Abstract
Purpose
Metastatic lung squamous cell carcinoma (LUSC) is one of the most common causes of cancer death worldwide. As yet, however, the molecular mechanism underlying LUSC metastasis remains elusive. In this study, we report a novel mechanism involving signaling interactions between FGF19 and GLI2 that could drive the progression of LUSC.
Methods
The expression of FGF19 in human LUSC samples was assessed by immunohistochemistry. The concentration of FGF19 in serum samples was assessed by ELISA. RNA sequencing, scratch wound-healing, trans-well, GO analysis, GSEA, luciferase reporter, Western blotting, immunofluorescence and immunohistochemistry assays, as well as an animal model were used to investigate the molecular mechanism underlying FGF19 driven LUSC progression. The therapeutic effect of a GLI2 inhibitor was determined using both in vitro cellular and in vivo animal experiments.
Results
We found that FGF19, a member of the fibroblast growth factor family, plays a crucial role in the invasion and metastasis of LUSC, and identified GLI2 as an important downstream effector of FGF19 involved in metastasis. Surprisingly, we found that FGF19 and GLI2 could reciprocally induce the expression of each other, and form a positive feedback loop to promote LUSC cell invasion and metastasis. These findings were corroborated by an association between a poor prognosis of LUSC patients and FGF19/GLI2 co-expression. In addition, we found that the GLI inhibitor GANT61 could effectively reduce FGF19-mediated LUSC invasion and metastasis.
Conclusion
Our data suggest that FGF19 may serve as a novel biomarker for predicting metastatic LUSC. Intervening with the FGF19-GLI2 feedback loop may be a strategy for the treatment of FGF19-driven LUSC metastasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung squamous cell carcinoma (LUSC) is one of the leading causes of cancer-associated death, with a rising incidence worldwide [1, 2]. Although the mortality rate of LUSC has been decreasing due to improvements in screening and treatment methods, the therapeutic options for metastatic LUSC are still limited. A deeper understanding of the molecular mechanisms underlying LUSC metastasis is critical for the development of better treatment modalities.
Signal feedback loops are essential components of the complex regulatory networks acting in cancer cells, and have been considered to be associated with cancer development, progression and metastasis [3,4,5]. LUSC, as a high incidence solid tumor, has been extensively studied in recent years regarding its signal transduction regulation. But, as yet feedback loop/crosstalk-related reports are still limited. Here, we revealed a novel regulatory feedback loop in LUSC which plays a key role in its progression and metastasis, implicating that intervention through this loop may have therapeutic effects.
The fibroblast growth factor 19 (FGF19) gene is highly conserved and has broad effects in various tissues ranging from fish to human [6, 7]. It has for instance been found that FGF19 regulates various metabolic processes, including gallbladder filling, hepatic lipid storage, glucose homeostasis and bile acid metabolism [8,9,10]. In addition, it has been found to promote the progression of several cancers such as colorectal adenoma, hepatoblastoma, prostate cancer and breast cancer [11,12,13,14]. FGF19 is an endocrine fibroblast growth factor that specifically binds and activates fibroblast growth factor receptor 4 (FGFR4) in conjunction with β-klotho [15, 16]. Increasing evidence indicates that a highly activated FGF19-FGFR4 signaling axis acts as an important driver of lung cancer development. Previously, we and others have shown that FGF19 may regulate multiple LUSC functions in a FGFR4-dependent manner, such as chemotherapy resistance, cell cycle progression and proliferation [17,18,19,20]. Recently, FGF19 has emerged as a key regulator of epithelial-mesenchymal transition (EMT), a critical event in cancer metastasis [21, 22]. FGF19/FGFR4 signaling can e.g. restrict and switch melatonin in head and neck cancer to promote metastasis [23]. These reports suggest that FGF19 may be an important regulatory factor promoting the invasion and metastasis of cancer cells. As yet, however, it remains to be investigated whether the FGF19 signaling pathway regulates LUSC metastasis.
The classical Hedgehog/GLI (HH/GLI) signaling pathway includes ligands (SHH, IHH and DHH), receptors (PTCH 1 and PTCH 2), transcription effectors (GLI1/2/3) and regulatory proteins. Their superior and subordinate relationships constitute complex regulatory cascades [24]. It has been reported that abnormal HH/GLI signaling may be associated with the occurrence of a variety of diseases including cancer [24, 25]. Alterations in this pathway may not only lead to an increase in HH/GLI signal transduction, but may also engage additional subsets of pathway interactions and atypical mechanisms of pathway activation, such as K-Ras and TGFβ activation in pancreatic cancer [26] and loss of SMARCB1 activity in malignant rhabdoid tumors [27] as well as TNFα via mTOR in esophageal cancer [28]. All of these are important contributors to disease progression.
Here, we show that FGF19 may play a crucial role in the invasion and metastasis of LUSC, and that GLI2 may serve as an important downstream effector of FGF19 involved in metastasis. We also found that GLI2 induces increased FGF19 expression in LUSC by elevating its transcription levels, thereby forming a positive feedback loop to facilitate LUSC metastasis. Administration of GANT61, a transcriptional inhibitor of GLI2, was found to effectively disrupt reciprocal FGF19-GLI2 signaling and suppress FGF19-mediated LUSC metastasis. Our results indicate that targeting GLI2 may be a promising option for the treatment of FGF19-high LUSC.
2 Materials and methods
2.1 Cell culture and reagents
SK-MES-1, H520, H1703 and HCC15 cells were purchased from the ATCC and cultured in accordance with the indicated protocols. All human cell lines have been authenticated using STR profiling within the last 3 years. All experiments were performed with mycoplasma-free cells. Recombinant human SHH protein (Peprotech, 100 − 45) was used to activate HH/GLI2. GANT61 (MedChemExpress, HY-13,901) and cyclopamine (TargetMol, T2825) were dissolved in DMSO, and then aliquoted and stored at − 80 °C for in vitro studies.
2.2 Lentivirus transduction and generation of stable cell lines
Lentivirus production was performed as previously described [29]. Tumor cells were transfected with lentivirus in 12-well plates at 40% confluency, after which stably transfected cells were sorted by flow cytometry.
