microstructure of proteins, thereby minimally affecting signaling and alternative pathways to modulate responses and facilitate cellular adjustments to the environment, especially tumor cells to the TME with high levels of lactate and other intermediate products.
9.2 Research focus
The discovery of lactylation provides a new opportunity to study lactate metabolism, but many questions remain unanswered.
Lactylation is considered a metabolic modulation mechanism in which lactate is the main substrate and is related to glycolysis, but the experimental evidence is insufficient. First, we must prove the necessary and sufficient relationship between lactate accumulation and lactylation. According to recent research, this relationship is sufficient to induce lactylation (no counterexample has been found [10] [26] [98]), but evidence showing nonenzymatic lactylation in which lactate is not a substrate has negated the hypothesis that lactate is the only substrate involved in lactylation [38]. Second, we still do not know the biochemical process through which lactyl-CoA is produced from lactate or the metabolic dynamics of lactyl-CoA. In particular, we must identify the transporter and synthase of lactyl-CoA. Studies aiming to identify the related metabolic mechanism of lactyl-CoA are important to understand the upstream modulatory mechanism of lactylation [10]. Lactylation and acetylation have been studied in parallel, and they use the same writer [10] and eraser [33]; therefore, an understanding of the respective contributions of carbon sources, such as glucose, as substrates in acetylation and lactylation dynamics will help us clarify the similarities and differences between the effects of acetylation and lactylation [122].
The most urgent problem to be solved requires a greater understanding of the downstream signaling pathways of lactylation. To date, these pathways have not been thoroughly researched. Moreover, the epigenetic modification known as lactylation that was initially discovered remains controversial, as some researchers believe that lactate and M2 gene expression are uncoupled. No specific connection between lactate and Arg1 expression during inflammatory glycolysis has been identified; therefore, lactylation does not upregulate Arg1 expression, and interleukin-6, a mediator of inflammation, drives Arg1 expression. Interestingly, the upregulation of Arg1 expression promotes nitric oxide production to drive glycolysis and accompanying lactylation [123]. This study provides us with a contrasting view and indicates that lactate is a substrate for lactylation. More research is needed to confirm the true effect of lactylation. Researchers have identified a writer [10] and eraser [33] of lactylation, and these two enzymes are also critical for acetylation. We propose that the identification of lactylases and delactylases will help us explore the mechanism favoring lactylation, which may help us explain the roles of ‘lactylation specific genes’ (namely, explain why histones at some genes undergo lactylation while others do not) through epigenetics [10]. Furthermore, readers initiate downstream signaling in lactylation, and elucidating reader mechanisms is the next step in lactylation research; the identification of domains, such as bromodomain in acetylation, serving as ‘radar’ that ‘sends’ signals of lactylation is desirable. Considering the similarity of writers and erasers involved in lactylation and acetylation, we speculate that the bromodomain is an important reader in lactylation, but the support for this speculation requires further research. Most importantly, we must consider the crosstalk among other lysine acylation processes in the signaling pathway, especially acetylation.
Based on research on acetylation writers (such as KAT), erasers (such as KDAC), readers (such as bromodomain), and large-scale sequencing of acetylation-related genes, we are able to precisely target the mechanism of KAT and KDAC activation/inhibition and identify the target sites by generating point mutations in proteins involved in acetylation. However, our understanding of lactylases, delactylases and the recognition domains in lactylation is limited. Research on the lactylation modulation has been very imprecise and mostly performed at the level of the substrate (lactate) using strategies such as exogenous lactate treatment and LDH activation/inhibition [10] [98]. However, these treatments affect the pathway too far upstream; we cannot exclude the possibility that metabolic bypasses outside the lactylation pathway affect experimental results and may even modulate the effect of therapies targeting lactylation. Hence, the results of many studies remain controversial, and confusion regarding causality persists [123]. If we want to target lactylation to precisely modulate its mechanism, we must map the lactylation atlas and identify specific enzymes and recognition domains to exclude crosstalk with other PTMs; this mapping exercise may help us better understand the mechanism of lactylation.
10 Conclusion
Lactate is always a core metabolite involved in cancer progression. In recent years, an increasing number of studies have been conducted on the effect and carcinogenic mechanism, mainly signaling, of lactate, which functions as a ‘general fuel’ in terms of the energy supply and activates downstream pathways through receptors, such as GPR81. The discovery of lactylation in 2019 revealed a novel mechanism: initial research on histones led to the explanation of lactylation as a metabolic mechanism that modulates the epigenome and plays role in the transformation of the inflammatory phenotype.
In this paper, the discovery process and modification mechanism of lactylation, the methods for site modification and the advances in understanding lactylation of nonhistone proteins are reviewed; the potential roles of lactylation in cancer progression and cell fate determination and the possible therapeutic strategies for targeting lysine lactylation are also discussed.
However, the existing research on lactylation is undeniably still at an initial stage, and many of the aforementioned questions urgently require answers. Lactylation will likely be the next hot topic in lactate research, and this research is expected to deepen and refine our understanding of the relationship between cancer progression and lactate metabolism. Studies exploring lactylation will be very interesting and important, and we look forward to discovering new and effective tumor therapeutic targets through this research.
