Abstact
Background
Non-small cell lung cancer (NSCLC) patients who do initially respond to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) may eventually develop resistance, which may at least partly be due to the acquisition of a secondary EGFR mutation (T790M). Additionally, it has been found that KRAS mutations may serve as poor prognostic biomarkers. Here, we aimed at establishing a suitable treatment regimen for the multi-target TKI sunitinib and the tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) in NSCLC-derived cells with or without EGFR and KRAS mutations.
Methods
Four NSCLC-derived cell lines with or without EGFR and KRAS mutations were exposed to different sunitinib and TRAIL treatment regimens. Alterations in cell viability, cell cycle distribution, apoptosis, phosphorylation of AKT and expression of the death receptors DR4 and DR5 were evaluated using CCK8, flow cytometry and Western blotting assays, respectively.
Results
A synergistic cytotoxic effect was observed in all four cell lines treated with sunitinib (1 nM) followed by TRAIL (100 ng/ml), as well as after simultaneous treatment with both agents. We found that sunitinib enhances TRAIL-induced G0/G1-phase cell cycle arrest and blocks TRAIL-triggered activation of AKT as the underlying mechanism. In contrast, we observed antagonistic effects when sunitinib was administered after TRAIL to the cell lines tested. A decreased DR4 and DR5 expression was found to be correlated with this antagonism.
Conclusion
From our data we conclude that administration of sunitinib followed by TRAIL, as well as a simultaneous administration of both agents, serve as favorable treatment regimens for NSCLC-derived cells, irrespective of their EGFR and/or KRAS mutation status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Until recently, the major treatment option for advanced non-small cell lung cancer (NSCLC) was platinum-based doublet chemotherapy. This treatment option has, however, resulted in only a limited increase in overall survival [1]. Currently, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) are applied to the treatment of NSCLC, and it has been found that a higher benefit from these TKIs can be obtained in patients harboring mutations in EGFR exons 18, 19 or 21 than in patients that are wild-type for the EGFR gene [2, 3]. Nevertheless, patients who do initially react to TKI-based therapy may eventually develop resistance, which may at least partly be attributed to the acquisition of a secondary mutation (T790M) in the EGFR gene [4, 5]. Additionally, it has been found that KRAS mutations occur in about 30 % of the NSCLC cases and that these mutations serve as poor prognostic biomarkers [6]. Clearly, efforts aimed at improving the survival rates of TKI-resistant patients are urgently needed.
Agents that simultaneously inhibit multiple targets may prevent the occurrence of TKI resistance [7]. Sunitinib, a multi-target TKI of the vascular endothelial growth factor receptors (VEGFR) -1, −2 and −3 and the platelet-derived growth factor receptors (PDGFR)-a and -b, is such an agent and it may exhibit both cytotoxic and anti-angiogenic effects in different cancers, depending on which of the receptors is inhibited [8]. Regardless the presence of KRAS mutations or the occurrence of a secondary T790M EGFR mutation in NSCLC cells, sunitinib may effectively induce lethality in these cells. Ongoing studies are aimed at determining how multi-target TKIs such as sunitinib can be applied, either alone or in combination with conventional chemotherapy regimens, to obtain better clinical outcomes in patients with NSCLC [9].
Additionally, sequential therapies are currently being explored as a promising approach to improve the outcome of patients with advanced cancers. The tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is emerging as a promising anticancer agent due to its selective cytotoxicity towards various types of cancer cells [10]. The majority of NSCLC cells is, however, resistant to TRAIL [11], implying that TRAIL by itself may be inappropriate for the treatment of NSCLC. TRAIL resistance is thought to result from the deregulation of cellular signaling pathways through ectopic up-regulation of the death receptors DR4 and DR5 and the phosphorylation of AKT [12]. Traditional chemotherapeutic agents may, however, enhance the lethality of TRAIL, which may subsequently result in the killing of TRAIL-resistant cells [13]. Associations between order-dependent reciprocities of the multi-target TKI sunitinib and TRAIL in NSCLC cells with diverse mutational statuses of the EGFR or KRAS genes have so far not been reported. Here, we assessed whether and how sunitinib may enhance TRAIL-triggered killing of NSCLC cells that either or not harbor EGFR or KRAS mutations.
2 Materials and methods
2.1 Cell cultures and reagents
The human lung cancer-derived cell lines H1299, A549, PC9 and H1975 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10 % fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) at 37 °C in an incubator with a humidified atmosphere containing 5 % CO2. Recombinant human TRAIL proteins were purchased from Abcam Corporation (Cambridge, MA, USA) and sunitinib (SU11248; Sutent™) was obtained from Pfizer (Pfizer Research, New York, NY, USA).