2.3 Tissue microarrays and peripheral blood samples
Tissue microarrays were obtained from commercial sources (Superchip). A total of 5 normal lung samples, 22 primary lung cancer samples and 7 metastatic lung cancer samples were included. In addition, 139 (10 normal and 129 NSCLC) peripheral blood samples were obtained from Shanghai Chest Hospital. All patients were informed of the study and consented to the use of samples for research purposes. The study was approved by the Research Ethics Committee of the School of Biomedical Engineering, Shanghai Jiao Tong University.
2.4 Western blot analysis
Cells were lysed for protein extraction after which Western blotting was performed as previously described [29]. The following antibodies were used in the Western blotting assays: anti-β-Actin (Proteintech, 66009-1-Ig), anti-FGF19 (R&D System, AF969), anti-GLI2 (Santa Cruz Biotechnology, sc-271,786), goat anti-mouse IgG-HRP (Sangon Biotech, D110087), goat anti-rabbit IgG-HRP (Sangon Biotech, D110058) and donkey anti-Goat IgG-HRP (Sangon Biotech, D110115). Other antibodies were obtained from Cell Signaling Technology.
2.5 Enzyme-linked immunosorbent assay (ELISA)
Blood samples were collected using a serum separator tube and allowed to clot for 30 min at room temperature before centrifugation at 1000 g for 15 min. Next, serum samples were collected and stored at − 80 ° C until use. The FGF19 concentration in the serum samples was assessed using an ELISA Kit (R&D SYSTEMS, Catalog No. DF1900) following the manufacturer’s instructions.
2.6 mRNA-seq analysis
Total RNA of stably transfected SK-MES-1 cells was extracted using RNAiso reagent (Takara, 9109), followed by treatment with RNase-free DNase I to remove contaminating genomic DNA. A Qubit® RNA Assay Kit in a Qubit®2.0 Flurometer (Life Technologies, CA, USA) was used to assess the quality and quantity of RNA. Sequencing libraries were prepared using a Hieff NGS™ MaxUp Dual-mode mRNA Library Prep Kit from Illumina® (YEASEN, 12301ES96) following the manufacturer’s instructions. Polymerase chain reaction (PCR) products were purified (AMPure XP system) and the library quality was assessed on an Agilent Bioanalyzer 2100 system. Paired-end sequencing was performed on HiSeq XTen sequencers (Illumina, San Diego, CA, USA). The quality of sequenced data was evaluated using FastQC (version 0.11.2). Raw reads were filtered by Trimmomatic (version 0.36) according to several steps: 1) Remove adaptor sequences if reads contain them, 2) Remove low quality bases from reads 3’ to 5’ (Q < 20), 3) Remove low quality bases from reads 5’ to 3’ (Q < 20), 4) Use a sliding window method to remove base values less than 20 of read tails (window size is 5 bp), 5) Remove reads with read lengths less than 35 nt and its pairing reads. The remaining clean data were used for further analysis. StringTie (version 1.3.3b) was used to calculate the expression values of the transcripts. DESeq2 (version 1.12.4) was used to determine differential gene expression, and genes were considered to be significant differentially expressed when p < 0.001 and |FoldChange| > 2.
2.7 Immunofluorescence (IF) and immunohistochemistry (IHC)
IF and IHC assays were performed as previously described [29]. Stained sections were stored at 4℃ until imaging and Image J software was used for analysis.
2.8 Scratch wound-healing migration assay
SK-MES-1 and H1703 cells were grown to a density of 90% after transfection or drug treatment. Next, they were scratched (‘wounded’) and cultured in fresh medium containing 1% serum (FBS). Photomicrographs were taken at the indicated time intervals after scratching.
2.9 Trans-well migration and invasion assays
SK-MES-1 and H1703 cells in medium supplemented with 1% FBS were seeded into a trans-well chamber (CORNING, 3422) or Matrigel-coated (CORNING, 356,231) membrane trans-well chamber. After incubation, migrated or invaded cells were stained as described before [30]. The stained cells were photographed and three randomly selected fields were counted.
2.10 Luciferase reporter assay
Cells were seeded in 24-well plates (5 × 103 cells/ well) and incubated for 24 h at 37 °C and 5% CO2. Next, the cells were transfected with a luciferase reporter plasmid using Lipofectamine 3000 (Invitrogen, USA). Subsequent luciferase activity was detected after cell lysis using a Dual Luciferase Assay (Promega, USA) according to the manufacturer’s instructions.
2.11 In vivo orthotopic model, metastasis model and bioluminescence imaging (BLI)
An orthotopic lung cancer model was constructed as previously described [18]. Twenty male BALB/c nude mice (5 weeks old) were used to construct the model. Next, the mice were randomly grouped and enrolled into treatment groups. GANT61 (MedChemExpress, HY-13,901), diluted in a 1% (v/v) solution of Tween-80 (Sigma, P1754), was given orally to the mice at 40 mg/kg body weight four times a week. Mouse body weights and survival periods were recorded. After 25 days, a BLI assay was performed. After imaging, lungs were excised and photographed, followed by GLI2 and Ki67 staining. For metastasis models, BALB/c nude mice (6 weeks old) were used. 1 × 106 SK-MES-1 cells, infected with LV-FGF19 or LV-Control were injected into the mice by tail vein injection. All the above animal experiments were approved by the Research Ethics Committee of the School of Biomedical Engineering, Shanghai Jiao Tong University.
2.12 TCGA, Oncomine and Kaplan-Meier plotter data analyses
Relative copy number and mRNA levels of FGF19 and GLI2 of provisional LUSC TCGA samples were downloaded from the Oncomine and cBioPortal databases. Linear regression and Spearman correlation analyses between mRNA levels of FGF19 and GLI2 were conducted. Prognostic values of FGF19 and GLI2 mRNA levels were analyzed by Kaplan-Meier survival curves of NSCLC patients, using the Kaplan-Meier Plotter tool (www.kmplot.com/analysis) [31].
2.13 Gene Set Enrichment Analysis (GSEA)
GSEA was performed using RNA-seq data and cBioPortal database data (TCGA, PanCancer Atlas). Genes were sorted according to the FGF19/GLI2 expression ratio, after which the top 50% and bottom 50% genes were selected for data analysis according to the official user guide of GSEA.