Data Availability
All data associated with this study are presented in the paper. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- PTM:
-
posttranslational modification
- MS:
-
mass spectrometry
- Kla:
-
lysine lactylation
- BMDM:
-
mouse bone marrow-derived macrophage
- lactyl-CoA:
-
lactyl-coenzyme A
- HDAC:
-
histone deacetylase
- LGSH:
-
lactyl-glutathione
- MGO:
-
methylglyoxal
- GLO:
-
glyoxalase
- GSH:
-
glutathione
- GLO 1&2:
-
glyoxalase I and II
- AI:
-
artificial intelligence
- HMGB1:
-
high mobility group box-1
- TME:
-
tumor microenvironment
- OXPHOS:
-
oxidative phosphorylation
- MCT:
-
monocarboxylate transporter
- TAM:
-
tumor-associated macrophage
- Treg:
-
regulatory T cell
- Kxla:
-
lactylation at the Lys x site (x is a number)
- PD-1:
-
programmed cell death protein 1
- YTHDF2:
-
YTH N6-methyladenosine RNA-binding protein 2
- GPR81:
-
G-protein-coupled receptor 81
- HyKxla:
-
lactylation at the Lys x site of histone y (x and y are numbers)
- iPSCs:
-
pluripotent stem cells
- HFSCs:
-
hair follicle stem cells
- PSC:
-
pluripotent stem cell
- Glis1:
-
Gli-like transcription factor 1
- LDH:
-
lactate dehydrogenase
- LDHA:
-
lactate dehydrogenase A
- CREB:
-
cAMP response element-binding protein
- KAT:
-
lysine acetyltransferase
- CBP:
-
CREB-binding protein
- Kac:
-
lysine acetylation
- NF-κB:
-
nuclear factor κB
- KDAC:
-
lysine deacetylase
References
O. Warburg, Science 123, 309–314 (1956). https://doi.org/10.1126/science.123.3191.309
J.D. Rabinowitz, S. Enerback, Nat. Metab. 2, 566–571 (2020). https://doi.org/10.1038/s42255-020-0243-4
W. Zhang, G. Wang, Z.G. Xu, H. Tu, F. Hu, J. Dai, Y. Chang, Y. Chen, Y. Lu, H. Zeng, Z. Cai, F. Han, C. Xu, G. Jin, L. Sun, B.S. Pan, S.W. Lai, C.C. Hsu, J. Xu, Z.Z. Chen, H.Y. Li, P. Seth, J. Hu, X. Zhang, H. Li, H.K. Lin, Cell 178, 176–189 e115 (2019). https://doi.org/10.1016/j.cell.2019.05.003
J. Roper, O.H. Yilmaz, Cell. Metab. 25, 993–994 (2017). https://doi.org/10.1016/j.cmet.2017.04.019
Y.S. Lee, T.Y. Kim, Y. Kim, S. Kim, S.H. Lee, S.U. Seo, B.O. Zhou, O. Eunju, K.S. Kim, M.N. Kweon, Exp. Mol. Med. 53, 1319–1331 (2021). https://doi.org/10.1038/s12276-021-00667-y
V. Scandella, M. Knobloch, Cell. Stem Cell. 25, 729–731 (2019). https://doi.org/10.1016/j.stem.2019.11.008
T. Narita, B.T. Weinert, C. Choudhary, Nat. Rev. Mol. Cell. Biol. 20, 156–174 (2019). https://doi.org/10.1038/s41580-018-0081-3
V.G. Allfrey, R. Faulkner, A.E. Mirsky, P. Natl. Acad. Sci. 51, 786–794 (1964). https://doi.org/10.1073/pnas.51.5.786
L. Sun, H. Zhang, P. Gao, Protein Cell. 13, 877–919 (2022). https://doi.org/10.1007/s13238-021-00846-7
D. Zhang, Z. Tang, H. Huang, G. Zhou, C. Cui, Y. Weng, W. Liu, S. Kim, S. Lee, M. Perez-Neut, J. Ding, D. Czyz, R. Hu, Z. Ye, M. He, Y.G. Zheng, H.A. Shuman, L. Dai, B. Ren, R.G. Roeder, L. Becker, Y. Zhao, Nature 574, 575–580 (2019). https://doi.org/10.1038/s41586-019-1678-1
J.R. Doherty, J.L. Cleveland, J. Clin. Invest. 123, 3685–3692 (2013). https://doi.org/10.1172/JCI69741
