Abstract
Background
Previously, we found that the Graffi murine leukemia virus (MuLV) is able to induce a wide spectrum of hematologic malignancies in vivo. Using high-density oligonucleotide microarrays, we established the gene expression profiles of several of these malignancies, thereby specifically focusing on genes deregulated in the lymphoid sub-types. We observed over-expression of a variety of genes, including Arntl2, Bfsp2, Gfra2, Gpm6a, Gpm6b, Nln, Fbln1, Bmp7, Etv5 and Celsr1 and, in addition, provided evidence that Fmn2 and Parm-1 may act as novel oncogenes. In the present study, we assessed the expression patterns of eight selected human homologs of these genes in primary human B-cell malignancies, and explored the putative oncogenic potential of GPM6A and GPM6B.
Methods
The gene expression levels of the selected human homologs were tested in human B-cell malignancies by semi-quantitative RT-PCR. The protein expression profiles of human GPM6A and GPM6B were analyzed by Western blotting. The localization and the effect of GPM6A and GPM6B on the cytoskeleton were determined using confocal and indirect immunofluorescence microscopy. To confirm the oncogenic potential of GPM6A and GPM6B, classical colony formation assays in soft agar and focus forming assays were used. The effects of these proteins on the cell cycle were assessed by flow cytometry analysis.
Results
Using semi-quantitative RT-PCR, we found that most of the primary B-cell malignancies assessed showed altered expression patterns of the genes tested, including GPM6A and GPM6B. Using confocal microscopy, we found that the GPM6A protein (isoform 3) exhibits a punctate cytoplasmic localization and that the GPM6B protein (isoform 4) exhibits a peri-nuclear and punctate cytoplasmic localization. Interestingly, we found that exogenous over-expression of both proteins in NIH/3T3 cells alters the actin and microtubule networks and induces the formation of long filopodia-like protrusions. Additionally, we found that these over-expressing NIH/3T3 cells exhibit anchorage-independent growth and enhanced proliferation rates. Cellular transformation (i.e., loss of contact inhibition) was, however, only observed after exogenous over-expression of GPM6B.
Conclusions
Our results indicate that several human homologs of the genes found to be deregulated in Graffi MuLV experimental mouse models may serve as candidate biomarkers for human B-cell malignancies. In addition, we found that GPM6A and GPM6B may act as novel oncogenes in the development of these malignancies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In 1957, Arnold Graffi identified the Graffi murine leukemia virus (MuLV) and showed that this virus can induce chloroleukemias with an incidence of 87 % in a particular strain of mice (Agnes-Bluhm) [1]. Several years ago, we showed that molecular clones of the virus are able to induce a wide spectrum of hematologic malignancies, including lymphoid (B- and T-cell) and non-lymphoid (erythroid, myeloid and megakaryoblastic) leukemias [2]. Using insertional mutagenesis assays, several proto-oncogenes were identified, including Cyclin D2, Fli-1, Spi1/Pu1, Pim-1 and c-Myc, that may act in the transformation initiation process [3–5]. The multistep nature of cancer development, however, implies the involvement of additional deregulated genes. To identify such genes implicated in lymphoid leukemia development, we previously established the expression profiles of three Graffi MuLV-induced T-cell leukemias, three Graffi MuLV-induced B-cell leukemias and one control for each type using oligonucleotide microarrays [6]. This approach allowed us to identify several candidate genes, including new potential oncogenes [6, 7, 19]. In particular, we found that the Fmn2 gene is over-expressed in murine B-cell leukemias and that its human homolog is over-expressed in human pre-B-ALLs, especially in pediatric cases carrying a t(12;21) translocation (TEL/AML1 rearrangement). In addition, we provided evidence for its role as a potential oncogene [6]. More recently, we also gained evidence for oncogenic potential of the Parm-1 gene, which is specifically over-expressed in CD8+ T-cell leukemias [7].
In the present study, we assessed the expression patterns of the human homologs of several candidate genes associated with Graffi MuLV-induced malignancies [6] in human B-cell leukemias, and found that several of them may serve as candidate biomarkers. In addition, we explored the putative oncogenic potential of the GPM6A and GPM6B genes. Both latter genes code for members of the proteolipid protein (PLP) family, which all contain four transmembrane domains and are highly conserved throughout evolution. While GPM6A has not been associated with cancer before, GPM6B has been reported to serve as a gene that may distinguish glioblastoma from meningioma [8], and to serve as a promising candidate marker for early stage ovarian cancer [9]. We found that both encoded proteins (i.e., splicing isoform 3 of GPM6A and splicing isoform 4 of GPM6B) are able to alter the actin and microtubule networks, to induce the anchorage-independent growth and to enhance the proliferative capacity of NIH/3T3 cells. We also found that, unlike GPM6A, exogenous over-expression of GPM6B increases the number of foci (defined by loss of contact inhibition of growth) in these cells.
2 Materials and methods
2.1 Human sample collection
Samples from 12 pediatric pre-B-ALL patients were obtained from Dr. Daniel Sinnet (Sainte-Justine Hospital, Montreal), and samples from 13 adult patients with different types of B-cell leukemia were obtained from the Quebec Leukemia Cell Bank. As a control (CH), peripheral blood mononuclear cells isolated from ten healthy adults were pooled. The mononuclear cells were isolated using a Ficoll--Hypaque gradient method. For more detailed information on each patient, see supplementary Table S1 of Charfi et al. [6]. The research protocol was approved by the ethics committees of all institutions involved.
2.2 Semi-quantitative RT-PCR
Total RNA (100 ng) was reverse transcribed using the Omniscript enzyme (QIAGEN, Mississauga, Canada) in conjunction with an oligo(dT) primer. Subsequent PCR reactions were performed using the Taq polymerase kit (Feldan) and 10 % of the reverse transcribed material (2 % for β-actin; 94 °C for 3 min, 94 °C for 45 s, 56 °C for 45 s, 72 °C for 30 s with a final extension at 72 °C for 10 min). For semi-quantitative analyses, the annealing temperature and the number of cycles were optimized for each of the selected genes (Table 1). PCR products were analyzed on agarose gels and band densities were quantified with Quantity One Image Software, using β-actin as an internal control.
2.3 Cell culture
Immortalized NIH/3T3 fibroblasts and Raji B-cells were obtained from the ATCC (Rockville, MD) and were grown in DMEM medium supplement with 10 % CS and in RPMI medium supplemented with 10 % FBS (Invitrogen, Carlslab, CA), respectively. 50 U penicillin and 50 μg streptomycin (Gibco, Invitrogen, Frederick, MD) were added to each culture medium.