2.14 Statistical analysis
GraphPad Prism 8 software was used for statistical analysis. All data are presented as mean ± SD. Paired or unpaired Student’s t-test or ANOVA were used to assess statistical significances between two groups. P < 0.05 was considered statistically significant.
3 Results
3.1 FGF19 is highly expressed in metastatic lung cancer patients, and its high expression is associated with a worse OS and PFS
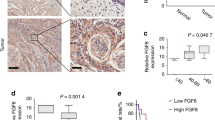
To assess the correlation of FGF19 protein expression in LUSC tissues with patients showing progression, we used LUSC tissue microarrays. Through IHC, we found that FGF19 was hardly expressed in normal lung tissues, but was highly expressed in lung cancer tissues. The FGF19 protein abundance correlated with the disease stage and metastasis status of the lung cancer patients (Fig. 1a-b). Since FGF19 is a secreted protein, detection of its serum level is a meaningful approach to assess its contribution to lung cancer malignancy. We found that higher FGF19 levels in patient sera were significantly associated with advanced lung cancer stages and metastasis (Fig. 1c), and that the level of free FGF19 protein in the sera of patients with metastasis was significantly higher than that of patients without metastasis (Fig. 1c, Supplemental Fig. S1). Next, we used TCGA data for gene set enrichment analysis (GSEA), and found that FGF19 concomitantly upregulated genes were enriched in the metastasis up and EMT cohorts (Fig. 1d). Furthermore, we found that high FGF19 mRNA expression was strongly associated with worse overall survival (OS) and progression-free survival (PFS) rates according to the Kaplan–Meier plotter database (KM plotter, http://kmplot.com/analysis/) (Fig. 1e, Supplemental Fig. S2). Taken together, these data indicate that high FGF19 expression is associated with malignant progression of LUSC.
FGF19 is highly expressed in metastatic LUSC patients, and its high expression is associated with a worse OS and PFS. (a) IHC staining of FGF19 in staged LUSC tissue samples. Scale bar: 200 μm. (b) Quantification of the percentage of FGF19 positive areas after IHC staining of LUSC tissue samples. (c) Comparison of serum FGF19 levels in patients (Serum from 10 normal subjects, 30 patients with stage I or II and 99 patients with stage III or IV; Among all patients, 69 patients with metastasis (M1+), 60 patients without metastasis (M0)). (d) Genes with concomitant elevated FGF19 expression in the LUSC cohort are enriched in metastasis up and EMT signatures. (e) OS and PFS plotted according to the expression level of FGF19 (KM plotter, http://kmplot.com/analysis/. Patients were split according to auto select best cut-off). Data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001
3.2 FGF19 enhances LUSC cell migration, invasion and metastasis
To further examine the role of FGF19 on LUSC progression, we first up-regulated FGF19 in LUSC cell lines (SK-MES-1 and H1703) by lentiviral transfection (Supplemental Fig. S3a). We found that exogenous up-regulation of FGF19 significantly increased the migratory and invasive abilities of both H1703 and SK-MES-1 cells (Fig. 2a, Supplemental Fig. S3b). Using an in vivo lung cancer metastasis model, we found that up-regulation of FGF19 significantly increased the incidence of lung metastases from LUSC cells and resulted in a larger number of intrapulmonary nodules. Relative to the control group, high FGF19 expression led to a rapid formation and progression of metastases, as well as a severe weight loss in mice (Fig. 2b-e). Consistently, using an orthotopic SK-MES-1 LV-FGF19 cell implantation assay, we found that FGF19 overexpression led to an increased number of metastatic nodules formed in multiple body organs and tissues (Fig. 2f-h, Supplemental Fig. S3c), an increased bioluminescence intensity (Fig. 2i) and an increase in pulmonary tumor lesions (Supplemental Fig. S3d), as well as a severe weight loss (Fig. 2j) and a shorter overall survival time (Fig. 2k). Collectively, these data indicate that FGF19 may enhance LUSC cell migration, invasion and metastasis.
FGF19 promotes LUSC cell migration, invasion and metastasis. (a) Migration and invasion of different groups of LUSC cells assessed by scratch wound-healing and trans-well assays. (b-e) In vivo lung cancer metastasis model (1 × 106 cells/50 µl PBS were injected through the tail vein of BALB/c nude mice). (b) Comparison of metastatic lung cancer and representative hematoxylin and eosin stained images of lung specimens from FGF19 overexpression group (LV-FGF19) and control group (LV-NC). (c) Statistics of the number of BALB/c nude mice with lung metastases. (d) Comparison of the number of pulmonary metastatic nodules in each group. (e) Statistics on body weight changes of mice. (f-k) In vivo orthotopic lung cancer model. (f) Comparison of growth of metastatic nodules formed by multiple organs and tissues in each group. (g-h) Representative hematoxylin and eosin stained images and solid tumors of metastatic nodules from each group. (i) Representative bioluminescence images of mice in each group 25 days after model construction. (j) Comparison of body weights and (k) overall survival times of mice in each group. Data are shown as mean ± SD. **p < 0.01; ***p < 0.001; ****p < 0.0001
3.3 FGF19 promotes LUSC cell invasion via GLI2-driven EMT
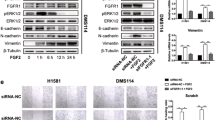
To understand how FGF19 promotes LUSC cell migration, invasion and metastasis, we examined transcriptome levels in FGF19 overexpressing LUSC (SK-MES-1 LV-FGF19) and control (SK-MES-1 LV-NC) cells by RNA-seq, and compared the differences between the two. Through GO enrichment analysis and GSEA, we found that FGF19 upregulation significantly promoted cell motility and migration, and that this was associated with EMT (Fig. 3a-b). We further explored the regulatory effect of FGF19 on the expression of key genes associated with EMT. Using immunoblotting, we found that FGF19 upregulation increased SNAIL, SLUG, ZEB1, N-cadherin and MMP3 expression, while decreasing E-cadherin and CLDN1 expression, illustrating the tumor promoting effect of FGF19 on EMT (Fig. 3c).