R. Aebersold, J.N. Agar, I.J. Amster, M.S. Baker, C.R. Bertozzi, E.S. Boja, C.E. Costello, B.F. Cravatt, C. Fenselau, B.A. Garcia, Y. Ge, J. Gunawardena, R.C. Hendrickson, P.J. Hergenrother, C.G. Huber, A.R. Ivanov, O.N. Jensen, M.C. Jewett, N.L. Kelleher, L.L. Kiessling, N.J. Krogan, M.R. Larsen, J.A. Loo, R.R. Ogorzalek Loo, E. Lundberg, M.J. MacCoss, P. Mallick, V.K. Mootha, M. Mrksich, T.W. Muir, S.M. Patrie, J.J. Pesavento, S.J. Pitteri, H. Rodriguez, A. Saghatelian, W. Sandoval, H. Schluter, S. Sechi, S.A. Slavoff, L.M. Smith, M.P. Snyder, P.M. Thomas, M. Uhlen, J.E. Van Eyk, M. Vidal, D.R. Walt, F.M. White, E.R. Williams, T. Wohlschlager, V.H. Wysocki, N.A. Yates, N.L. Young and B. Zhang, Nat Chem Biol 14, 206–214 (2018). https://doi.org/10.1038/nchembio.2576
B.R. Sabari, D. Zhang, C.D. Allis, Y. Zhao, Nat. Rev. Mol. Cell. Biol. 18, 90–101 (2017). https://doi.org/10.1038/nrm.2016.140
Z.A. Wang, P.A. Cole, Cell. Chem. Biol. 27, 953–969 (2020). https://doi.org/10.1016/j.chembiol.2020.07.002
Y. Chen, R. Sprung, Y. Tang, H. Ball, B. Sangras, S.C. Kim, J.R. Falck, J. Peng, W. Gu, Y. Zhao, Mol. Cell. Proteomics 6, 812–819 (2007). https://doi.org/10.1074/mcp.M700021-MCP200
A.F. Kebede, A. Nieborak, L.Z. Shahidian, S. Le Gras, F. Richter, D.A. Gomez, M.P. Baltissen, G. Meszaros, H.F. Magliarelli, A. Taudt, R. Margueron, M. Colome-Tatche, R. Ricci, S. Daujat, M. Vermeulen, G. Mittler, R. Schneider, Nat. Struct. Mol. Biol. 24, 1048–1056 (2017). https://doi.org/10.1038/nsmb.3490
Z. Zhang, M. Tan, Z. Xie, L. Dai, Y. Chen, Y. Zhao, Nat. Chem. Biol. 7, 58–63 (2011). https://doi.org/10.1038/nchembio.495
Y. Yang, G.E. Gibson, Neurochem Res. 44, 2346–2359 (2019). https://doi.org/10.1007/s11064-019-02780-x
M. Tan, H. Luo, S. Lee, F. Jin, J.S. Yang, E. Montellier, T. Buchou, Z. Cheng, S. Rousseaux, N. Rajagopal, Z. Lu, Z. Ye, Q. Zhu, J. Wysocka, Y. Ye, S. Khochbin, B. Ren, Y. Zhao, Cell 146, 1016–1028 (2011). https://doi.org/10.1016/j.cell.2011.08.008
J. Wan, H. Liu, J. Chu, H. Zhang, J. Cell. Mol. Med. 23, 7163–7169 (2019). https://doi.org/10.1111/jcmm.14650
L. Dai, C. Peng, E. Montellier, Z. Lu, Y. Chen, H. Ishii, A. Debernardi, T. Buchou, S. Rousseaux, F. Jin, B.R. Sabari, Z. Deng, C.D. Allis, B. Ren, S. Khochbin, Y. Zhao, Nat. Chem. Biol. 10, 365–370 (2014). https://doi.org/10.1038/nchembio.1497
H. Huang, S. Tang, M. Ji, Z. Tang, M. Shimada, X. Liu, S. Qi, J.W. Locasale, R.G. Roeder, Y. Zhao, X. Li, Mol Cell 70, 663–678 e666 (2018). https://doi.org/10.1016/j.molcel.2018.04.011
H. Huang, D. Zhang, Y. Wang, M. Perez-Neut, Z. Han, Y.G. Zheng, Q. Hao, Y. Zhao, Nat. Commun. 9, 3374 (2018). https://doi.org/10.1038/s41467-018-05567-w
Z. Tang, W.Y. Chen, M. Shimada, U.T. Nguyen, J. Kim, X.J. Sun, T. Sengoku, R.K. McGinty, J.P. Fernandez, T.W. Muir, R.G. Roeder, Cell 154, 297–310 (2013). https://doi.org/10.1016/j.cell.2013.06.027
L.T. Izzo, K.E. Wellen, Nature 574, 492–493 (2019). https://doi.org/10.1038/d41586-019-03122-1
G. Notarangelo, M.C. Haigis, Immunity 51, 980–981 (2019). https://doi.org/10.1016/j.immuni.2019.11.008
S. Sokalingam, G. Raghunathan, N. Soundrarajan, S.G. Lee, PLoS One 7, e40410 (2012). https://doi.org/10.1371/journal.pone.0040410
R. Bischoff, H. Schluter, J Proteom. 75, 2275–2296 (2012). https://doi.org/10.1016/j.jprot.2012.01.041
D.J. Slade, V. Subramanian, J. Fuhrmann, P.R. Thompson, Biopolymers 101, 133–143 (2014). https://doi.org/10.1002/bip.22256
C. Azevedo, A. Saiardi, Adv. Biol. Regul. 60, 144–150 (2016). https://doi.org/10.1016/j.jbior.2015.09.008