2.4 Confocal and indirect immunofluorescence microscopy
For protein localization, Raji and NIH/3T3 cells (107) were transiently transfected with 15 μg plasmids by electroporation using the Gene Pulser System (Bio-Rad) and the polyfect reagent (Qiagen), respectively. For both cell lines, pEGFP-N1 (control vector) (Clontech Laboratories), GFP-tagged GPM6A (GPM6A-GFP) and GFP-tagged GPM6B (GPM6B-GFP) constructs were used.
Protein localization was assessed by confocal microscopy 48 h after transfection. For cell surface membrane co-localization analyses (CellMask™ Plasma Membrane Stains (Invitrogen)), Raji cells were pelleted 48 h after transfection, washed in phosphate-buffered saline (PBS) and transferred for 30 min at 37 °C to glass slides coated with polylysine. NIH/3T3 cells were directly plated on glass coverslips before transfection. After 48 h, cells were fixed for 20 min with 4 % paraformaldehyde, washed with PBS, permeabilized for 5 min with 0.1 % Triton X-100 in PBS, blocked for 1 h in PBS with 10 % goat serum, 10 % BSA and 0.1 % Triton X-100, and incubated overnight with primary anti-α-tubulin antibody (1/2000, Sigma). After three washes with PBS, coverslips were incubated with an Alexa Fluor 568-conjugated secondary antibody (1/1000, Invitrogen-Molecular Probes) for 1 h and washed three times with PBS. Actin filament staining was performed after cell fixation for 20 min with 4 % paraformaldehyde, followed by PBS washes and permeabilization for 5 min with 0.1 % Triton X-100 in PBS. Next, the cells were incubated 1 h in PBS with 1 % bovine serum albumin, washed twice with PBS and then incubated for 20 min with 0.3 M phalloidin coupled to AlexaFluor-555 (Invitrogen). Finally, after α-tubulin and actin filament staining, coverslips were mounted onto slides using Prolong Gold antifade reagent (Invitrogen) and imaged by confocal microscopy.
For the indirect immunofluorescence assays, Raji and NIH/3T3 cells were transfected with the same plasmids and cultured in dishes containing glass coverslips. After 48 h, the coverslips were transferred to a coverslip-cell chamber containing medium for imaging.
2.5 Western blot analysis
For Western blot analyses, NIH/3T3 cells were homogenized in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 10 mM sodium pyrophosphate, 100 mM sodium fluoride, 1.5 mM MgCl2, 1 mM EGTA, 200 μM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 % glycerol, and 1 % Triton X-100) supplemented with a complete protease inhibitor cocktail (Roche Molecular Diagnostics, Mannheim, Germany). Cells were incubated in this lysis buffer for 30 min at 4 °C, and centrifuged at 15,000×g for 10 min at 4 °C. The resulting cell lysates were boiled for 5 min and the proteins (30 μg) were separated in 10 % SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. These membranes were subsequently blocked for 1 h in buffer (PBS, 0.1 % Tween 20 [PBS-T] with 5 % wt/vol nonfat dry milk) and incubated overnight at 4 °C with an anti-GFP primary antibody (1:1000; Santa Cruz Biotechnology). As loading control an anti-β-actin (Sigma-Aldrich, St. Louis, MO) antibody was used. Next, the membranes were incubated for 1 h with horseradish peroxidase–conjugated secondary antibodies diluted in blocking buffer and the resulting signals were revealed using the Immobilon Western HRP reagent (Millipore, Billerica, USA).
2.6 Colony formation assay in soft agar
Anchorage-independent growth was determined as previously described [6, 10]. Briefly, NIH/3T3 cells were transiently transfected with 2.5 μg pEGFP-N1, GPM6A-GFP or GPM6B-GFP expression vectors. After 48 h, 1 × 104 cells were mixed with melted 0.3 % agarose in DMEM medium and seeded in six-well plates on top of a 0.6 % agarose base layer containing the same medium. The top layer was covered with 1.5 ml of DMEM. Cells were fed twice a week for 4 weeks and, using an inversion microscope, the colonies were counted.
2.7 Focus forming assay
To assess focus formation, NIH/3T3 cells were transfected with 2.5 μg of either plasmid expressing pEGFP-N1, GPM6A-GFP or GPM6B-GFP. Two days later, cells were split 1:3 and placed in DMEM medium containing 10 % CS. After 4 weeks of growth, the dishes were stained with 0.2 % methylene blue and 50 % methanol and photographed, after which foci formation was quantified.
2.8 Cell cycle analysis
For cell cycle analyses, flow cytometry was performed using a FACScan flow cytometer (Becton Dickinson). Briefly, 1 × 106 cells were pelleted by centrifugation, resuspended in 0.2 ml PBS, added to 4 ml of ice-cold 70 % ethanol and incubated overnight at 4 °C. Next, the cells were centrifuged, re-suspended in propidium iodide (40 μg/ml)-RNase (100 μg/ml) staining solution for 30 min at 37 °C and, finally, analyzed by flow cytometry for DNA content. The percentages of cells in different phases of the cell cycle were determined using the ModFit software.