2.2 Cell viability assay
Cell viability was assessed by a cell counting Kit-8 (CCK8) assay (Dojindo, Kumamoto, Japan). To this end, cells were seeded in 96-well plates, cultured overnight and, subsequently, treated with sunitinib and/or TRAIL at various concentrations (0, 0.01, 0.1, 1, 2, 4 nM and 0, 1, 10, 100, 500, 1000 ng/ml, respectively). Twenty-four hours later, 10 μl CCK-8 solution was added to each well. After 4 h of incubation, the optical density (OD) of each well was determined at 450 nm. The IC50 value, indicating the concentration resulting in 50 % inhibition of the maximal cell growth, was determined from corresponding dose-response curves as reported before [14]. This assay was independently repeated in three experiments.
2.3 Schedule-dependent drug treatment
Cell viabilities were evaluated after three different schedules of sunitinib and TRAIL treatment. In the first schedule, cells were concurrently treated with sunitinib and TRAIL for 48 h. In the second schedule, cells were pretreated with sunitinib for 24 h, followed by a washout with PBS and an additional exposure to TRAIL for 24 h. In the third schedule, a reverse sequence of TRAIL and sunitinib treatment was performed. In the single drug tests, the minimum drug concentrations that exhibited significant differences compared to the control were recorded. In the combination drug tests, the concentration of each drug was based on the minimum drug concentration recorded in the corresponding single drug test. The ratios of the minimum concentrations were used to establish the combination drug concentrations. The combination drug concentrations were calculated using 0.25, 0.5, 1 and 2 times the minimum concentration of each drug. The combination index (CI) values of interactions between sunitinib and TRAIL were calculated according to the Chou -Talalay equation using the CompuSyn software package (ComboSyn, Inc., Paramus, NJ, USA). CI values <1, 1 and >1 indicate synergism, additive effect, and antagonism, respectively [15].
2.4 Cell cycle analyses
1 × 105 cells/well were seeded in 6-well plates and, subsequently, treated with sunitinib and TRAIL as single agents and in different sequence combinations at concentrations calculated as described above. At the end of each exposure the cells were collected and washed with PBS, after which the cells were fixed in 70 % ethanol and stored at 4 °C overnight. DNA staining was performed using a solution containing propidium iodide (50 μg/ml) and RNase (100 μg/ml) for 30 min at room temperature. Next, the cells were analyzed by flow cytometry using a BD FACS Calibur (BD, CA, USA), and the resulting DNA histograms were quantified using the Modifit software package (Verity Software House, Turramurra, New South Wales, Australia).
2.5 Apoptosis assays
For apoptosis analysis by flow cytometry, Annexin V assays were performed using an Annexin V: FITC Apoptosis Detection Kit II (BD, CA, USA). Briefly, cells were treated with sunitinib and TRAIL as single agents and in different sequence combinations at concentrations as described above. After a 20 h incubation, the cells were harvested, washed twice in ice-cold PBS and stained with 5 μl Annexin V-FITC and 10 μl propidium iodide (50 μg/ml) for 15 min at room temperature in the dark. The population of Annexin V-positive cells was evaluated by flow cytometry (FACS Calibur, BD, CA, USA) using 488 nm laser excitation.
2.6 Antibodies and Western blotting
Anti-AKT, anti-phospho-AKT, anti-DR4, anti-DR5 and anti-GAPDH antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Goat anti-mouse and goat anti-rabbit IgG were purchased from Cell Signaling Technology (Beverly, MA, USA). Cells (5 × 105/well) were treated with sunitinib and TRAIL as single agents and in different sequence combinations during the desired time. Next, the cells were washed in ice-cold PBS and scraped off in lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 1 % Triton X-100, 0.5 % sodium deoxycholate, 0.1 % SDS and one Complete Mini Protease Inhibitor Cocktail tablet (Roche, Indianapolis, IN, USA). The resulting lysates were centrifuged at 12,000×g for 30 min at 4 °C after which the supernatants were collected. Then total proteins were extracted and protein concentrations were determined by Bradford assay. 100 μg protein from each sample was loaded on a 12 % sodium dodecyl sulfate-polyacrylamide gel and subjected to electrophoresis (SDS-PAGE). The separated proteins were transferred to PVDF membranes (IPVH00010, Millipore), after which the membranes were blocked with 5 % nonfat milk in TBS-T buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05 % Tween 20) and probed with the above-mentioned primary antibodies. Subsequently, the blots were incubated with horseradish-conjugated secondary antibodies and protein bands were detected using a SuperSignal West Pico chemiluminescence substrate (Pierce, Rockford, IL, USA). The optical densities of the bands were determined using the LabWork software package (UVP Laboratory Products, CA, USA). All experiments were repeated at least three times.
2.7 Statistical analyses
The results are expressed as means with standard deviation (SD). In order to assess statistically significant differences between the studied variables, we performed one-way analysis of variance (ANOVA). Tukey’s test was used for post-hoc analyses. P values < 0.05 were defined as statistically significant.