Z. Kaczmarska, E. Ortega, A. Goudarzi, H. Huang, S. Kim, J.A. Marquez, Y. Zhao, S. Khochbin, D. Panne, Nat. Chem. Biol. 13, 21–29 (2017). https://doi.org/10.1038/nchembio.2217
E.M. Flynn, O.W. Huang, F. Poy, M. Oppikofer, S.F. Bellon, Y. Tang, A.G. Cochran, Structure 23, 1801–1814 (2015). https://doi.org/10.1016/j.str.2015.08.004
C. Moreno-Yruela, D. Zhang, W. Wei, M. Baek, W. Liu, J. Gao, D. Dankova, A.L. Nielsen, J.E. Bolding, L. Yang, S.T. Jameson, J. Wong, C.A. Olsen, Y. Zhao, Sci. Adv. 8, eabi6696 (2022). https://doi.org/10.1126/sciadv.abi6696
E.L. Varner, S. Trefely, D. Bartee, E. von Krusenstiern, L. Izzo, C. Bekeova, R.S. O’Connor, E.L. Seifert, K.E. Wellen, J.L. Meier, N.W. Snyder, Open. Biol. 10, 200187 (2020). https://doi.org/10.1098/rsob.200187
A.M. James, C.L. Smith, A.C. Smith, A.J. Robinson, K. Hoogewijs, M.P. Murphy, Trends Biochem. Sci. 43, 921–932 (2018). https://doi.org/10.1016/j.tibs.2018.07.002
G.R. Wagner, R.M. Payne, J. Biol. Chem. 288, 29036–29045 (2013). https://doi.org/10.1074/jbc.M113.486753
G.R. Wagner, D.P. Bhatt, T.M. O’Connell, J.W. Thompson, L.G. Dubois, D.S. Backos, H. Yang, G.A. Mitchell, O.R. Ilkayeva, R.D. Stevens, P.A. Grimsrud, M.D. Hirschey, Cell Metab 25, 823–837 e828 (2017). https://doi.org/10.1016/j.cmet.2017.03.006
D.O. Gaffney, E.Q. Jennings, C.C. Anderson, J.O. Marentette, T. Shi, A.M. Schou Oxvig, M.D. Streeter, M. Johannsen, D.A. Spiegel, E. Chapman, J.R. Roede, J.J. Galligan, Cell. Chem. Biol. 27, 206–213 e206 (2020). https://doi.org/10.1016/j.chembiol.2019.11.005
X. Jia, T. Chang, T.W. Wilson, L. Wu, PLoS One 7, e36610 (2012). https://doi.org/10.1371/journal.pone.0036610
E.L. Jacobson, P.U. Giacomoni, M.J. Roberts, G.T. Wondrak, M.K. Jacobson, J. Photochem. Photobiol., B 63, 141–147 (2001). https://doi.org/10.1016/s1011-1344(01)00211-1
J.J. Galligan, J.A. Wepy, M.D. Streeter, P.J. Kingsley, M.M. Mitchener, O.R. Wauchope, W.N. Beavers, K.L. Rose, T. Wang, D.A. Spiegel, L.J. Marnett, Proc. Natl. Acad. Sci. U S A 115, 9228–9233 (2018). https://doi.org/10.1073/pnas.1802901115
N. Rabbani, P.J. Thornalley, Kidney Int. 93, 803–813 (2018). https://doi.org/10.1016/j.kint.2017.11.034
C.G. Schalkwijk, C.D.A. Stehouwer, Physiol. Rev. 100, 407–461 (2020). https://doi.org/10.1152/physrev.00001.2019
C. Antognelli, F. Baldracchini, V.N. Talesa, E. Costantini, A. Zucchi, E. Mearini, Cancer J. 12, 222–228 (2006). https://doi.org/10.1097/00130404-200605000-00011
C. Antognelli, V.N. Talesa, Int. J. Mol. Sci. 19, (2018). https://doi.org/10.3390/ijms19020415
A. Luengo, K.L. Abbott, S.M. Davidson, A.M. Hosios, B. Faubert, S.H. Chan, E. Freinkman, L.G. Zacharias, T.P. Mathews, C.B. Clish, R.J. DeBerardinis, C.A. Lewis, and M.G. Vander Heiden, Nat Commun 10, 5604 (2019). https://doi.org/10.1038/s41467-019-13419-4
C.B. Lebrilla, L.K. Mahal, Curr. Opin. Chem. Biol. 13, 373–374 (2009). https://doi.org/10.1016/j.cbpa.2009.08.002
B.M. Zee, B.A. Garcia, Essays Biochem. 52, 147–163 (2012). https://doi.org/10.1042/bse0520147
S.C. Kim, R. Sprung, Y. Chen, Y. Xu, H. Ball, J. Pei, T. Cheng, Y. Kho, H. Xiao, L. Xiao, N.V. Grishin, M. White, X.J. Yang, Y. Zhao, Mol. Cell. 23, 607–618 (2006). https://doi.org/10.1016/j.molcel.2006.06.026
M. Gao, N. Zhang, W. Liang, Front. Microbiol. 11, 594743 (2020). https://doi.org/10.3389/fmicb.2020.594743