3 Results
3.1 Microarray data validation in human B-cell leukemias
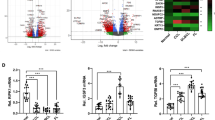
Extensive studies on the Graffi murine leukemia retrovirus have revealed a multipotent retrovirus that can induce the full spectrum of leukemias in mice [2, 11, 12]. Taking advantage of these characteristics, and to gain insight into the malignancy signature of lymphoid leukemias induced by the Graffi MuLV, we previously used high-density oligonucleotide microarrays to derive and compare the transcriptome of three T-cell leukemias (T1 (CD4+CD8+), T2 (CD4−CD8+) and T3 (CD4+CD8−)) and three B-cell leukemias (B1 and B2 (CD45R+CD19+), B3 (CD45R+lowCD19+Sca1+)) induced by this virus vs. appropriate controls (non-leukemic B-cell (CB1 (CD45R+CD19+)) and T-cell (CT1 (CD4+CD8+)) populations) to identify genes whose expression was either increased (putative oncogenes) or reduced (putative tumor suppressor genes) in a specific leukemia type when compared to controls (NCBI GEO: GSE12581) [6, 13]. This analysis, allowed us to identify a number of genes that are now considered as specific markers for B-cell leukemias (Arntl2, Bfsp2, Gfra2, Gpm6a, Gpm6b), T-cell leukemias (Nln, Fbln1, Bmp7) or both leukemias (Etv5 and Celsr1) induced by the Graffi MuLV [6]. We also showed that the Fmn2 and Parm-1 genes exhibit oncogenic potential [6, 7]. In the present study, we measured the expression levels of the human counterparts of several of the lymphoid leukemia-associated genes induced by the Graffi MuLV (Gpm6a, Gpm6b, Arntl2, Bfsp2, Kif4, 2010007H06Rik, Celsr1 and Etv5) in human B-cell leukemias (Fig. 1). Using semi-quantitative RT-PCR, no expression was detected in control samples (CH, see section 2) for the human homologues of Gpm6b, Arntl2, Kif4, 2010007H06Rik, Celsr1 and Etv5, whereas a very weak expression was detected for the human homologues of Gpm6a and Bfsp2. In contrast, expression of each of these genes was detected in the majority of the human B-cell leukemias tested. Indeed, among the 25 tested cases, 24 (96 %) expressed the Gpm6a, Kif4, 2010007H06Rik, Etv5 or Celsr1 homologues, 20 (80 %) expressed the Arntl2 homologue, 19 (76 %) expressed the Bfsp2 homologue and 15 (60 %) expressed the Gpm6b homologue. Furthermore, our analysis revealed that human GPM6A was specifically over-expressed in a mantel cell lymphoma (MCL) and a chronic lymphocytic leukemia (CLL), and that human GPM6B was specifically over-expressed in pediatric and adult pre-B-ALLs. The human homologues of Arntl2, Kif4, 2010007H06Rik and Etv5 were over-expressed in all tested leukemia types, whereas the Bfsp2 homologue was expressed in the MCL and adult pre-B-ALLs, and the Celsr1 homologue was expressed in the pediatric pre-B-ALLs. For the Gpm6a, Arntl2, Kif4, 2010007H06Rik, Etv5 and Bfsp2 homologues a larger number of cases will be necessary to confirm an association with MCL and/or CLL.
Analysis of selected genes differentially expressed in murine and human lymphoid leukemia samples. Semi-quantitative RT-PCR analysis was performed on pediatric pre-B ALLs (lanes 1–12), adult pre-B ALLs (lanes 13–19), Burkitt leukemias (lanes 20–21), one mantel cell lymphoma (lane 22), one follicular lymphoma (lane 23), one B-cell prolymphocytic leukemia (lane 24) and one chronic lymphocytic leukemia (lane 25). Values were compared to the respective control (CH). Information relevant for each patient has been provided in the supplementary data of ref 6. RT-PCRs were performed in triplicate for each gene. The actin gene was used as internal control (see section 2) and the expression level in each leukemia sample is presented as a gene/actin density ratio. Statistical analysis was performed using T-test with Welch correction and p < 0.05 was considered to be significant (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001)
3.2 Identification of GPM6A and GPM6B specific isoforms in human B-cell leukemias
Similar to the murine Gpm6a and Gpm6b genes, its respective human homologues exhibit many spliced isoforms, which are more closely related to GPM6B than to GPM6A (Online Resource Fig. 1a and b, [14]). To determine which isoforms are over-expressed in human B-cell leukemias compared to control cells, RT-PCR analysis was used in conjuction with different primers to distinguish the multiple isoforms. By doing so, we found that GPM6A isoform 3 (NM_201592) was the only isoform which is highly expressed in several of the B-cell leukemias tested (Fig. 1). In addition, we found that GPM6A isoform 2 (NM_001261447) was over-expressed in only one of the B-cell leukemia samples tested (B-cell prolymphocytic leukemia) (data not shown). We also noticed that GPM6B isoform 4 (NM_001001994) was the only highly expressed isoform in human B-cell leukemias (Fig. 1).
3.3 In silico analysis and expression profiling of human GPM6A and GPM6B proteins
Using the NCBI PROTEIN database, we found that GPM6A isoform 3 and GPM6B isoform 4 encode proteins of 267aa (NP_963886) and 246aa (NP_001001994) resulting in predicted molecular weights of 29.9 kDa and 26.8 kDa, respectively. These proteins are members of the proteolipid protein (PLP) family, which also includes the closely related PLP1 protein (proteolipid protein 1). The amino acid sequences of GPM6A isoform 3 and of GPM6B isoform 4 were compared to the amino acid sequence of DM20, a splice isoform of PLP1 (NCBI protein BLAST program). We found that the amino acid sequences of human GPM6A isoform 3 and of human GPM6B isoform 4 share 51 % identity (Online Resource Fig. 2), while human GPM6A isoform 3 and human GPM6B isoform 4 were found to share 43 % and 57 % identity with DM20, respectively (Online Resource Fig. 2b and c), suggesting that these human proteins are highly related.
Computer modeling using the TMRPres2D software package (http://bioinformatics.biol.uoa.gr/TMRPres2D/download.jsp) revealed that both human GPM6A isoform 3 and human GPM6B isoform 4 proteins contain 4 trans-membrane α-helical domains (tetraspan; two extracellular and one cytoplasmic loops). In addition, these proteins were found to contain a short amino terminus and a carboxyl terminal tail, both predicted to be in the cytoplasmic space (Fig. 2a). Bio-informatic analyses using different programs revealed inconsistent signal peptide predictions. Some programs, such as the Signal-3L program [15], predicted a signal peptide of 36 aa for GPM6A and of 41 aa for GPM6B, while others predicted no signal peptide for the two proteins.
GPM6A and GPM6B 2-dimensional protein structures and protein expression profiles. a Transmembrane region topologies of GPM6A and GPM6B as predicted using HMMTOP. The data were used as input into TMRPres2D software to generate a 2-D model showing four transmembrane domains and short N- and C-terminal cytoplasmic tails. b Immunoblotting of lysates from NIH/3T3 cells transiently transfected with expression vectors of GPM6A-GFP (isoform 3 of Gpm6a) or c of GPM6B-GFP (isoform 4 of Gpm6b) using an anti-GFP antibody. Images were cropped to eliminate the signal of the GFP alone
To determine the molecular weights and expression profiles of human GPM6A and GPM6B, the cDNA of their corresponding isoforms was amplified from human leukemias and fused with GFP. Lysate from NIH/3T3 cells transfected with GPM6A-GFP or GPM6B-GFP constructs were analysed by Western blotting using an anti-GFP antibody. For GPM6A, we observed a band of approximately 50 kDa (Fig. 2b), indicating that the molecular weight of GPM6A, including the GFP-tag, is slightly lower than the predicted 56.9 kDa. For GPM6B-GFP, the protein profile was more complex and showed several bands. The most intense band at about 50 kDa is very likely the monomeric form after signal peptide cleavage (Fig. 2c). The intensity of the higher molecular weight bands, i.e., >230 kDa and at 100 kDa, decreased at higher concentrations of DTT (results not shown). The fact that the molecular weights of GPM6A and of the smallest GPM6B band were lower than that predicted for the entire proteins, strongly suggests the presence of N-terminal signal peptides in these two proteins.