3 Results
3.1 Schedule-dependent anti-proliferative activity of sunitinib and TRAIL in EGFR-dependent and EGFR-independent NSCLC cells
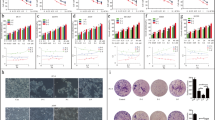
CCK8 analyses were carried out to assess the cytotoxicity of sunitinib and TRAIL as sole drugs and in three different sequential combinations on EGFR-independent H1299 (wild-type EGFR/wild-type KRAS), A549 (wild-type EGFR/mutant G12S KRAS) and H1975 (mutant T790M + L858R EGFR/wild-type KRAS) cells and EGFR-dependent PC9 (mutant delE746-A750 EGFR/wild-type KRAS) cells. The respective IC50 values obtained after sunitinib and TRAIL treatment are listed in Table 1. We observed a difference in sunitinib sensitivity between PC9 and the other three cell lines, whereas only minor differences were observed in TRAIL responsiveness between all four cell lines. The respective cell viabilities are depicted in Fig. 1a and b.
Effects of sunitinib and TRAIL on the proliferation of NSCLC-derived cells in vitro. Cells were treated with the indicated concentrations of sunitinib a or TRAIL b alone for 24 h, after which the proliferation of the cells was determined by CCK8 assay. Each data point results from three independent measurements. c–f Administration of sunitinib and TRAIL in three different sequences reveals sequence-dependent anti-proliferative effects in NSCLC-derived cells. The cells were incubated in three sequential combinations of suntinib and TRAIL at a constant ratio. The concentrations applied were 0–2 nM for sunitinib and 0–200 ng/ml for TRAIL. The proliferation of the cells was determined by CCK8 assay. *significantly lower than that observed in TRAIL → sunitinib combinatorial treatments; P < 0.05. g–j Combination index (CI) values of each drug fraction calculated using the Chou-Talalay method in g H1299, h A549, i H1975 and j PC9 cells following the different exposure sequences. Cross S + T, plus S-T, and circle T-S refer to concurrent administration of sunitinib and TRAIL, sunitinib followed by TRAIL, and TRAIL followed by sunitinib, respectively. In the four cell lines a CI < 1 was detected at every drug concentration with the sequence of sunitinib plus TRAIL and sunitinib followed by TRAIL
Next, we evaluated schedule-dependent interactions of sunitinib and TRAIL in three different sequential administration regimens. In order to explore the cytotoxicity of sunitinib and TRAIL when administered concurrently, the above-mentioned NSCLC-derived cells were treated with 1 nM sunitinib and 100 ng/ml TRAIL for 48 h. We found that this combination induced a significant reduction in viable cells compared to the untreated controls (P < 0.05) (Fig. 1c–f). A synergistic effect was seen in all four cell lines tested (CI values <1) (Fig. 1g–j) (see additional data in a supplementary table). Similar synergistic results were seen when the cells were treated with sunitinib alone for 24 h and subsequently with TRAIL alone for 24 h (CI values <1) (Fig. 1c–j) (see additional data in a supplementary table). However, a reversed combination of TRAIL followed by sunitinib resulted in antagonistic effects (CI values >1) (Fig. 1c–j) (see additional data in a supplementary table). These results indicate that stronger anti-proliferative effects of TRAIL are achieved through a schedule-dependent combination with sunitinib, regardless the EGFR and/or KRAS mutation statuses of the cells.
3.2 Schedule-dependent apoptosis induction by sunitinib and TRAIL in EGFR-dependent and EGFR-independent NSCLC cells
Next, we set out to assess whether the reduced viabilities observed in the different NSCLC-derived cell lines subjected to single, sequential or concurrent exposures of 1 nM sunitinib and 100 ng/ml TRAIL were due to apoptosis. Through flow cytometry analyses, we found that TRAIL exerted its anti-proliferative activity mainly via apoptosis in the H1299, A549 and H1975 cells (Fig. 2a–c), but not in the PC9 cells (Fig. 2d), whereas exposure to sunitinib rarely resulted in apoptosis in the different cells tested (Fig. 2a–d), indicating that apoptosis is not the main cause of sunitinib-induced cytotoxicity. However, the concurrent administration of sunitinib and TRAIL was found to lead to a more profound apoptotic rate than single-drug administration in all four cell lines. Similar synergistic interactions were also observed when TRAIL was administered after sunitinib. In contrast, converse effects were seen when TRAIL was administered before sunitinib (Fig. 2). Therefore, we conclude that exposure to sunitinib followed by TRAIL, as well as their concurrent administration, exerts synergistic effects, whereas a reversed sequence of TRAIL followed by sunitinib exerts antagonistic effects in all four cell lines tested. These results appear to be independent of the EGFR and KRAS mutation statuses of the cells, i.e., in all cases the sequential administration of sunitinib followed by TRAIL, as well as the concurrent administration of both drugs, was found to be superior to the administration of TRAIL followed by sunitinib.