P. Jiang, W. Ning, Y. Shi, C. Liu, S. Mo, H. Zhou, K. Liu, Y. Guo, Comput. Struct. Biotechnol. J. 19, 4497–4509 (2021). https://doi.org/10.1016/j.csbj.2021.08.013
Y. Guo, W. Ning, P. Jiang, S. Lin, C. Wang, X. Tan, L. Yao, D. Peng, Y. Xue, Cells 9 (2020). https://doi.org/10.3390/cells9051266
Y. Yang, R. Heffernan, K. Paliwal, J. Lyons, A. Dehzangi, A. Sharma, J. Wang, A. Sattar, Y. Zhou, in Prediction of Protein Secondary Structure, ed. by Y. Zhou, A. Kloczkowski, E. Faraggi and Y. Yang (Springer, 2017), p. 55–63
L. Li, K. Chen, T. Wang, Y. Wu, G. Xing, M. Chen, Z. Hao, C. Zhang, J. Zhang, B. Ma, Z. Liu, H. Yuan, Z. Liu, Q. Long, Y. Zhou, J. Qi, D. Zhao, M. Gao, D. Pei, J. Nie, D. Ye, G. Pan, X. Liu, Nat. Metab. 2, 882–892 (2020). https://doi.org/10.1038/s42255-020-0267-9
J. Ma, S.H. Fong, Y. Luo, C.J. Bakkenist, J.P. Shen, S. Mourragui, L.F.A. Wessels, M. Hafner, R. Sharan, J. Peng, T. Ideker, Nat. Cancer 2, 233–244 (2021). https://doi.org/10.1038/s43018-020-00169-2
Y. Zhang, R. Xie, J. Wang, A. Leier, T.T. Marquez-Lago, T. Akutsu, G.I. Webb, K.C. Chou, J. Song, Brief. Bioinform 20, 2185–2199 (2019). https://doi.org/10.1093/bib/bby079
N. Wan, N. Wang, S. Yu, H. Zhang, S. Tang, D. Wang, W. Lu, H. Li, D.G. Delafield, Y. Kong, X. Wang, C. Shao, L. Lv, G. Wang, R. Tan, N. Wang, H. Hao, H. Ye, Nat. Methods 19, 854–864 (2022). https://doi.org/10.1038/s41592-022-01523-1
X. Wu, W.A. Tao, Nat. Methods 19, 793–794 (2022). https://doi.org/10.1038/s41592-022-01536-w
K. Yang, M. Fan, X. Wang, J. Xu, Y. Wang, F. Tu, P.S. Gill, T. Ha, L. Liu, D.L. Williams, C. Li, Cell. Death Differ. 29, 133–146 (2022). https://doi.org/10.1038/s41418-021-00841-9
L. Ippolito, A. Morandi, E. Giannoni, P. Chiarugi, Trends Biochem. Sci. 44, 153–166 (2019). https://doi.org/10.1016/j.tibs.2018.10.011
D. Hanahan, R.A. Weinberg, Cell 144, 646–674 (2011). https://doi.org/10.1016/j.cell.2011.02.013
T.P. Brown, P. Bhattacharjee, S. Ramachandran, S. Sivaprakasam, B. Ristic, M.O.F. Sikder, V. Ganapathy, Oncogene 39, 3292–3304 (2020). https://doi.org/10.1038/s41388-020-1216-5
N. Raghunand, R.A. Gatenby, R.J. Gillies, Br J Radiol 76 Spec No 1, S11-22 (2003). https://doi.org/10.1259/bjr/12913493
M.G. Vander Heiden, L.C. Cantley, C.B. Thompson, Science 324, 1029–1033 (2009). https://doi.org/10.1126/science.1160809
M.S. Nakazawa, B. Keith, M.C. Simon, Nat. Rev. Cancer 16, 663–673 (2016). https://doi.org/10.1038/nrc.2016.84
C. Corbet, O. Feron, Nat. Rev. Cancer 17, 577–593 (2017). https://doi.org/10.1038/nrc.2017.77
K. Fischer, P. Hoffmann, S. Voelkl, N. Meidenbauer, J. Ammer, M. Edinger, E. Gottfried, S. Schwarz, G. Rothe, S. Hoves, K. Renner, B. Timischl, A. Mackensen, L. Kunz-Schughart, R. Andreesen, S.W. Krause, M. Kreutz, Blood 109, 3812–3819 (2007). https://doi.org/10.1182/blood-2006-07-035972
E. Persi, M. Duran-Frigola, M. Damaghi, W.R. Roush, P. Aloy, J.L. Cleveland, R.J. Gillies, E. Ruppin, Nat. Commun. 9, 2997 (2018). https://doi.org/10.1038/s41467-018-05261-x
T.M. Ashton, W.G. McKenna, L.A. Kunz-Schughart, G.S. Higgins, Clin. Cancer Res. 24, 2482–2490 (2018). https://doi.org/10.1158/1078-0432.CCR-17-3070
S.Y. Lunt, M.G. Vander Heiden, Annu. Rev. Cell. Dev. Biol. 27, 441–464 (2011). https://doi.org/10.1146/annurev-cellbio-092910-154237
M.V. Liberti, J.W. Locasale, Trends Biochem. Sci. 41, 211–218 (2016). https://doi.org/10.1016/j.tibs.2015.12.001