3.4 Sub-cellular localization of human GPM6A and GPM6B and their effects on the cytoskeleton
Since the sub-cellular localization of a protein may directly be linked to its function, we set out to investigate the localization of the GPM6A and GPM6B proteins. Using confocal microscopy, we found that GPM6A exhibits a punctated cytoplasmic localization and that GPM6B exhibits a perinuclear and punctated cytoplasmic localization (Fig. 3a). Despite the presence of four transmembrane domains, plasma membrane localization was not noted in our confocal analyses using the CellMask marker to stain the plasma membrane (Fig. 3b). Raji cells were used to confirm these results. Although we were not able to successfully express the GPM6A-GFP construct, we confirmed GPM6B-GFP localization in these B lymphoblastic cells (Fig. 3b).
Sub-cellular localization of the GPM6A and GPM6B proteins and their effect on the cytoskeleton. a NIH/3T3 cells were transfected with GPM6A-GFP or GPM6B-GFP constructs and visualized with a laser-scanning confocal microscope (Bio-Rad MRC-1024 ES) equipped with a Nikon TE-300 camera and a Plan Apochromat 60× (NA 1.40) oil objective (Nikon). Images were digitally acquired using Laser Sharp software Version 3.2 (Bio-Rad). b Raji cells were transfected with a GPM6B-GFP construct, stained with the plasma membrane marker CellMask and visualized using confocal microscopy. c Both proteins were also tested for their induction of protrusions using indirect immunofluorescence microscopy. Arrows indicate long cellular protrusions. d α-tubulin labeling with anti-α-tubulin antibody. e Actin labeling with AlexaFluor-555-conjugated phalloidin. Images were taken with an Eclipse Ti fluorescent microscope (Nikon) equipped with a 20 × 0.75-numerical-aperture objective lens. Images were captured as 8-bit tagged image file format files with a Scion CFW-1608C camera. Data are representatives of three independent experiments. The GFP vector alone was used as a control
By indirect immunofluorescence, we observed that NIH/3T3 cells expressing GPM6A-GFP or GPM6B-GFP show long extensions when compared to control cells (Fig. 3c), possibly corresponding to filopodia-like protrusions. These observations prompted us to test the effect of GPM6A and GPM6B over-expression on actin filaments and the cellular microtubule network. As shown in Fig. 3d and e, cells over-expressing GPM6A or GPM6B showed a loss of actin stress fibers, characteristic of a disruption of the actin and microtubule networks, when compared to control GFP-transfected cells.
3.5 Human GPM6A and GPM6B induce anchorage-independent growth
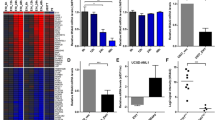
Since the GPM6A and GPM6B genes were found to be specifically over-expressed in both murine and human B-cell leukemias, we next investigated their putative oncogenic potential. To this end, we first studied the impact of exogenous GPM6A and GPM6B over-expression on the anchorage-independent growth of NIH/3T3 cells, which is considered to be a classic assay [16–18] to demonstrate the oncogenic potential of certain genes. We found that the number of colonies formed in soft agar was significantly higher in both GPM6A (Fig. 4a) and GPM6B (Fig. 4b) over-expressing cells compared to controls (i.e., NIH/3T3 cells either untransfected or transfected with the empty GFP vector). These results suggest that over-expression of both the GPM6A or GPM6B genes promotes anchorage-independent growth of NIH/3T3 cells.
Effect of GPM6A and GPM6B protein expression on anchorage-independent growth and foci formation. NIH/3T3 cells were transiently transfected with GFP empty vector, a GPM6A-GFP or b GPM6B-GFP expression vectors. Transfected or control cells (104) were plated in soft agar as described in section 2. After 4 weeks, the number of colonies was scored. The results represent the average of three independent experiments. Statistical analysis was performed using One-way ANOVA; p < 0.05 was considered to be significant. Cells were observed with an optical microscope (Ernst Leitz, 6MBH Wetzlar) and representative fields were photographed using a numerical camera (Nikon coolpix 4500) (original magnification × 40). c NIH/3T3 cells were transfected with either a GFP empty vector (left) or with GPM6B-GFP (right), split, and maintained in normal conditions as described in section 2. Foci started to appear after 20 days and were photographed at day 30
3.6 Human GPM6B increases the number of transforming foci
We also tested the effect of the GPM6A and GPM6B genes on NIH/3T3 transformation using a focus-forming assay. Our results showed that, unlike GPM6A, the number of large foci formed in GPM6B over-expressing cells was higher than that in control cells (Fig. 4c) suggesting that cells over-expressing GPM6B exhibit loss of contact inhibition of growth.
3.7 Human GPM6A and GPM6B induce proliferation
To further characterize the oncogenic properties of the GPM6A and GPM6B genes, we next tested their ability to induce the proliferation of synchronized NIH/3T3 cells. Our FACS analyses indicate that GPM6A-GFP or GPM6B-GFP transfected NIH/3T3 cells show an increase in the percentage of cells in S phase of about 2 and 1.8 fold, respectively, when compared to control cells (Fig. 5a and b), suggesting that over-expression of GPM6A and GPM6B may confer a growth advantage. The results from the soft agar assays, the cell cycle analyses and the confocal microscopy analyses were confirmed using Myc-tagged proteins to exclude a possible induction of oncogenic potential via mis-localization of the proteins due to the GFP-tag.
Effect of (a) GPM6A and (b) GPM6B proteins on NIH/3T3 cell proliferation. Synchronized NIH/3T3 cells either untransfected or transfected with an empty vector (GFP), GPM6A-GFP or GPM6B-GFP constructs, were fixed at 72 h post-transfection, stained with propidium iodide and analyzed for cell cycle phase distribution. For diagrams, values were normalized relative to control cells. Statistical analysis was performed using T-test with Welch correction and p < 0.05 was considered to be significant (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). Results represent the average of at least 3 independent experiments
Taken together, our data indicate that GPM6A and GPM6B may promote the transformation and proliferation of NIH/3T3 cells.