FGF19 is involved in EMT. (a-b) RNA-seq analysis of SK-MES-1 LV-FGF19 versus SK-MES-1 LV-Control cells. (a) GO enrichment indicates that upregulation of FGF19 significantly facilitates LUSC cell motility and migration. (b) GSEA analysis showing metastasis up and EMT signatures according to FGF19 expression levels in LUSC cells. (c) Expression of EMT-associated proteins in LUSC cells after treatment for different durations with recombinant human FGF19 or in FGF19 stable expressing cells and control cells
As an extracellular factor, FGF19 does not directly affect the expression of EMT-related genes. Therefore, we next explored which factors could mediate the regulation of EMT-related genes by FGF19. Using KEGG analysis, we found that the Hedgehog signaling pathway, which is known to be involved in cancer-related pathways, was significantly affected by FGF19 expression in LUSC, and also that GLI2 may play an important regulatory role in it (Fig. 4a-b). GLI2 is known to be in volved in promoting cancer metastasis [32,33,34] and, therefore, we hypothesized that GLI2 may also be involved in FGF19-driven LUSC metastasis.
FGF19 promotes LUSC cell invasion via GLI2-driven EMT. (a) KEGG analysis of cancer-related pathways enriched in samples with high FGF19 expression in the LUSC cohort. (b) Heat maps generated from the TCGA LUSC patient dataset. Heatmap showing Hedgehog pathway gene expression profiles, with GL2 being the most differentially expressed gene. (c) CNA of FGF19 and GLI2 are common in LUSC Weiss Lung and TCGA Lung 2 cohorts. Expression of FGF19 and GLI2 in the Oncomine database with LUSC cohorts in the Hou Lung dataset. (d) Linear regression and Pearson correlation analysis of data from TCGA’s LUSC cohort indicate that FGF19 positively correlates with GLI2 mRNA expression. (e) SK-MES-1 and H520 cells were treated with recombinant human FGF19 (25 ng/ml), after which GLI2 expression was found to increase in a time-dependent manner. (f) Western blotting and (g) immunofluorescence assays were used to detect GLI2 expression in LUSC cell lines overexpressing FGF19 (GFP). (h) Genes concomitant with elevated GLI2 expression in the LUSC cohort are enriched in metastasis up and EMT signature maps. (i-j) Expression of EMT-associated proteins in (i) GLI2 overexpressing LUSC cells and control cells, and (j) in cells treated with FGF19, FGF19 and GANT61, or DMSO. (k) OS and PFS plotted according to the expression level of GLI2. Data are shown as mean ± SD and compared by unpaired t-test. **p < 0.01, ***p < 0.001 and ****p < 0.0001
To gain further insight into the relationship between FGF19 and GLI2, we first analyzed gene copy number and mRNA expression profiles of these two genes in a LUSC cohort using the Oncomine database. Compared with normal lung tissue, we found that the gene copy numbers of both FGF19 and GLI2 were significantly increased in LUSC tissues in the Weiss Lung dataset and in the TCGA Lung2 dataset (Fig. 4c). Concordantly, we found that the mRNA expression levels of both FGF19 and GLI2 were increased in the LUSC cohort of the Hou Lung dataset (Fig. 4c). Moreover, we found that these two genes showed a significant co-expression (Fig. 4d). We also found that rhFGF19-treated LUSC cells exhibited significantly upregulated GLI2 (Fig. 4e) and SMO (Supplemental Fig. S4) expression levels, classic mediators of the Hedgehog pathway. Moreover, FGF19-induced GLI2 expression could be rescued by the FGFR4 inhibitor BLU9931, indicating that FGFR4 may mediate the regulation of GLI2 by FGF19 (Supplemental Fig. S5). Additionally, we observed a higher GLI2 expression in SK-MES-1 LV-FGF19 cells compared to control SK-MES-1 LV-NC cells (Fig. 4f-g).
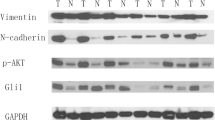
Subsequently, we set out to determine the potential role of GLI2 in LUSC cell metastasis. GSEA plots for GLI2 in LUSC revealed that metastasis up and EMT cohorts were enriched in GLI2 up genes (Fig. 4 h), which was consistent with the enrichment analysis for FGF19. Therefore, we speculated that GLI2 may also have a regulatory effect on EMT-related genes in LUSC. Using Western blotting we found that the up-regulation of GLI2 enhanced the expression of SLUG, N-cadherin and MMP3, and inhibited the expression of E-cadherin, ZO-1 and CLDN1 (Fig. 4i). Furthermore, we found that the GLI2 inhibitor GANT61 significantly inhibited the up-regulation of EMT-related proteins by FGF19 (Fig. 4j). We next analyzed the effect of GLI2 expression on patient survival using Kaplan-Meier plots and found that higher GLI2 levels were associated with shorter OS and PFS rates (Fig. 4k) compared to lower GLI2 levels. Collectively, these findings suggest that FGF19 may promote LUSC cell invasion via GLI2-driven EMT.
3.4 GLI2 is involved in transcriptional regulation of FGF19, and FGF19 and GLI2 co-expression positively correlates with a poor prognosis
Previously it has been reported that FGF15 (expressed in Mus musculus, orthologous to human FGF19) expression requires sonic hedgehog signaling mediated by the transcription factor GLI2 in the medial region of the diencephalon and midbrain [35], which encouraged us to further investigate the relationship between GLI2 and FGF19. To this end, we first activated the sonic hedgehog signaling pathway by the ligand SHH. Using Western blotting we found that the FGF19 protein level in LUSC cells was significantly increased after 24 h of treatment and that this process could be prevented by the GLI2 inhibitor GANT61 (Fig. 5a) and the SMO inhibitor cyclopamine (Supplemental Fig. S6). Exogenous addition of recombinant SHH could also significantly upregulate the activity of a FGF19 promoter-controlled luciferase reporter gene in cells (Fig. 5b). Moreover, we found that the expression of FGF19 in H520 and HCC15 cells (with a high endogenous expression of FGF19) was significantly decreased after GANT61 treatment (Fig. 5c). These finding indicate that activation of the sonic hedgehog pathway may promote the expression of FGF19.