T.P. Brown, V. Ganapathy, Pharmacol. Ther. 206, 107451 (2020). https://doi.org/10.1016/j.pharmthera.2019.107451
S. Pavlides, D. Whitaker-Menezes, R. Castello-Cros, N. Flomenberg, A.K. Witkiewicz, P.G. Frank, M.C. Casimiro, C. Wang, P. Fortina, S. Addya, R.G. Pestell, U.E. Martinez-Outschoorn, F. Sotgia, M.P. Lisanti, Cell. Cycle 8, 3984–4001 (2009). https://doi.org/10.4161/cc.8.23.10238
O.R. Colegio, N.Q. Chu, A.L. Szabo, T. Chu, A.M. Rhebergen, V. Jairam, N. Cyrus, C.E. Brokowski, S.C. Eisenbarth, G.M. Phillips, G.W. Cline, A.J. Phillips, R. Medzhitov, Nature 513, 559–563 (2014). https://doi.org/10.1038/nature13490
M. Certo, C.H. Tsai, V. Pucino, P.C. Ho, C. Mauro, Nat. Rev. Immunol. 21, 151–161 (2021). https://doi.org/10.1038/s41577-020-0406-2
J. Gu, J. Zhou, L. Lu, Ann. Oncol 32, (2021). https://doi.org/10.1016/j.annonc.2021.08.264
J. Yu, P. Chai, M. Xie, S. Ge, J. Ruan, X. Fan, R. Jia, Genome Biol. 22, 85 (2021). https://doi.org/10.1186/s13059-021-02308-z
R. Perez-Tomas, I. Perez-Guillen, Cancers (Basel) 12, (2020). https://doi.org/10.3390/cancers12113244
A.P. Halestrap, M.C. Wilson, IUBMB Life 64, 109–119 (2012). https://doi.org/10.1002/iub.572
V.V. Lao, W.M. Grady, Nat. Rev. Gastroenterol. Hepatol. 8, 686–700 (2011). https://doi.org/10.1038/nrgastro.2011.173
S.J. Hogg, P.A. Beavis, M.A. Dawson, R.W. Johnstone, Nat. Rev. Drug Discov 19, 776–800 (2020). https://doi.org/10.1038/s41573-020-0077-5
E. Gangoso, B. Southgate, L. Bradley, S. Rus, F. Galvez-Cancino, N. McGivern, E. Guc, C.A. Kapourani, A. Byron, K.M. Ferguson, N. Alfazema, G. Morrison, V. Grant, C. Blin, I. Sou, M.A. Marques-Torrejon, L. Conde, S. Parrinello, J. Herrero, S. Beck, S. Brandner, P.M. Brennan, P. Bertone, J.W. Pollard, S.A. Quezada, D. Sproul, M.C. Frame, A. Serrels, S.M. Pollard, Cell 184, 2454–2470 e2426 (2021). https://doi.org/10.1016/j.cell.2021.03.023
S.N. Shapira, H.R. Christofk, Trends Cell. Biol. 30, 566–576 (2020). https://doi.org/10.1016/j.tcb.2020.04.004
J.G. Ryall, T. Cliff, S. Dalton, V. Sartorelli, Cell. Stem Cell. 17, 651–662 (2015). https://doi.org/10.1016/j.stem.2015.11.012
A. Flores, J. Schell, A.S. Krall, D. Jelinek, M. Miranda, M. Grigorian, D. Braas, A.C. White, J.L. Zhou, N.A. Graham, T. Graeber, P. Seth, D. Evseenko, H.A. Coller, J. Rutter, H.R. Christofk, W.E. Lowry, Nat. Cell. Biol. 19, 1017–1026 (2017). https://doi.org/10.1038/ncb3575
Y.S. Lee, T.Y. Kim, Y. Kim, S.H. Lee, S. Kim, S.W. Kang, J.Y. Yang, I.J. Baek, Y.H. Sung, Y.Y. Park, S.W. Hwang, E. O, K.S. Kim, S. Liu, N. Kamada, N. Gao, M.N. Kweon, Cell Host Microbe 24, 833–846 e836 (2018). https://doi.org/10.1016/j.chom.2018.11.002
L. Yang, L. Gao, T. Nickel, J. Yang, J. Zhou, A. Gilbertsen, Z. Geng, C. Johnson, B. Young, C. Henke, G.R. Gourley, J. Zhang, Circ. Res. 121, 1251–1262 (2017). https://doi.org/10.1161/CIRCRESAHA.117.311819
S. Tohyama, F. Hattori, M. Sano, T. Hishiki, Y. Nagahata, T. Matsuura, H. Hashimoto, T. Suzuki, H. Yamashita, Y. Satoh, T. Egashira, T. Seki, N. Muraoka, H. Yamakawa, Y. Ohgino, T. Tanaka, M. Yoichi, S. Yuasa, M. Murata, M. Suematsu, K. Fukuda, Cell. Stem Cell. 12, 127–137 (2013). https://doi.org/10.1016/j.stem.2012.09.013
Q. Tian, L.Q. Zhou, Cells 11, 548 (2022). https://doi.org/10.3390/cells11030548
P.D. Gregory, K. Wagner, W. Horz, Exp. Cell. Res. 265, 195–202 (2001). https://doi.org/10.1006/excr.2001.5187