Sunitinib enhances TRAIL-triggered apoptosis in a schedule-dependent manner in EGFR-dependent and EGFR-independent NSCLC-derived cells. H1299 (a), A549 (b), H1975 (c) and PC9 (d) cells were treated with sunitinib (1 nM), TRAIL (100 ng/ml), or three different sunitinib-TRAIL sequences for 48 h. Untreated cells served as controls. Apoptosis was evaluated using Annexin V-FITC and PI staining in conjunction with flow cytometry. Total apoptosis was defined as early apoptosis plus late apoptosis (i.e., Annexin V-positive cell population). The values are presented as mean ± SD of three independent experiments. *Sunitinib + TRAIL and sunitinib → TRAIL groups relative to TRAIL alone group, P < 0.05. **Sunitinib + TRAIL and sunitinib → TRAIL groups relative to TRAIL → sunitinib group, P < 0.01 using one-way ANOVA + Tukey’s test
3.3 Sunitinib enhances TRAIL-induced G0/G1-phase cell cycle arrest in a schedule-dependent manner
As a next step, we set out to assess the cell cycle distributions of the different NSCLC-derived cell lines subjected to single, sequential or concurrent exposures to sunitinib and TRAIL. We found that the H1299, A549 and H1975 cells, which are insensitive to EGFR TKIs, underwent a G0/G1 arrest when treated with TRAIL alone (P < 0.05) (Fig. 3a–c), whereas the PC9 cells, which are sensitive to EGFR TKIs, remained unaffected by TRAIL (Fig. 3d). We also found that sunitinib treatment resulted in a considerable increase of cells in the G0/G1-phase of the cell cycle in both EGFR-independent H1299 and A549 and EGFR-dependent PC9 cells (Fig. 3a, b, d), but not in EGFR-independent H1975 cells (Fig. 3c). However, administration of sunitinib concurrently or before TRAIL resulted in a clear increase in cells in the G0/G1-phase of the cell cycle, whereas sunitinib administered after TRAIL only marginally affected cell cycle progression compared to TRAIL or sunitinib administration alone in all four cell lines tested (Fig. 3a-d). These results, which are consistent with the above apoptosis results, indicate that a schedule-dependent combination of sunitinib and TRAIL most likely induces apoptosis through a G0/G1-phase cell cycle arrest.
Flow cytometric analysis to determine alterations in cell cycle distributions in NSCLC-derived cell lines. Cells were treated with vehicle control, 1 nM sunitinib, 100 ng/ml TRAIL, both agents concurrently, sunitinib followed by TRAIL, or TRAIL followed by sunitinib for 48 h. Columns in the diagram depict the cell cycle phase distributions in H1299 (a), A549 (b), H1975 (c) and PC9 (d) cells following the exposure schedules as stated above. Results are presented as means ± SD of three independent experiments
3.4 Sunitinib blocks TRAIL-triggered activation of AKT in EGFR-dependent and EGFR-independent NSCLC cells
As a downstream signaling pathway of EGFR, the PI3K/AKT axis plays a crucial role in conferring growth, metastasis and chemoresistance capacities to NSCLC cells [16, 17]. To dissect whether AKT activity affects TRAIL sensitivity, we assessed the phosphorylation status of AKT after single, sequential and concurrent administration of sunitinib and TRAIL in the NSCLC-derived cells. Through Western blot analyses, we found that 1 nM sunitinib treatment significantly increased the levels of phosphorylated AKT in the H1975 and PC9 cells (Fig. 4c, d), but not in the H1299 and A549 cells (Fig. 4a, b), as compared to controls. Besides, we found that in all four cell lines the levels of phosphorylated AKT significantly increased when the cells were exposed to TRAIL alone as compared to untreated cells (Fig. 4a–d). This increase in phosphorylated AKT was, however, abrogated when sunitinib was added. A significant down-regulation of phosphorylated AKT was observed when sunitinib was added before or simultaneously with TRAIL in all cell lines tested (Fig. 4a–d). In contrast, when TRAIL was added before sunitinib, we found that the phosphorylated AKT levels were not down-regulated (Fig. 4a–d). In contrast to the controls, no overt changes in the total AKT levels were observed. Collectively, these results suggest that sunitinib may enhance TRAIL-induced apoptosis through blocking the activation of the AKT survival pathway. It should be noted that, in order to reach this result, the treatment sequence is important, i.e., sunitinib should be administered before TRAIL.