4 Discussion
Since it was first discovered that the Graffi MuLV is able to induce a large spectrum of hematologic malignancies in vivo, including lymphoid leukemias, we have shown that this viral system may serve as an appropriate model to gain further insights into leukemia initiation and progression and to identify and characterize new oncogenes and suitable biomarkers [2, 12]. We previously analyzed the gene expression profiles and malignancy signatures of different leukemia types (T- and B-cell, myeloid, erythroid, and megakaryoblastic) using high-density oligonucleotide microarrays [6, 13]. These analyses already allowed us to identify new candidate markers and potential proto-oncogenes such as the Fmn2 and Parm-1 genes [6, 7].
In the present study, we analyzed the expression profiles of several recently identified markers specific for lymphoid leukemias induced by the Graffi MuLV, including four genes specific to B-cell leukemia, two genes common to B- and T-cell leukemias [6], and two newly identified genes (Kif4 and 2010007H06Rik). This analysis revealed over-expression of the human homologues of the Gpm6a, Gpm6b, Arntl2, Bfsp2, Kif4, 2010007H06Rik genes (B-cell leukemia-specific genes), and the human homologues of the Celsr1 and Etv5 genes (genes common to B- and T-cell leukemias) in human B-cell leukemias [6, 19].
Arntl2 pt?>is a member of the basic Helix-Loop-Helix-PAS (bHLH-PAS) family of transcription factors [20], known for its involvement in the control of the circadian rhythm [21]. Interestingly, a relationship between tumorigenesis and the disruption of the circadian rhythm has previously been suggested [22]. Human ARNTL2 has been found to be up-regulated in primary colorectal cancers and in one highly proliferative colon cancer cell line. It has been suggested that this gene may be used as a predictor of tumor invasiveness and cancer aggressiveness [23]. Bfsp2 is a member of the intermediate filament (IF) family and is specifically expressed in lens cells [24]. Like the IF proteins, human BFSP2 plays a major role in the organization of cytoskeleton proteins in these cells [25] and it has been demonstrated that mutations in this gene may cause human cataracts [26]. The Kif4 gene encodes a microtubule-based molecular motor that belongs to the kinesin-like protein family. This gene has been shown to be implicated in many cellular processes, including mitosis and DNA damage response [27, 28]. Loss of the Kif4 gene has been reported in 35 % of human cancers originating from several tissues, including three out of six tested leukemic cell lines (two T-cell leukemias, three non-lymphoid leukemias and one lymphoma of undetermined cellular origin) [29]. The discrepancy with our results may be explained by the fact that the expression level of the Kif4 gene in the previous study was not compared to control cells, such as e.g. peripheral blood mononuclear cells. Also, our preliminary results show that the mouse Kif4 cDNA, cloned from a B-cell leukemia induced by the Graffi MuLV, is slightly different from that listed in the NCBI data base (NM_008446) (data not shown). Thus, it is possible that leukemic B-cells may over-express a novel transcript of Kif4, which may be different from that being expressed at low levels in other human malignancies. For 2010007H06Rik (AK084978), no function has been assigned yet in either mouse or human, but a human ortholog has recently been characterized (C11orf92). This gene appears to be over-expressed in stage III serous ovarian carcinomas, a tumor stage with a poor prognosis [30]. Celsr1, a component of the mammalian planar cell polarity (PCP) signalling pathway [31], is a member of the cadherin family and was shown to be expressed in gastrointestinal tumors [32]. Our microarray data showed that Celsr1 is over-expressed in both murine T- and B-cell leukemias. However, RT-PCR validation showed that this gene was specific to T-cell leukemias induced by the Graffi MuLV [6]. Interestingly, assessment of the expression level of the human homologue of this gene in human B-cell leukemias revealed a good agreement with our microarray data (Fig. 1). Finally, Etv5 is a member of the Ets family of transcription factors. In the present report, this gene was considered as a positive control since it had previously been reported to be associated with B-CLL [33]. Indeed, according to our data, the human homologue of this gene appears to be over-expressed in human B-cell leukemias.
While GPM6B appears to be up-regulated in multiple types of human B-cell leukemias, GPM6A seems to be more restricted to MCL and CLL. As for BFSP2, it will be important to further substantiate the modulation of its expression in these specific types of B-cell malignancies.
Overall, our results strongly suggest that Gpm6a, Gpm6b, Arntl2, Bfsp2, Kif4, 2010007H06Rik, Celsr1 and Etv5 may serve as candidate biomarkers for murine and human B-lymphoid malignancies and, in addition, that these genes may represent potential proto-oncogenes.
Among the above eight genes studied and validated in human B-cell leukemias, we further characterized Gpm6a and Gpm6b, which were found to be specifically associated with murine and human B-cell leukemias. The two proteins encoded by these genes are members of the PLP family, which is considered to be a major structural component of the brain myelin sheath. Both human GPM6A and GPM6B are expressed in neurons but GPM6B is also expressed in oligodendrocytes [34, 35]. As yet, very little is known about the biological functions exerted by this protein family. However, in addition to their role in myelin formation, they are suspected to serve as housekeeping proteins implicated in intracellular trafficking [34, 36]. Unlike GPM6A, whose implication in the development of tumors has not been demonstrated yet, GPM6B is expressed at a relatively higher level in glioblastomas (very aggressive and with a poor prognosis) than in meningiomas. Thus, GPM6B is considered to be a predictive gene discriminating between these two types of tumors [8, 37]. Here, we report for the first time that GPM6A isoform 3 and GPM6B isoform 4 are over-expressed in human B-cell leukemias. The GPM6A and GPM6B proteins have been shown to be neuronal homologs of the PLP/DM20 protein [38, 39]. Indeed, we noted that the human GPM6A isoform 3, the human GPM6B isoform 4, PLP1 and DM20 are evolutionary linked. The PLPs are known to constitute four transmembrane domain proteins with both N- and C-termini located in the cytosolic compartment [40–42]. As GPM6A and GPM6B encode different glycoprotein isoforms, we wanted to ensure that our human B-cell leukemia-specific isoforms shared similar characteristics. Indeed, TMRPres2D software revealed the same features for GPM6A isoform 3 and GPM6B isoform 4.
Protein expression profiling in transfected NIH/3T3 cells revealed the absence of post-translational modifications in the GPM6A protein. In contrast, a complex protein expression profile for GPM6B was observed, which may result from the oligomeric state of this protein. This finding is not surprising since many PLPs have the ability to form oligomers [43, 44].