GLI2 is involved in transcriptional regulation of FGF19, and the expression of FGF19 and GLI2 positively correlates with a poor prognosis. (a) The HH/GLI2 signaling pathway is activated by recombinant human SHH (50 ng/ml, 24 h) and inhibited by GANT61 (10 µM, 24 h). Expression of FGF19 and GLI2 in LUSC cells detected by Western blotting. (b) Relative luciferase activity assays performed on cells transfected with plasmids containing the FGF19 promoter after 24 h of SHH treatment. (c) Protein level of FGF19 evaluated by Western blotting after cells (with high endogenous expression of FGF19) were treated with GANT61 (10 µM, 24 h). (d) FGF19 protein expression in GLI2-overexpressing cells and control cells. (e) Prediction of GLI2 binding motif and five potential GLI2-specific nucleotide-binding sites in the promoter region of the FGF19 gene using the JASPAR database. (f) Relative activity of luciferase measured in LUSC cells co-transfected with GF19 promoter and GLI2 expression constructs. (g) High FGF19 and high GLI2 mRNA levels are associated with a shorter OS and PFS in patients, evaluated by Kaplan–Meier Plotter (https://kmplot.com/analysis/). The total mRNA level is presented as the sum of FGF19 and GLI2 mRNA levels at a 1:1 ratio. Data are shown as mean ± SD and compared by unpaired t-test. **p < 0.01, ***p < 0.001 and ****p < 0.0001
Next, we explored whether GLI2 may play a critical role in this regulatory relationship. Using Western blotting we found that up-regulation of GLI2 expression significantly promoted the expression of FGF19 (Fig. 5d). To next investigate whether GLI2 acts as a direct transcription factor of FGF19 expression, we analyzed the promoter sequence of FGF19 and detected, through sequence alignment, five putative GLI2 binding motifs (Fig. 5e, Supplemental Fig. S7). Consistent with this, we found that overexpression of GLI2 led to increased luciferase activity using a reporter construct containing the FGF19 promoter (Fig. 5f). In addition, higher FGF19 and GLI2 mRNA levels (patients split by median values) were found to be associated with shorter OS and PFS rates (Fig. 5 g). In summary, these data indicate that GLI2 may promote FGF19 expression and that their co-expression is associated with a poor prognosis.
3.5 The GLI2 inhibitor GANT61 suppresses FGF19-mediated LUSC migration, invasion and metastasis
Above, we found that a reciprocal FGF19-GLI2 loop may promote LUSC invasion and metastasis by regulating EMT. Next, we set out to explore whether the GLI2 inhibitor GANT61 can block this positive feedback loop and affect FGF19-mediated LUSC migration, invasion and metastasis. We found that GANT61 (10 µM/ml) significantly inhibited the migration and invasion of LUSC cells with a high FGF19 expression using both scratch wound-healing and trans-well experiments (Fig. 6a-b). In addition, we used an in vivo orthotopic lung cancer model to further determine the inhibitory effect of GANT61 (50 mg/kg, orally administered, 4 days on/3 days off) on the metastasis of FGF19 overexpressing LUSC cells. We found that GANT61 could significantly inhibit the growth and metastasis of FGF19 overexpressing LUSC cells in mice (Fig. 6c-g). Since the malignancy of the cancer cells decreased, the weight loss of the mice was alleviated (Fig. 6 h) and the survival time was prolonged (Fig. 6i). Subsequent IHC staining confirmed that the expression levels of GLI2 and Ki67 in lung tumor lesions were significantly decreased upon GANT61 treatment compared with the control group (Fig. 6j). These results suggest that GANT61 can effectively inhibit FGF19-driven LUSC migration, invasion and metastasis by disrupting the reciprocal FGF19-GLI2 loop.
GANT61 inhibits LUSC cell metastasis in vivo. (a) Migration and (b) invasion of GANT61 cells and DMSO treated cells assessed by scratch wound-healing and trans-well assays. (c-j) Evaluation of GANT61 efficacy using an orthotopic lung cancer model. (c) Experimental timeline for the orthotopic lung cancer model. (d) Bioluminescence images of BALB/c nude mice of the GANT61 and control groups after 25 days of model construction. (e) Comparison of metastatic nodules from each group. Tumors are indicated by arrows. (f) Statistics of the number of BALB/c nude mice with lung metastases. Comparisons of (g) lung tumors, (h) body weights and (i) overall survival times of mice in each group. (j) IHC staining of GLI2 and Ki67 in tumor sections. Scale bar: 200 μm. Data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, calculated by two-way ANOVA test. Scale bar: 50 μm. H&E, hematoxylin and eosin. (k) Schematic diagram of this work: A potential mechanism by which the reciprocal FGF19-GLI2 signaling pathway is involved in LUSC metastasis. A reciprocal FGF19-GLI2 circuit promotes LUSC metastasis via EMT. The use of GLI2 inhibitors effectively disrupts the FGF19-GLI2 loop and inhibits LUSC metastasis
4 Discussion
We found that a high FGF19 expression is positively associated with advanced stages in LUSC, and significantly promotes the emergence of more aggressive LUSC phenotypes and the occurrence of metastases. FGF19 was identified as a novel biomarker for predicting the malignant progression and metastasis of LUSC, and GLI2 was identified as an important downstream effector of FGF19-induced metastasis. In addition, we found that GLI2 could induce FGF19 expression in LUSC cells by elevating its transcription level, thereby forming a positive feedback loop to facilitate LUSC metastasis. Administration of GANT61, a transcriptional inhibitor of GLI2, effectively disrupted reciprocal FGF19-GLI2 signaling and suppressed FGF19-mediated LUSC metastasis.