Effects of sunitinib and TRAIL as single agents and in different exposure combinations on the expression of phosphorylated AKT/AKT, DR4 and DR5 in H1299 (a), A549 (b), H1975 (c) and PC9 (d) cells. Cells were exposed to vehicle control, 1 nM sunitinib, 100 ng/ml TRAIL, both agents concurrently, sunitinib followed by TRAIL, or TRAIL followed by sunitinib for 48 h. The proteins were detected by Western blotting using the corresponding antibodies. C, S, T, S + T, S-T and T-S refer to control, sunitinib, TRAIL, concurrent administration of both drugs, sunitinib followed by TRAIL and TRAIL followed by sunitinib, respectively. The figures are representative of three independent experiments
3.5 Expression of DR4 and DR5 is involved in the schedule-dependent synergistic effects of sunitinib and TRAIL
The expression of TRAIL death receptors is essential for the transmission of death signals from their ligands. Western blotting was applied to explore the effects of sunitinib and TRAIL on the expression of the death receptors DR4 and DR5 when NSCLC-derived cells were exposed to these agents in different sequences. As shown in Fig. 4a–d, a considerable decrease in DR4 and DR5 expression was noted when the cells were exposed to TRAIL alone in the four cell lines tested, as well as after exposure to sunitinib alone in the EGFR-dependent PC9 cells (Fig. 4d) and the EGFR-independent H1975 cells (Fig. 4c). Besides, a sequential treatment of TRAIL followed by sunitinib elicited a significant down-regulation of DR4 expression compared to TRAIL treatment alone in the H1975 and PC9 cells (Fig. 4c, d), and almost the same levels of DR5 expression compared to single TRAIL treatment in all cell lines tested (Fig. 4a–d). However, no reduction in DR4 and DR5 expression was observed when sunitinib was administered before or simultaneously with TRAIL. Consistent with the apoptosis induction observed above, these results indicate that the sequential administration of sunitinib followed by TRAIL, or the concurrent administration of sunitinib and TRAIL, blocks the TRAIL-induced down-regulation of DR4 and DR5. The reverse administration, i.e., TRAIL followed by sunitinib, does not block the TRAIL-induced down-regulation of DR4 and DR5.
4 Discussion
Although platinum-based chemotherapy has in the past improved the survival of patients with advanced NSCLC, it has clearly reached its limits [18, 19]. In order to further improve survival, recent studies have focused on the design of targeted and apoptosis-inducing therapies, which may more selectively induce cancer cell death while minimizing the damage to normal tissues. Here, we explored the putative synergistic effects of sunitinib and TRAIL in NSCLC-derived H1299, A549 and H1975 cells harboring EGFR-independent mutations and in NSCLC-derived PC9 cells harboring an EGFR dependent mutation. In addition, we assessed whether the sensitivity to sunitinib and TRAIL was associated with the EGFR and/or KRAS mutation statuses of the respective cells. Finally, we set out to define the optimal schedule of sunitinib and TRAIL treatment in EGFR-dependent and EGFR-independent NSCLC-derived cells.
Proliferation, apoptosis and cell cycle analyses were performed to assess schedule-dependent synergistic interactions between sunitinib and TRAIL. In a single-agent test, PC9 cells harboring an exon19delE746-A750 EGFR-dependent mutation (wild-type KRAS) were found to be most sensitive to sunitinib in the proliferation assay. This marked efficacy of sunitinib was not observed in NSCLC-derived cells harboring EGFR-independent mutations. In contrast, only minor differences were found in cancer cells with EGFR-dependent and EGFR-independent mutations with respect to the anti-proliferative activity of TRAIL. These diversities in cytotoxicity may at least partly be due to other, as yet unknown, (epi)genetic features of the cells. We did find, however, that the schedule-dependent synergy of both drugs does not rely on the mutation statuses of the EGFR and KRAS genes. The sequence-dependent administration of sunitinib followed by TRAIL, as well as the simultaneous administration of both agents, was found to act synergistically with respect to cytostatic activity and apoptosis-inducing effect in all NSCLC-derived cell lines tested, irrespective of their EGFR and KRAS mutation status. In contrast, we observed drug antagonism after TRAIL treatment followed by sunitinib treatment in all NSCLC-derived cell lines tested.
Currently, putative synergistic effects between TRAIL and different TKIs have been addressed in only a handful preclinical studies. Shrader et al. showed that gefitinib interacts with TRAIL to cause high levels of apoptosis through XIAP expression induction, which is mediated by AKT in EGFR-dependent bladder cancer cells [20]. Rosato et al. found that sorafinib enhances TRAIL-induced apoptosis via down-regulation of Mcl-1 and cFLIPL expression [21]. However, even though Ding et al. assessed interactions between sunitinib and TRAIL in SW620 and 95-D cells and showed a synergistic effect of both drugs [22], no sequence-related cytotoxicity of these drugs was noted and the mechanism underlying its synergism has so far remained unclear. We found that changes in cell cycle distribution and growth signaling pathways may explain the observed effects. Our data showed that TRAIL causes an increase in G0/G1-phase cell cycle arrest concurrently with or after sunitinib administration in the four cell lines tested. Conversely, we found that sunitinib failed to cause any G0/G1-phase arrest after TRAIL treatment followed by S-phase entry, which is crucial for cellular proliferation. Therefore, we conclude that sunitinib may promote TRAIL-induced apoptosis via G0/G1-phase cell cycle arrest and that this synergy is sequence-dependent.
AKT activation is known to contribute to cancer development and metastasis [23] and, in addition, to therapy resistance [24, 25]. In the present study, we found that aberrant activation of AKT is involved in the acquisition of TRAIL resistance in the four NSCLC-derived cell lines tested, irrespective of their EGFR and KRAS mutation status. Conventional chemotherapy in conjunction with AKT kinase inhibition has been found to delay chemotherapy resistance. Zhang et al. found that sunitinib can reverse multidrug resistance of cisplatin-resistant A549 cells by inhibition of AKT phosphorylation [26]. Our data show that sunitinib can lead to decreased AKT activation and enhanced cytotoxicity when administered concurrently with or before TRAIL, not only in EGFR-dependent but also in EGFR-independent NSCLC-derived cells. Thus, sunitinib may suppress TRAIL-induced activation of AKT, thereby inhibiting cell survival and promoting apoptosis.