We also determined the intracellular localization of these proteins. We showed that GPM6A-GFP is granularly scattered in the cytoplasm and that GPM6B-GFP has a perinuclear and punctate cytoplasmic localization. Despite their predicted four transmembrane domains, these proteins were not found to be localized at the plasma membrane, whereas most four transmembrane domain proteins of this family are located at the cell membrane or at the endoplasmic reticulum [45–48]. However, perinuclear and cytoplasmic localization has been reported for other members of this family, such as PMP22CD and TMEM225 [45, 49]. Moreover, as for human GPM6B, the murine homologue exhibits many isoforms, with two promoters and alternative exons yielding more than 8 GPM6B proteins and polypeptides. Werner et al. showed that 6 isoforms are tetraspan membrane proteins that differ by 4 highly conserved N- and C-terminal domains (α, β, ψ, and ω domains). When the sub-cellular localizations of the latter were determined in transfected MDCK cells, it was found that the N-terminal β-domain of GPM6B was responsible for the predominant plasma membrane localization by stabilizing the tetraspan proteolipid at the cell surface. Moreover, the authors demonstrated that isoforms lacking this β-domain were more abundant in intracellular compartments [39]. This result is related to the fact that μ1A may serve as a specific binding partner of the β-domain, which is a component of tetrameric adaptor complexes relevant for sub-cellular sorting of membrane proteins in clathrin-coated vesicles [50, 51]. Interestingly, the GPM6B isoform 4 lacks the β-domain which, in turn, may explain its predominant intracellular localization. It is also important to note that PLP/DM20 does not have a β-like domain. DM20 is able to reach the plasma membrane only when it is co-expressed with the full-length PLP isoform, which possesses a putative signal sequence in its cytoplasmic loop [52, 53].
Our indirect immunofluorescence data also showed that the GPM6A and GPM6B proteins may induce the formation of long filamentous structures, possibly representing filopodia-like protrusions. In fact, GPM6A from both rat and zebrafish has been shown to induce filopodia formation [54–57] and it was demonstrated that all murine GPM6B isoforms, except those lacking the 4 transmembrane domains, were able to induce filopodia formation [58]. These observations may be relevant since filopodia formation has been implicated in cancer cell migration and adhesion [59, 60]. Thus, the observed over-expression of these proteins may contribute to tumor development through the induction of cell migration. Our data are supported by those of Li et al. who showed that the TAPP2 protein regulates leukemic B-cell migration through the reorganization of their cytoskeleton in response to chemo-attractant stimulation [61]. Also, Scielzo et al. uncovered a role of the HS1 protein in migration, trafficking and homing of leukemic B-cells through impaired cytoskeletal remodeling [62].
We also showed that GPM6A and GPM6B may have oncogenic properties, i.e., exogenous expression of both GPM6A and GPM6B conferred anchorage-independent growth to NIH/3T3 cells. In addition we found that, unlike GPM6A, GPM6B over-expression increases the number of foci in these cells, which is indicative for a loss of contact inhibition of growth. We also found that each of these genes induced the proliferation of NIH/3T3 cells, again suggesting a transforming potential.
In conclusion, we identified a set of genes that may be considered as candidate biomarkers for human B-lymphoid malignancies, although this has to be confirmed in larger patient cohorts. We also showed for the first time that GPM6A isoform 3 and GPM6B isoform 4 may play a role in the development of human B-cell malignancies and, as such, may act as novel potential proto-oncogenes. Obviously, additional research is needed to firmly establish their role in human B-cell leukemia development.
References
A. Graffi, Chloroleukemia of mice. Ann. N. Y. Acad. Sci. 68, 540–558 (1957)
V. Voisin, C. Barat, T. Hoang, E. Rassart, Novel insights into the pathogenesis of the Graffi murine leukemia retrovirus. J. Virol. 80, 4026–4037 (2006)
C. Denicourt, E. Edouard, E. Rassart, Oncogene activation in myeloid leukemias by Graffi murine leukemia virus proviral integration. J. Virol. 73, 4439–4442 (1999)
C. Denicourt, C.A. Kozak, E. Rassart, Gris1, a new common integration site in Graffi murine leukemia virus-induced leukemias: overexpression of a truncated cyclin D2 due to alternative splicing. J. Virol. 77, 37–44 (2003)
C. Denicourt, P. Legault, F.A. McNabb, E. Rassart, Human and mouse cyclin D2 splice variants: transforming activity and subcellular localization. Oncogene 27, 1253–1262 (2008)
C. Charfi, V. Voisin, L.C. Levros Jr., E. Edouard, E. Rassart, Gene profiling of Graffi murine leukemia virus-induced lymphoid leukemias: identification of leukemia markers and Fmn2 as a potential oncogene. Blood 117, 1899–1910 (2011)
C. Charfi, L.C. Levros Jr., E. Edouard, E. Rassart, Characterization and identification of PARM-1 as a new potential oncogene. Mol. Cancer 12, 84 (2013)
X. Castells, J.J. Acebes, S. Boluda, A. Moreno-Torres, J. Pujol, M. Julia-Sape, A.P. Candiota, J. Arino, A. Barcelo, C. Arus, Development of a predictor for human brain tumors based on gene expression values obtained from two types of microarray technologies. OMICS 14, 157–164 (2010)
P. Urban, M. Bilecova-Rabajdova, Z. Stefekova, A. Ostro, M. Marekova, Overview of potential oncomarkers for detection of early stages of ovarian cancer. Klin. Onkol. 24, 106–111 (2011)
S. Landais, S. Landry, P. Legault, E. Rassart, Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 67, 5699–5707 (2007)