Previously, we identified FGF19 as a prognostic marker and potential driver gene in patients with lung squamous cell carcinoma by sequencing clinical samples [36], and found that abnormally activated FGF19/FGFR4 signaling drives LUSC progression [17]. Through association analysis of key genes, we elucidated the oncogenic role of FGF19 in LUSC and provided insight into how co-amplification of its neighboring gene CCND1 synergistically functioned to promote cancer growth [18]. Here we show, by further exploring the molecular mechanism of FGF19 in LUSC, that FGF19 is also involved in the emergence of the LUSC metastatic phenotype. We found that FGF19 expression is closely related to the metastatic phenotype in patients by IHC staining of LUSC tissues, the detection of FGF19 serum levels and the analysis of TCGA data. We identified a novel FGF19 downstream gene, GLI2, which is also involved in LUSC metastasis. GLI2, a transcription factor belonging to the Gli-Kruppel family [37], is known to be one of the mediators in Hedgehog signaling [38]. Hedgehog/GLI2 signaling has been reported to play an important role in enhancing hepatic carcinogenesis, chemo-resistance, metastasis and EMT [39]. Previously, it has also been found that increased GLI2 stabilization may promote breast cancer cell anoikis resistance and metastasis [32] and that GLI2 overexpression may result in enhanced metastasis and progression of osteosarcomas [33]. Concordantly, aberrant activation of GLI2 has been found to be crucial for the migration and invasion ovarian cancer cells [34]. These studies suggest that GLI2 may act as an intrinsic activation driver that contributes to the development of metastases in multiple cancer types. We found that GLI2, as an important downstream effector of FGF19, is involved in LUSC metastasis. Overexpression of FGF19 induced the expression of GLI2 which, in turn, increased the expression of FGF19 and formed a positive feedback loop to promote LUSC metastasis.
Signal feedback regulation is known to underlie robust dynamics and phenotype switching in complex regulatory networks and to dictate cell fate and phenotype changes [40]. Feedback-loop signal regulation has a dual role in cancer progression [41]. Positive feedback regulation leads to mutual regulation of upstream and downstream key molecules to form a signal amplification cascade, while negative feedback regulation manifests itself as mutual inhibition of expression after signal receival of upstream and downstream molecules. These regulatory feedback mechanisms can promote cancer development by inducing cell differentiation, drug resistance, proliferation and metastasis, but the effects may also be opposite, depending on the key molecules involved in feedback regulation. Detailed analysis of regulatory mechanisms underlying feedback loops may be used to target cancer key signals in order to achieve better therapeutic benefits. Along this line, therapeutic strategies aimed at targeting the FGF19-GLI2 reciprocal loop may be helpful in the treatment of LUSC. Strategies for targeting FGF19-GLI2 positive feedback may include the use FGF19 neutralizing antibodies, FGFR4 tyrosine kinase inhibitors or GLI2 inhibitors. Among these strategies, targeting GLI2 stands out for its feasibility, as it is the most straightforward one. Here, we show that GANT61 can potently reduce FGF19-induced LUSC cell invasion and metastasis by inhibiting EMT, implicating the effectiveness of interrupting a key node of the FGF19-GLI2 reciprocal loop to repress LUSC metastasis.
There are still some questions that remain to be addressed. For example, while directly targeting GLI2 to disrupt the FGF19-GLI2 feedback loop may have promising therapeutic effects, we did not compare this option with other strategies like targeting FGF19 or the FGF19-specific receptor FGFR4, or dual targeting of FGF19 and GLI2. It is worth mentioning that previously we found that FGFR4 inhibitors alone failed to achieve a significant tumor inhibitory effect, but that combined targeting of FGF19-FGFR4 and CCND1-CDK4/6 achieved significant therapeutic effects [18]. It has been reported that CCND1 is one of the downstream effector molecules of GLI2 [42, 43]. This may be an important reason for the better effect of co-targeting. We previously reported that FGF19 expression in LUSC was closely associated with smoking and endoplasmic reticulum stress in patients [17]. Endoplasmic reticulum stress has been reported to activate Sonic Hedgehog signaling [44, 45]. Therefore, GLI2 may be a link to explain ER stress-induced FGF19 upregulation, but this suggestion requires further investigation.
In summary, we report the involvement of FGF19 in LUSC invasion and metastatis, and that this process is dependent on a FGF19-GLI2 reciprocal loop. We propose the use of GANT61 as a therapeutic option based on the finding that GANT61 could effectively disrupt the FGF19-GLI2 loop and significantly inhibit the occurrence of EMT, thereby inhibiting LUSC migration, invasion and metastasis. This feedback loop may provide further insight into the malignant progression of advanced LUSC and provide a rationale for the development of future anticancer drugs (Fig. 6k).
Data availability
The RNA-seq data generated in this study are available in GEO under accession number GSE207588. Other data that support the findings of this study are available from the corresponding author upon request.