TRAIL appears to bind the death receptors DR4 and DR5 to enhance the killing of cancer cells. Some antitumor agents require DR4 and/or DR5 to render cancer cells more sensitive to TRAIL [27, 28]. Absence of DR4/DR5 on the cell surface results in a loss of the capacity to generate a death-inducing signaling complex which, consequently, may attenuate TRAIL-triggered apoptosis in cancer-derived cells [29, 30]. Although we have not noted any up-regulation of DR4 and/or DR5, we found that DR4 and DR5 are down-regulated in all four cell lines tested after exposure to TRAIL alone and to TRAIL followed by sunitinib, in accordance with the above described cytotoxic effects and apoptosis induction.
Genetic heterogeneity, such as the occurrence of different mutations in the EGFR and KRAS genes, has frequently been observed in patients with advanced NSCLC. Previous studies have indicated that patients with EGFR exon 19 and exon 21 mutations may show a better response to single-targeted EGFR inhibitors than those wild-type for the EGFR gene [31, 32]. Some Phase III clinical trials have shown, however, that the response rates to gefitinib and erlotinib were typically ~10 %. These trials were conducted in non-selected populations, thus indicating that the majority of patients failed to respond to these drugs [33]. By comparison, patients with EGFR-independent mutations and KRAS mutations have commonly been found to not benefit from single-targeted EGFR TKIs. Therefore, the sequential administration of the multi-target TKI sunitinib and TRAIL as explored in our current study may represent a useful alternative, even for NSCLC patients harboring different EGFR and/or KRAS mutations.
In conclusion, we show here for the first time that sunitinib followed by TRAIL treatment, as well as the concurrent administration of both agents, may serve as an efficacious treatment option for NSCLC patients, independent of the mutation statuses of the EGFR and KRAS genes.
References
N. Watanabe, S. Niho, K. Kirita, S. Umemura, S. Matsumoto, K. Yoh, H. Ohmatsu, K. Goto, Vinorelbine and cisplatin in patients with advanced non-small cell lung cancer with interstitial pneumonia. Anticancer Res. 35, 1697–1701(2015)
Y. Togashi, H. Hayashi, K. Okamoto, S. Fumita, M. Terashima, M. A. de Velasco, K. Sakai, Y. Fujita, S. Tomida, K. Nakagawa, K. Nishio, Chronic nicotine exposure mediates resistance to EGFR-TKI in EGFR-mutated lung cancer via an EGFR signal. Lung Cancer 88, 16–23 (2015)
S. N. Bichev, D. M. Marinova, Y. G. Slavova, A. S. Savov, Epidermal growth factor receptor mutations in East European non-small cell lung cancer patients. Cell. Oncol. 38, 145–153 (2015)
F. Nurwidya, F. Takahashi, A. Murakami, I. Kobayashi, M. Kato, T. Shukuya, K. Tajima, N. Shimada, K. Takahashi, Acquired resistance of non-small cell lung cancer to epidermal growth factor receptor tyrosine kinase inhibitors. Respir. Investig. 52, 82–91 (2014)
K. J. Gotink, M. Rovithi, R. R. de Haas, R. J. Honeywell, H. Dekker, D. Poel, K. Azijli, G. J. Peters, H. J. Broxterman, H. M. Verheul, Cross-resistance to clinically used tyrosine kinase inhibitors sunitinib, sorafenib and pazopanib. Cell. Oncol. 38, 119–129 (2015)
F.A. Shepherd, C. Domerg, P. Hainaut, P.A. Jänne, J.P. Pignon, S. Graziano, J.Y. Douillard, E. Brambilla, T. Le Chevalier, L. Seymour, A. Bourredjem, G. Le Teuff, R. Pirker, M. Filipits, R. Rosell, R. Kratzke, B. Bandarchi, X. Ma, M. Capelletti, J.C. Soria, M.S. Tsao, Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 31, 2173–2181 (2013)
I. Nakachi, K. Naoki, K. Soejima, I. Kawada, H. Watanabe, H. Yasuda, S. Nakayama, S. Yoda, R. Satomi, S. Ikemura, H. Terai, T. Sato, A. Ishizaka, The combination of multiple receptor tyrosine kinase inhibitor and mammalian target of rapamycin inhibitor overcomes erlotinib resistance in lung cancer cell lines through c-Met inhibition. Mol. Cancer Res. 8, 1142–1151 (2010)