M. Ru, C. Shustik, E. Rassart, Graffi murine leukemia virus: molecular cloning and characterization of the myeloid leukemia-inducing agent. J. Virol. 67, 4722–4731 (1993)
E. Edouard, C. Charfi, V. Voisin, C. Denicourt, C. Barat, E. Rassart, The Graffi murine leukemia virus: a multipotent retrovirus ideal for the study and characterization of lymphoid and non lymphoid leukemias. Transw. Res. Netw. 8, 23–47 (2012)
V. Voisin, P. Legault, D.P. Ospina, Y. Ben-David, E. Rassart, Gene profiling of the erythro- and megakaryoblastic leukaemias induced by the Graffi murine retrovirus. BMC Med. Genomics 3, 2 (2010)
F. Sievers, A. Wilm, D. Dineen, T.J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Soding, J.D. Thompson, D.G. Higgins, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
K.C. Chou, H.B. Shen, Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem. Biophys. Res. Commun. 357, 633–640 (2007)
N.H. Colburn, W.F. Bruegge, J.R. Bates, R.H. Gray, J.D. Rossen, W.H. Kelsey, T. Shimada, Correlation of anchorage-independent growth with tumorigenicity of chemically transformed mouse epidermal cells. Cancer Res. 38, 624–634 (1978)
V.H. Freedman, S.I. Shin, Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell 3, 355–359 (1974)
S.I. Shin, V.H. Freedman, R. Risser, R. Pollack, Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl. Acad. Sci. U. S. A. 72, 4435–4439 (1975)
C. Charfi, E. Edouard, E. Rassart, Lymphoid leukemias induced by the murine retrovirus Graffi. http://www.biomed.uqam.ca/rassart/microarray2.html. Accessed 14 November 2013
M. Ikeda, W. Yu, M. Hirai, T. Ebisawa, S. Honma, K. Yoshimura, K.I. Honma, M. Nomura, cDNA cloning of a novel bHLH-PAS transcription factor superfamily gene, BMAL2: its mRNA expression, subcellular distribution, and chromosomal localization. Biochem. Biophys. Res. Commun. 275, 493–502 (2000)
S. Jones, An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226 (2004)
Y. Yasuniwa, H. Izumi, K.Y. Wang, S. Shimajiri, Y. Sasaguri, K. Kawai, H. Kasai, T. Shimada, K. Miyake, E. Kashiwagi, G. Hirano, A. Kidani, M. Akiyama, B. Han, Y. Wu, I. Ieiri, S. Higuchi, K. Kohno, Circadian disruption accelerates tumor growth and angio/stromagenesis through a Wnt signaling pathway. PLoS ONE 5, e15330 (2010)
G. Mazzoccoli, V. Pazienza, A. Panza, M.R. Valvano, G. Benegiamo, M. Vinciguerra, A. Andriulli, A. Piepoli, ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J. Cancer Res. Clin. Oncol. 138, 501–511 (2012)
S.D. Georgatos, F. Gounari, S. Remington, The beaded intermediate filaments and their potential functions in eye lens. BioEssays 16, 413–418 (1994)
M.D. Perng, Q. Zhang, R.A. Quinlan, Insights into the beaded filament of the eye lens. Exp. Cell Res. 313, 2180–2188 (2007)
M.A. Aldahmesh, A.O. Khan, J. Mohamed, F.S. Alkuraya, Novel recessive BFSP2 and PITX3 mutations: insights into mutational mechanisms from consanguineous populations. Genet. Med. 13, 978–981 (2011)
G. Wu, L. Zhou, L. Khidr, X.E. Guo, W. Kim, Y.M. Lee, T. Krasieva, P.L. Chen, A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle 7, 2013–2020 (2008)
Y. Kurasawa, W.C. Earnshaw, Y. Mochizuki, N. Dohmae, K. Todokoro, Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237–3248 (2004)
M. Mazumdar, J.H. Lee, K. Sengupta, T. Ried, S. Rane, T. Misteli, Tumor formation via loss of a molecular motor protein. Curr. Biol. 16, 1559–1564 (2006)
Y.S. Kim, J.D. Hwan, S. Bae, D.H. Bae, W.A. Shick, Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer 10, 576 (2010)
L.L. Yates, C. Schnatwinkel, J.N. Murdoch, D. Bogani, C.J. Formstone, S. Townsend, A. Greenfield, L.A. Niswander, C.H. Dean, The PCP genes Celsr1 and Vangl2 are required for normal lung branching morphogenesis. Hum. Mol. Genet. 19, 2251–2267 (2010)
M. Katoh, Comparative integromics on non-canonical WNT or planar cell polarity signaling molecules: transcriptional mechanism of PTK7 in colorectal cancer and that of SEMA6A in undifferentiated ES cells. Int. J. Mol. Med. 20, 405–409 (2007)
C. Korz, A. Pscherer, A. Benner, D. Mertens, C. Schaffner, E. Leupolt, H. Dohner, S. Stilgenbauer, P. Lichter, Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood 99, 4554–4561 (2002)
A.W. Fjorback, H.K. Muller, O. Wiborg, Membrane glycoprotein M6B interacts with the human serotonin transporter. J. Mol. Neurosci. 37, 191–200 (2009)
S. Olinsky, B.T. Loop, A. DeKosky, B. Ripepi, W. Weng, J. Cummins, S.L. Wenger, Y. Yan, C. Lagenaur, V. Narayanan, Chromosomal mapping of the human M6 genes. Genomics 33, 532–536 (1996)
N.L. Nadon, S. Miller, K. Draeger, M. Salvaggio, Myelin proteolipid DM20: evidence for function independent of myelination. Int. J. Dev. Neurosci. 15, 285–293 (1997)
X. Castells, J.M. Garcia-Gomez, A. Navarro, J.J. Acebes, O. Godino, S. Boluda, A. Barcelo, M. Robles, J. Arino, C. Arus, Automated brain tumor biopsy prediction using single-labeling cDNA microarrays-based gene expression profiling. Diagn. Mol. Pathol. 18, 206–218 (2009)
Y. Yan, C. Lagenaur, V. Narayanan, Molecular cloning of M6: identification of a PLP/DM20 gene family. Neuron 11, 423–431 (1993)
H. Werner, L. Dimou, M. Klugmann, S. Pfeiffer, K.A. Nave, Multiple splice isoforms of proteolipid M6B in neurons and oligodendrocytes. Mol. Cell. Neurosci. 18, 593–605 (2001)
J.L. Popot, D. Pham Dinh, A. Dautigny, Major myelin proteolipid: the 4-alpha-helix topology. J. Membr. Biol. 120, 233–246 (1991)
T. Weimbs, W. Stoffel, Proteolipid protein (PLP) of CNS myelin: positions of free, disulfide-bonded, and fatty acid thioester-linked cysteine residues and implications for the membrane topology of PLP. Biochemistry 31, 12289–12296 (1992)
A. Gow, A. Gragerov, A. Gard, D.R. Colman, R.A. Lazzarini, Conservation of topology, but not conformation, of the proteolipid proteins of the myelin sheath. J. Neurosci. 17, 181–189 (1997)