References
F. Bray, J. Ferlay, I. Soerjomataram, R.L. Siegel, L.A. Torre, A. Jemal, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424 (2018)
R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020)
C. Cheng, D. Liu, Z. Liu, M. Li, Y. Wang, B. Sun, R. Kong, H. Chen, G. Wang, L. Li, J. Hu, Y. Li, H. Chen, Z. Zhao, T. Zhang, S. Zhu, S. Pan, Positive feedback regulation of lncRNA TPT1-AS1 and ITGB3 promotes cell growth and metastasis in pancreatic cancer. Cancer Sci. 113(9), 2986–3001 (2022)
M. Ghomlaghi, A. Hart, N. Hoang, S. Shin, L.K. Nguyen, Feedback, crosstalk and competition: ingredients for Emergent Non-Linear Behaviour in the PI3K/mTOR Signalling Network. Int. J. Mol. Sci. 22(13), 6944 (2021)
S. Zhao, Y. Mi, B. Zheng, P. Wei, Y. Gu, Z. Zhang, Y. Xu, S. Cai, X. Li, D. Li, Highly-metastatic colorectal cancer cell released miR-181a-5p-rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J. Extracell. Vesicles 11, e12186 (2022)
M. Katoh, M. Katoh, Evolutionary conservation of CCND1-ORAOV1-FGF19-FGF4 locus from zebrafish to human. Int. J. Mol. Med. 12, 45–50 (2003)
B. Angelin, T.E. Larsson, M. Rudling, Circulating fibroblast growth factors as metabolic regulators–a critical appraisal. Cell. Metab. 16, 693–705 (2012)
T. Inagaki, M. Choi, A. Moschetta, L. Peng, C.L. Cummins, J.G. McDonald, G. Luo, S.A. Jones, B. Goodwin, J.A. Richardson, R.D. Gerard, J.J. Repa, D.J. Mangelsdorf, S.A. Kliewer, Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell. Metab. 2, 217–225 (2005)
M. Choi, A. Moschetta, A.L. Bookout, L. Peng, M. Umetani, S.R. Holmstrom, K. Suino-Powell, H.E. Xu, J.A. Richardson, R.D. Gerard, D.J. Mangelsdorf, S.A. Kliewer, Identification of a hormonal basis for gallbladder filling. Nat. Med. 12, 1253–1255 (2006)
E. Somm, F.R. Jornayvaz, Fibroblast growth factor 15/19: from Basic Functions to therapeutic perspectives. Endocr. Rev. 39, 960–989 (2018)
H.I. Sussman, Apical or cervical? J. Am. Dent. Assoc. 118, 684 (1989)
X. Zhao, F. Xu, N.P. Dominguez, Y. Xiong, Z. Xiong, H. Peng, C. Shay, Y. Teng, FGFR4 provides the conduit to facilitate FGF19 signaling in breast cancer progression. Mol. Carcinog. 57, 1616–1625 (2018)
H. Nagamatsu, J. Teishima, K. Goto, H. Shikuma, H. Kitano, K. Shoji, S. Inoue, A. Matsubara, FGF19 promotes progression of prostate cancer. Prostate 75, 1092–1101 (2015)
D. Wang, J. Zhang, Z. Li, J. Han, Y. Gao, M. Chen, Y. Li, Upregulation of fibroblast growth factor 19 is Associated with the initiation of colorectal adenoma. Dig. Dis. 37, 214–225 (2019)
Y. Liu, M. Cao, Y. Cai, X. Li, C. Zhao, R. Cui, Dissecting the role of the FGF19-FGFR4 signaling pathway in Cancer Development and Progression. Front. Cell. Dev. Biol. 8, 95 (2020)
H.K. Ho, S. Pok, S. Streit, J.E. Ruhe, S. Hart, K.S. Lim, H.L. Loo, M.O. Aung, S.G. Lim, A. Ullrich, Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J. Hepatol. 50, 118–127 (2009)
F. Li, Z. Li, Q. Han, Y. Cheng, W. Ji, Y. Yang, S. Lu, W. Xia, Enhanced autocrine FGF19/FGFR4 signaling drives the progression of lung squamous cell carcinoma, which responds to mTOR inhibitor AZD2104. Oncogene 39, 3507–3521 (2020)
Y. Zhang, T. Wu, F. Li, Y. Cheng, Q. Han, X. Lu, S. Lu, W. Xia, FGF19 is Coamplified with CCND1 to promote proliferation in lung squamous cell carcinoma and their combined inhibition shows improved efficacy. Front. Oncol. 12, 846744 (2022)
X. Zhang, M. Kong, Z. Zhang, S. Xu, F. Yan, L. Wei, J. Zhou, FGF19 genetic amplification as a potential therapeutic target in lung squamous cell carcinomas. Thorac. Cancer 8, 655–665 (2017)
J. Chen, J. Shao, A. Shen, X. Zhu, X. Zhang, H. Sun, S. Wei, Y. Ling, Enhanced expression of FGF19 predicts poor prognosis in patients with non-small cell lung cancer. J. Thorac. Dis. 13, 1769–1784 (2021)
H. Zhao, F. Lv, G. Liang, X. Huang, G. Wu, W. Zhang, L. Yu, L. Shi, Y. Teng, FGF19 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells by modulating the GSK3beta/beta- catenin signaling cascade via FGFR4 activation. Oncotarget 7, 13575–13586 (2016)
L. Gao, X. Wang, Y. Tang, S. Huang, C.A. Hu, Y. Teng, FGF19/FGFR4 signaling contributes to the resistance of hepatocellular carcinoma to sorafenib. J. Exp. Clin. Cancer Res. 36, 8 (2017)
L. Lang, Y. Xiong, N. Prieto-Dominguez, R. Loveless, C. Jensen, C. Shay, Y. Teng, FGF19/FGFR4 signaling axis confines and switches the role of melatonin in head and neck cancer metastasis. J. Exp. Clin. Cancer Res. 40, 93 (2021)
A.N. Sigafoos, B.D. Paradise, M.E. Fernandez-Zapico, Hedgehog/GLI signaling pathway: transduction, regulation, and implications for Disease. Cancers (Basel) 13(14), 3410 (2021)
E. Peer, S. Tesanovic, F. Aberger, Next-generation Hedgehog/GLI pathway inhibitors for Cancer Therapy. Cancers (Basel) 11(4), 538 (2019)
O. Nolan-Stevaux, J. Lau, M.L. Truitt, G.C. Chu, M. Hebrok, M.E. Fernandez-Zapico, D. Hanahan, GLI1 is regulated through smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 23, 24–36 (2009)
Z. Jagani, E.L. Mora-Blanco, C.G. Sansam, E.S. McKenna, B. Wilson, D. Chen, J. Klekota, P. Tamayo, P.T. Nguyen, M. Tolstorukov, P.J. Park, Y.J. Cho, K. Hsiao, S. Buonamici, S.L. Pomeroy, J.P. Mesirov, H. Ruffner, T. Bouwmeester, S.J. Luchansky, J. Murtie, J.F. Kelleher, M. Warmuth, W.R. Sellers, C.W. Roberts, M. Dorsch, Loss of the tumor suppressor Snf5 leads to aberrant activation of the hedgehog-gli pathway. Nat. Med. 16, 1429–1433 (2010)