B. Escudier, C. Porta, P. Bono, T. Powles, T. Eisen, C. N. Sternberg, J. E. Gschwend, U. De Giorgi, O. Parikh, R. Hawkins, E. Sevin, S. Négrier, S. Khan, J. Diaz, S. Redhu, F. Mehmud, D. Cella, Randomized, controlled, double-blind, cross-over trial assessing treatment preference for pazopanib versus sunitinib in patients with metastatic renal cell carcinoma: PISCES study. J. Clin. Oncol. 32, 1412–1418 (2014)
C. J. Tai, C. K. Wang, C. J. Tai, C. Tzao, Y. C. Lien, C. C. Hsieh, C. I. Hsieh, H. C. Wu, C. H. Wu, C. C. Chang, R. J. Chen, H. Y. Chiou, Evaluation of safety and efficacy of salvage therapy with sunitinib, docetaxel (tyxane) and cisplatinum followed by maintenance vinorelbine for unresectable/metastatic nonsmall cell lung cancer: stage 1 of a Simon 2 stage clinical trial. Medicine 94, e2303 (2015)
D. Seyman, A. D. Yalcin, N. Oztoprak, G. E. Genc, N. S. Ozen, F. Kizilates, H. Berk, S. Gumuslu, Soluble TRAIL levels decreased in chronic hepatitis C treatment with pegylated interferon α plus ribavirin: association with viral responses. Int. J. Clin. Exp. Med. 7, 5650–5656 (2014)
Y. Lan, X. Liu, R. Zhang, K. Wang, Y. Wang, Z. C. Hua, Lithium enhances TRAIL-induced apoptosis in human lung carcinoma A549 cells. Biometals 26, 241–254 (2013)
H. Zhuang, W. Jiang, X. Zhang, F. Qiu, Z. Gan, W. Cheng, J. Zhang, S. Guan, B. Tang, Q. Huang, X. Wu, X. Huang, W. Jiang, Q. Hu, M. Lu, Z. C. Hua, Suppression of HSP70 expression sensitizes NSCLC cell lines to TRAIL-induced apoptosis by upregulating DR4 and DR5 and downregulating c-FLIP-L expressions. J. Mol. Med. 91, 219–235 (2013)
R. Sriraksa, T. Limpaiboon, TRAIL in combination with subtoxic 5-FU effectively inhibit cell proliferation and induce apoptosis in cholangiocarcinoma cells. Asian Pac. J. Cancer Prev. 16, 6991–6996 (2015)
F. Pan, J. Tian, X. Zhang, Y. Zhang, Y. Pan, Synergistic interaction between sunitinib and docetaxel is sequence dependent in human non-small lung cancer with EGFR TKIs-resistant mutation. J. Cancer Res. Clin. Oncol. 137, 1397–1408 (2011)
B. Saigal, B. S. Glisson, F. M. Johnson, Dose-dependent and sequence-dependent cytotoxicity of erlotinib and docetaxel in head and neck squamous cell carcinoma. Anti-Cancer Drugs 19, 465–475 (2008)
H. Makinoshima, M. Takita, K. Saruwatari, S. Umemura, Y. Obata, G. Ishii, S. Matsumoto, E. Sugiyama, A. Ochiai, R. Abe, K. Goto, H. Esumi, K. Tsuchihara, Signaling through the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) axis is responsible for aerobic glycolysis mediated by glucose transporter in epidermal growth factor receptor (EGFR)-mutated lung adenocarcinoma. J. Biol. Chem. 290, 17495–17504 (2015)
N. Ishida, T. Fukazawa, Y. Maeda, T. Yamatsuji, K. Kato, K. Matsumoto, T. Shimo, N. Takigawa, J. A. Whitsett, Y. Naomoto, A novel PI3K inhibitor iMDK suppresses non-small cell lung cancer cooperatively with A MEK inhibitor. Exp. Cell Res. 335, 197–206 (2015)
C. Gridelli, P. Maione, A. Rossi, M. L. Ferrara, M. A. Bareschino, C. Schettino, P. C. Sacco, F. Ciardiello, Potential treatment options after first-line chemotherapy for advanced NSCLC: maintenance treatment or early second-line? Oncologist 14, 137–147 (2009)
G. Genestreti, F. Grossi, C. Genova, M. A. Burgio, A. Bongiovanni, G. Gavelli, M. Bartolotti, M. Di Battista, G. Cavallo, A. A. Brandes, Third- and further-line therapy in advanced non-small-cell lung cancer patients: an overview. Future Oncol. 10, 2081–2096 (2014)
M. Shrader, M. S. Pino, L. Lashinger, M. Bar-Eli, L. Adam, C. P. Dinney, D. J. McConkey, Gefitinib reverses TRAIL resistance in human bladder cancer cell lines via inhibition of AKT-mediated X-linked inhibitor of apoptosis protein expression. Cancer Res. 67, 1430–1435 (2007)
R. R. Rosato, J. A. Almenara, S. Coe, S. Grant, The multikinase inhibitor sorafenib potentiates TRAIL lethality in human leukemia cells in association with Mcl-1 and cFLIPL downregulation. Cancer Res. 67, 9490–9500 (2007)
W. Ding, T. Cai, H. Zhu, R. Wu, C. Tu, L. Yang, W. Lu, Q. He, B. Yang, Synergistic antitumor effect of TRAIL in combination with sunitinib in vitro and in vivo. Cancer Lett. 293, 158–166 (2010)
Y. Safdari, M. Khalili, M. A. Ebrahimzadeh, Y. Yazdani, S. Farajnia, Natural inhibitors of PI3K/AKT signaling in breast cancer: emphasis on newly-discovered molecular mechanisms of action. Pharmacol. Res. 93C, 1–10 (2014)