Y. Feng, Y. Xiong, T. Qiao, X. Li, L. Jia, Y. Han, Cancer Med. 7, 6124–6136 (2018). https://doi.org/10.1002/cam4.1820
G. Lagana, D. Barreca, A. Calderaro, E. Bellocco, Curr. Med. Chem. 26, 3242–3252 (2019). https://doi.org/10.2174/0929867324666170209103444
H. Shim, C. Dolde, B.C. Lewis, C.S. Wu, G. Dang, R.A. Jungmann, R. Dalla-Favera, C.V. Dang, Proc. Natl. Acad. Sci. U S A 94, 6658–6663 (1997). https://doi.org/10.1073/pnas.94.13.6658
C.V. Dang, J.W. Kim, P. Gao, J. Yustein, Nat. Rev. Cancer 8, 51–56 (2008). https://doi.org/10.1038/nrc2274
M.L. Short, D. Huang, D.M. Milkowski, S. Short, K. Kunstman, C.J. Soong, K.C. Chung, R.A. Jungmann, Biochem. J. 304(Pt 2), 391–398 (1994). https://doi.org/10.1042/bj3040391
Y.H. Zhao, M. Zhou, H. Liu, Y. Ding, H.T. Khong, D. Yu, O. Fodstad, M. Tan, Oncogene 28, 3689–3701 (2009). https://doi.org/10.1038/onc.2009.229
G.A. Brooks, Cell. Metab. 27, 757–785 (2018). https://doi.org/10.1016/j.cmet.2018.03.008
H. Hagihara, H. Shoji, H. Otabi, A. Toyoda, K. Katoh, M. Namihira, T. Miyakawa, Cell. Rep. 37, 109820 (2021). https://doi.org/10.1016/j.celrep.2021.109820
R. Eckner, M.E. Ewen, D. Newsome, M. Gerdes, J.A. DeCaprio, J.B. Lawrence, D.M. Livingston, Genes Dev. 8, 869–884 (1994). https://doi.org/10.1101/gad.8.8.869
Z.X. He, B.F. Wei, X. Zhang, Y.P. Gong, L.Y. Ma, W. Zhao, Eur. J. Med. Chem. 209, 112861 (2021). https://doi.org/10.1016/j.ejmech.2020.112861
R. Dutta, B. Tiu, K.M. Sakamoto, Mol. Genet. Metab. 119, 37–43 (2016). https://doi.org/10.1016/j.ymgme.2016.06.013
A.R. Waddell, H. Huang, D. Liao, Cancers (Basel) 13, (2021). https://doi.org/10.3390/cancers13122872
O. Witt, H.E. Deubzer, T. Milde, I. Oehme, Cancer Lett. 277, 8–21 (2009). https://doi.org/10.1016/j.canlet.2008.08.016
T.C.S. Ho, A.H.Y. Chan, A. Ganesan, J. Med. Chem. 63, 12460–12484 (2020). https://doi.org/10.1021/acs.jmedchem.0c00830
Nat. Med. 20, 795–795 (2014). https://doi.org/10.1038/nm.3658
A.L. Lightner, T. Chan, Stem Cell. Res. Ther. 12, 39 (2021). https://doi.org/10.1186/s13287-020-02092-w
H. Clevers, K.M. Loh, R. Nusse, Science 346, 1248012 (2014). https://doi.org/10.1126/science.1248012
J.C. Schell, D.R. Wisidagama, C. Bensard, H. Zhao, P. Wei, J. Tanner, A. Flores, J. Mohlman, L.K. Sorensen, C.S. Earl, K.A. Olson, R. Miao, T.C. Waller, D. Delker, P. Kanth, L. Jiang, R.J. DeBerardinis, M.P. Bronner, D.Y. Li, J.E. Cox, H.R. Christofk, W.E. Lowry, C.S. Thummel, J. Rutter, Nat. Cell. Biol. 19, 1027–1036 (2017). https://doi.org/10.1038/ncb3593
E.M. Holgersen, S. Gandhi, Y. Zhou, J. Kim, B. Vaz, J. Bogojeski, M. Bugno, Z. Shalev, K. Cheung-Ong, J. Goncalves, M. O’Hara, K. Kron, M. Verby, M. Sun, B. Kakaradov, A. Delong, D. Merico, A.G. Deshwar, Nucleic Acid Ther 31, 392–403 (2021). https://doi.org/10.1089/nat.2020.0921
L.M. Smith, N.L. Kelleher, and P. Consortium for Top Down. Nat. Methods 10, 186–187 (2013). https://doi.org/10.1038/nmeth.2369
J.M.P. Desterro, M.S. Rodriguez, R.T. Hay, Mol. Cell 2, 233–239 (1998). https://doi.org/10.1016/s1097-2765(00)80133-1
W. Gu, R.G. Roeder, Cell 90, 595–606 (1997). https://doi.org/10.1016/s0092-8674(00)80521-8
J.C. Black, A. Mosley, T. Kitada, M. Washburn, M. Carey, Mol. Cell 32, 449–455 (2008). https://doi.org/10.1016/j.molcel.2008.09.018