Y. Wang, Q. Ding, C.J. Yen, W. Xia, J.G. Izzo, J.Y. Lang, C.W. Li, J.L. Hsu, S.A. Miller, X. Wang, D.F. Lee, J.M. Hsu, L. Huo, A.M. Labaff, D. Liu, T.H. Huang, C.C. Lai, F.J. Tsai, W.C. Chang, C.H. Chen, T.T. Wu, N.S. Buttar, K.K. Wang, Y. Wu, H. Wang, J. Ajani, M.C. Hung, The crosstalk of mTOR/S6K1 and hedgehog pathways. Cancer Cell. 21, 374–387 (2012)
W. Ji, Y. Yu, Z. Li, G. Wang, F. Li, W. Xia, S. Lu, FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the hedgehog pathway. Oncotarget 7, 15118–15134 (2016)
L. Wang, H. Yang, Z. Lei, J. Zhao, Y. Chen, P. Chen, C. Li, Y. Zeng, Z. Liu, X. Liu, H.T. Zhang, Repression of TIF1gamma by SOX2 promotes TGF-beta-induced epithelial-mesenchymal transition in non-small-cell lung cancer. Oncogene 35, 867–877 (2016)
B. Gyorffy, P. Surowiak, J. Budczies, A. Lanczky, Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8, e82241 (2013)
P. Gupta, N. Gupta, N.M. Fofaria, A. Ranjan, S.K. Srivastava, HER2-mediated GLI2 stabilization promotes anoikis resistance and metastasis of breast cancer cells. Cancer Lett. 442, 68–81 (2019)
H. Nagao-Kitamoto, M. Nagata, S. Nagano, S. Kitamoto, Y. Ishidou, T. Yamamoto, S. Nakamura, A. Tsuru, M. Abematsu, Y. Fujimoto, M. Yokouchi, S. Kitajima, T. Yoshioka, S. Maeda, S. Yonezawa, S. Komiya, T. Setoguchi, GLI2 is a novel therapeutic target for metastasis of osteosarcoma. Int. J. Cancer 136, 1276–1284 (2015)
H. Zhang, Y. Wang, T. Chen, Y. Zhang, R. Xu, W. Wang, M. Cheng, Q. Chen, Aberrant activation of hedgehog signalling promotes cell migration and invasion via matrix metalloproteinase-7 in ovarian cancer cells. J. Cancer 10, 990–1003 (2019)
H. Saitsu, M. Komada, M. Suzuki, R. Nakayama, J. Motoyama, K. Shiota, M. Ishibashi, Expression of the mouse Fgf15 gene is directly initiated by sonic hedgehog signaling in the diencephalon and midbrain. Dev. Dyn. 232, 282–292 (2005)
Q. Tan, F. Li, G. Wang, W. Xia, Z. Li, X. Niu, W. Ji, H. Yuan, Q. Xu, Q. Luo, J. Zhang, S. Lu, Identification of FGF19 as a prognostic marker and potential driver gene of lung squamous cell carcinomas in chinese smoking patients. Oncotarget 7, 18394–18402 (2016)
D. Javelaud, V.I. Alexaki, S. Dennler, K.S. Mohammad, T.A. Guise, A. Mauviel, TGF-beta/SMAD/GLI2 signaling axis in cancer progression and metastasis. Cancer Res. 71, 5606–5610 (2011)
Y. Katoh, M. Katoh, Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int. J. Mol. Med. 22, 271–275 (2008)
X. Chen, S. Lingala, S. Khoobyari, J. Nolta, M.A. Zern, J. Wu, Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol. 55, 838–845 (2011)
B.N. Kholodenko, J.F. Hancock, W. Kolch, Signalling ballet in space and time. Nat. Rev. Mol. Cell. Biol. 11, 414–426 (2010)
W. Gu, H. Shen, L. Xie, X. Zhang, J. Yang, The role of feedback loops in targeted therapy for pancreatic cancer. Front. Oncol. 12, 800140 (2022)
E.J. Tolosa, M.G. Fernandez-Barrena, E. Iguchi, A.L. McCleary-Wheeler, R.M. Carr, L.L. Almada, L.F. Flores, R.E. Vera, G.W. Alfonse, D.L. Marks, T.L. Hogenson, A.M. Vrabel, I.P. Horn, A.N. Koenig, S.L. Safgren, A.N. Sigafoos, M. Erkan, P.A. Romecin-Duran, A. Sarabia Gonzalez, B. Zhou, D. Javelaud, V. Marsaud, R.P. Graham, A. Mauviel, S.F. Elsawa, M.E. Fernandez-Zapico, GLI1/GLI2 functional interplay is required to control Hedgehog/GLI targets gene expression. Biochem. J. 477, 3131–3145 (2020)
Y. Katoh, M. Katoh, Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 9, 873–886 (2009)
X. Yang, W. Sun, X. Jing, Q. Zhang, H. Huang, Z. Xu, C/EBP homologous protein promotes Sonic hedgehog secretion from type II alveolar epithelial cells and activates hedgehog signaling pathway of fibroblast in pulmonary fibrosis. Respir Res. 23, 86 (2022)
A. Saito, S. Kanemoto, Y. Zhang, R. Asada, K. Hino, K. Imaizumi, Chondrocyte proliferation regulated by secreted luminal domain of ER stress transducer BBF2H7/CREB3L2. Mol. Cell. 53, 127–139 (2014)
Acknowledgements
The authors thank to all those who financed the study.
Funding
This work was supported by grants from the Science and Technology Commission of Shanghai Municipality (21ZR1433100), the State Key Laboratory of Oncogenes and Related Genes (KF2122) and SJTU funding (YG2022ZD016).
Author information
Authors and Affiliations
Contributions
Conception and design: Y.Z. and W.X.; Data acquisition and analysis: Y.Z., T.W., Y.W., Z.C., J.C. and W.X.; writing and original draft preparation: Y.Z., T.W. and W.X.; critical review and editing: Y.Z., T.W. and W.X.; studies related to clinical samples: Y.W., S.L.; supervision: W.X.; funding acquisition: W.X. All authors have seen and approved the final draft of the manuscript before submission. The work reported has been performed by the authors, unless clearly specified in the text.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This work was approved by the Research Ethics Committee of the School of Biomedical Engineering, Shanghai Jiao Tong University (Shanghai, China). All patients were informed of the study and consented to the use of samples for research purposes. All animal experiments were performed following the regulations and internal biosafety and bioethics guidelines of the Research Ethics Committee of the School of Biomedical Engineering, Shanghai Jiao Tong University (Shanghai, China).
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Wu, T., Wang, Y. et al. Reciprocal FGF19-GLI2 signaling induces epithelial-to-mesenchymal transition to promote lung squamous cell carcinoma metastasis. Cell Oncol. 46, 437–450 (2023). https://doi.org/10.1007/s13402-022-00760-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-022-00760-y