T. C. Hour, S. D. Chung, W. Y. Kang, Y. C. Lin, S. J. Chuang, A. M. Huang, W. J. Wu, S. P. Huang, C. Y. Huang, Y. S. Pu, EGFR mediates docetaxel resistance in human castration-resistant prostate cancer through the Akt-dependent expression of ABCB1 (MDR1). Arch. Toxicol. 89, 591–605 (2015)
E. Piovan, J. Yu, V. Tosello, D. Herranz, A. Ambesi-Impiombato, A. C. Da Silva, M. Sanchez-Martin, A. Perez-Garcia, I. Rigo, M. Castillo, S. Indraccolo, J. R. Cross, E. de Stanchina, E. Paietta, J. Racevskis, J. M. Rowe, M. S. Tallman, G. Basso, J. P. Meijerink, C. Cordon-Cardo, A. Califano, A. A. Ferrando, Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell 24, 766–776 (2013)
K. Zhang, X. Wang, H. Wang, Effect and mechanism of Src tyrosine kinase inhibitor sunitinib on the drug-resistance reversal of human A549/DDP cisplatin-resistant lung cancer cell line. Mol. Med. Rep. 10, 2065–2072 (2014)
A. Kisim, H. Atmaca, B. Cakar, B. Karabulut, C. Sezgin, S. Uzunoglu, R. Uslu, B. Karaca, Pretreatment with AT-101 enhances tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of breast cancer cells by inducing death receptors 4 and 5 protein levels. J. Cancer Res. Clin. Oncol. 138, 1155–1163 (2012)
G. Pasello, L. Urso, M. Silic-Benussi, M. Schiavon, I. Cavallari, G. Marulli, N. Nannini, F. Rea, V. Ciminale, A. Favaretto, Synergistic antitumor activity of recombinant human Apo2L/tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in combination with carboplatin and pemetrexed in malignant pleural mesothelioma. J. Thorac. Oncol. 9, 1008–1017 (2014)
Y. Zhang, B. Zhang, TRAIL resistance of breast cancer cells is associated with constitutive endocytosis of death receptors 4 and 5. Mol. Cancer Res. 6, 1861–1871 (2008)
J. J. Chen, H. C. Shen, L. A. Rivera Rosado, Y. Zhang, X. Di, B. Zhang, Mislocalization of death receptors correlates with cellular resistance to their cognate ligands in human breast cancer cells. Oncotarget 3, 833–842 (2012)
O. Arrieta, A. F. Cardona, L. Corrales, A. D. Campos-Parra, R. Sánchez-Reyes, E. Amieva-Rivera, J. Rodríguez, C. Vargas, H. Carranza, J. Otero, N. Karachaliou, H. Astudillo, R. Rosell, CLICaP, the impact of common and rare EGFR mutations in response to EGFR tyrosine kinase inhibitors and platinum-based chemotherapy in patients with non-small cell lung cancer. Lung Cancer 87(169–175) (2015)
V. Noronha, A. Joshi, A. Gokarn, V. Sharma, V. Patil, A. Janu, N. Purandare, A. Chougule, N. Jambhekar, K. Prabhash, The importance of brain metastasis in EGFR mutation positive NSCLC patients. Chemother. Res. Pract. 2014, 856156 (2014)
X. Chen, Y. Liu, O. D. Røe, Y. Qian, R. Guo, L. Zhu, Y. Yin, Y. Shu, Gefitinib or erlotinib as maintenance therapy in patients with advanced stage non-small cell lung cancer: a systematic review. PLoS one 8, e59314 (2013)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Grant Support
This study was supported by a research Grant from the Postdoctoral Research Fund (Grant No. LBH-Z12159) provided by the Postdoctoral Management Office of Heilongjiang Province, China, and a research Grant from the Natural Science Foundation of Heilongjiang Province, China (Grant No. H201311) provided by the committee of Natural Science Foundation of Heilongjiang Province. This study was also supported by a research grant from the PhD Research Fund (Grant No. BS2012–16) provided by the Second Affiliated Hospital of Harbin Medical University, Heilongjiang, China. We thank all participants who contributed to in this study.
Conflicts of interest
none to declare.
Additional information
Yong-Xia Bao and Xiao-Dan Zhao contributed equally to this work.
Electronic supplementary material
ESM 1
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Bao, YX., Zhao, XD., Deng, HB. et al. Schedule-dependent cytotoxicity of sunitinib and TRAIL in human non-small cell lung cancer cells with or without EGFR and KRAS mutations. Cell Oncol. 39, 343–352 (2016). https://doi.org/10.1007/s13402-016-0278-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-016-0278-4