A.M. Schneider, I.R. Griffiths, C. Readhead, K.A. Nave, Dominant-negative action of the jimpy mutation in mice complemented with an autosomal transgene for myelin proteolipid protein. Proc. Natl. Acad. Sci. U. S. A. 92, 4447–4451 (1995)
E. Swanton, A. Holland, S. High, P. Woodman, Disease-associated mutations cause premature oligomerization of myelin proteolipid protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 102, 4342–4347 (2005)
S. Yang, W. Wang, C. Lei, Q. Liu, F. Xu, X. Xing, H. Chen, J. Liu, S. Wu, M. Wang, Localization and characterization of rat transmembrane protein 225 specifically expressed in testis. DNA Cell Biol. 30, 9–16 (2011)
M. Namiki, M. Kitamura, E. Buczko, M.L. Dufau, Rat testis P-450(17)alpha cDNA: the deduced amino acid sequence, expression and secondary structural configuration. Biochem. Biophys. Res. Commun. 157, 705–712 (1988)
J. Haugstetter, T. Blicher, L. Ellgaard, Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J. Biol. Chem. 280, 8371–8380 (2005)
M. Schweneker, A.S. Bachmann, K. Moelling, JM4 is a four-transmembrane protein binding to the CCR5 receptor. FEBS Lett. 579, 1751–1758 (2005)
M. Wang, G. Luo, F. Li, S. Wu, J. Jiang, C. Huang, Isolation and characterization of a novel four-transmembrane protein PMP22CD specifically expressed in the testis. Int. J. Mol. Sci. 7, 425–437 (2006)
H. Ohno, J. Stewart, M.C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, J.S. Bonifacino, Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269, 1872–1875 (1995)
J.S. Bonifacino, E.C. Dell’Angelica, Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 145, 923–926 (1999)
B.D. Trapp, A. Nishiyama, D. Cheng, W. Macklin, Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 137, 459–468 (1997)
M.P. Sinoway, K. Kitagawa, S. Timsit, G.A. Hashim, D.R. Colman, Proteolipid protein interactions in transfectants: implications for myelin assembly. J. Neurosci. Res. 37, 551–562 (1994)
J. Alfonso, M.E. Fernandez, B. Cooper, G. Flugge, A.C. Frasch, The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc. Natl. Acad. Sci. U. S. A. 102, 17196–17201 (2005)
B. Fuchsova, M.E. Fernandez, J. Alfonso, A.C. Frasch, Cysteine residues in the large extracellular loop (EC2) are essential for the function of the stress-regulated glycoprotein M6a. J. Biol. Chem. 284, 32075–32088 (2009)
M.A. Brocco, M.E. Fernandez, A.C. Frasch, Filopodial protrusions induced by glycoprotein M6a exhibit high motility and aids synapse formation. Eur. J. Neurosci. 31, 195–202 (2010)
K.Y. Huang, G.D. Chen, C.H. Cheng, K.Y. Liao, C.C. Hung, G.D. Chang, P.P. Hwang, S.Y. Lin, M.C. Tsai, K.H. Khoo, M.T. Lee, C.J. Huang, Phosphorylation of the zebrafish M6Ab at serine 263 contributes to filopodium formation in PC12 cells and neurite outgrowth in zebrafish embryos. PLoS ONE 6, e26461 (2011)
M.E. Fernandez, J. Alfonso, M.A. Brocco, A.C. Frasch, Conserved cellular function and stress-mediated regulation among members of the proteolipid protein family. J. Neurosci. Res. 88, 1298–1308 (2010)
F. Xue, D.M. Janzen, D.A. Knecht, Contribution of filopodia to cell migration: a mechanical link between protrusion and contraction. Int. J. Cell Biol. 2010, 507821 (2010)
A. Arjonen, R. Kaukonen, J. Ivaska, Filopodia and adhesion in cancer cell motility. Cell Adhes. Migr. 5, 421–430 (2011)
H. Li, S. Hou, X. Wu, S. Nandagopal, F. Lin, S. Kung, A.J. Marshall, The tandem PH domain-containing protein 2 (TAPP2) regulates chemokine-induced cytoskeletal reorganization and malignant B cell migration. PLoS ONE 8, e57809 (2013)
C. Scielzo, M.T. Bertilaccio, G. Simonetti, A. Dagklis, E. ten Hacken, C. Fazi, M. Muzio, V. Caiolfa, D. Kitamura, U. Restuccia, A. Bachi, M. Rocchi, M. Ponzoni, P. Ghia, F. Caligaris-Cappio, HS1 has a central role in the trafficking and homing of leukemic B cells. Blood 116, 3537–3546 (2010)
Acknowledgments
We thank Dr. Daniel Sinnet for providing the pediatric tumor samples, Bertrand Fournier for his help with the statistical analysis and Denis Flipo for his help with the confocal microscopy analyses. This work was supported by the Canadian Institutes of Health Research, grant MOP-37994 (ER). CC is a recipient of a studentship from the Tunisia Government and Fondation UQAM.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource Fig. 1
Alignment of deduced amino acid sequences for GPM6A isoforms together and for GPM6B isoforms together. Alignment a of the 5 Homo sapiens isoforms of GPM6A (NP_005268 (isoform 1), NP_963885 (isoform 1), NP_001248376 (isoform 2), NP_963886 (isoform 3) and NP_001248377 (isoform 4)) and b of the 4 homosapiens isoforms of GPM6B protein (NP_001001995 (isoform 1), NP_005269 (isoform 3), NP_001001996 (isoform 2) and NP_001001994 (isoform 4)) using the Clustal Omega software. Stars indicate a perfect match between all aligned isoforms and dashed lines indicate gaps. (PDF 30 kb)
Online Resource Fig. 2
Alignment of GPM6A, GPM6B and DM20 proteins. The amino acid residues of GPM6A isoform 3, GPM6B isoform 4 and DM20 proteins were aligned in pairs to determine their identity percentage. a Alignment of GPM6A isoform 3 and GPM6B isoform 4. b Alignment of DM20 and GPM6A isoform 3. c Alignment of DM20 and GPM6B isoform 4. Boxes: N- and C- cytoplasmic tails, dotted lines: transmembrane domains, lines: extracellular loops, double lines: intracellular loop. (PDF 48 kb)
Rights and permissions
About this article
Cite this article
Charfi, C., Edouard, E. & Rassart, E. Identification of GPM6A and GPM6B as potential new human lymphoid leukemia-associated oncogenes. Cell Oncol. 37, 179–191 (2014). https://doi.org/10.1007/s13402-014-0171-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-014-0171-y