Y. Sun, X. Jiang, S. Chen, N. Fernandes, B.D. Price, Proc. Natl. Acad. Sci. U S A 102, 13182–13187 (2005). https://doi.org/10.1073/pnas.0504211102
J. Schimmel, K.M. Larsen, I. Matic, M. van Hagen, J. Cox, M. Mann, J.S. Andersen, A.C. Vertegaal, Mol. Cell. Proteomics 7, 2107–2122 (2008). https://doi.org/10.1074/mcp.M800025-MCP200
J. Lee, Y. Lee, M.J. Lee, E. Park, S.H. Kang, C.H. Chung, K.H. Lee, K. Kim, Mol. Cell. Biol. 28, 6056–6065 (2008). https://doi.org/10.1128/MCB.00583-08
E. Gallagher, M. Gao, Y.C. Liu, M. Karin, Proc. Natl. Acad. Sci. U S A 103, 1717–1722 (2006). https://doi.org/10.1073/pnas.0510664103
T. Hunter, Mol. Cell. 28, 730–738 (2007). https://doi.org/10.1016/j.molcel.2007.11.019
F. Huang, D. Kirkpatrick, X. Jiang, S. Gygi, A. Sorkin, Mol. Cell. 21, 737–748 (2006). https://doi.org/10.1016/j.molcel.2006.02.018
E. Beurel, S.F. Grieco, R.S. Jope, Pharmacol. Ther. 148, 114–131 (2015). https://doi.org/10.1016/j.pharmthera.2014.11.016
L.D. Vu, K. Gevaert, I. De Smet, Trends Plant. Sci. 23, 1068–1080 (2018). https://doi.org/10.1016/j.tplants.2018.09.004
M.V. Liberti, J.W. Locasale, Trends Biochem. Sci. 45, 179–182 (2020). https://doi.org/10.1016/j.tibs.2019.12.004
S. Dichtl, L. Lindenthal, L. Zeitler, K. Behnke, D. Schlosser, B. Strobl, J. Scheller, K.C. El Kasmi, P.J. Murray, Sci. Adv. 7, (2021). https://doi.org/10.1126/sciadv.abg3505
J. Gu, J. Zhou, Q. Chen, X. Xu, J. Gao, X. Li, Q. Shao, B. Zhou, H. Zhou, S. Wei, Q. Wang, Y. Liang, L. Lu, Cell. Rep. 39, 110986 (2022). https://doi.org/10.1016/j.celrep.2022.110986
D. Yang, J. Yin, L. Shan, X. Yi, W. Zhang, Y. Ding, iScience 25, 104630 (2022). https://doi.org/10.1016/j.isci.2022.104630
X. Meng, J.M. Baine, T. Yan, S. Wang, J. Agric. Food Chem. 69, 8287–8297 (2021). https://doi.org/10.1021/acs.jafc.1c00760
Acknowledgements
We thank Professor Yu Shi, Professor You-Hong Cui, and Professor Liang Yi for their constructive suggestions. We also thank Si-yi Zhang for providing language assistance.
Funding
This study was supported by grants from the National Key Research and Development Program of China (2022YFA1104704 to SCY), Major Projects of the National Natural Science Foundation of China (U22A20325 to SCY), General Projects of the National Natural Science Foundation of China (82273419 to JW), and Major Projects of Technological Innovation and Application Development Foundation in Chongqing (CSTB2022TIAD-STX0012 to SCY).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Jun-han Wang, Ling Mao and Shi-cang Yu contributed significantly to the analyses and manuscript preparation. The first draft of the manuscript was written by Jun-han Wang and Ling Mao. Jun Wang, Ling Mao, Xiao Zhang, Min Wu and Qian Wen participated in the analysis through constructive discussions. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Competing interests
All authors have no conflicts of interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jun-han Wang and Ling Mao contributed equally to this study.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Jh., Mao, L., Wang, J. et al. Beyond metabolic waste: lysine lactylation and its potential roles in cancer progression and cell fate determination. Cell Oncol. 46, 465–480 (2023). https://doi.org/10.1007/s13402-023-00775-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00775-z