Abstract
Out of 61 fungal isolates, Geotrichum candidum GAD1 isolated from a marine algal sample belonging to the coastline of Abo Qir, Alexandria, Egypt, that was molecularly identified with accession no. MN638741 is the most potent strain producing cellulase degrading carboxymethylcellulose salt (CMC). Dox medium was optimized using OFAT design with CMC as the sole carbon source giving high productivity of cellulase enzyme with incubation period 6 days at 180 rpm, at pH 7, and 30 °C. Ammonium sulfate of fraction 50% gave the highest specific activity 209.1 U/mg and gave a single protein band (72 KDa) using SDS–PAGE. Maximum purified cellulase activity (49.78 U/mL) was reported at pH 7, 45 °C, and 10% salinity and was stable between (40–60 °C) and (pH 4.0–10.0). In addition, different inhibitors and activators were tested, and subsequently, it was reported that all tested heavy metal ions (Fe2+, Co2+, Mg2+, Ca2+, Na+, Mn2+, and K+) were significantly affected, especially, Fe2+ (10 mM) giving a double-fold increase in cellulase activity, while EDTA, petroleum ether, and n-Hexane exerted a suppression action. The kinetic Vmax and Km for CMC degradation were 238.09 µM/min and 0.42 mg/mL, respectively. Optimized purified cellulase approaching bioethanol production from pretreated rice straw and CMC as substrates was achieved with maximum bioethanol yield of 11.9 g g−1 and 13.2 g g−1, respectively, and conversion efficiency by GC–MS; 90.4% and 87.4%, respectively. Conclusively, the presented cellulase profile could be applied as an efficient alternative biofuel source as renewable energy under different industrial scales.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Although microbes are considered as deteriorating bioagents as in the case of post-harvest infections of crops [1], sometimes microorganisms are considered as sources of valuable compounds such as PUFAs from the oleaginous fungi [2], lipids [3], and biodiesel [4]. As well, microbes may act as biodegraders for toxic substances such as PAHs [5] and producers for intracellular and extracellular enzymes. Enzymes are proteins biomolecules that act as a biocatalyst (the compound that speeds the reaction process without introducing in the reaction) to a biological or chemical reaction [6]. Many factors were induced in the enzyme activity as temperature, substrate concentrations, pH, inhibitor, and cofactor so, it's essential to adjust this factor to increase the enzyme activity and to determine if this enzyme is appropriate for the production of certain industries or not.
Enzymes play an important role in the industrial world especially cellulase. Cellulase enzymes are the most important industrial enzyme due to their versatile applications in various industries such as bioethanol production, paper, food industries, and the detergent industry. Cellulose can hydrolyze by synergistic actions of cellulases (endo-βeta-1,4 glucanase, cellobiohydrolase, and βeta-D-glucosidase) into glucose units [7].
In previous studies, cellulase was produced by actinomycetes, fungi, and bacteria but fungi are the most common production of cellulase [8] because cellulase produced by fungi is structurally less complicated, more desirable, and resistant to hard ecological conditions. Cellulase productivity of fungal origin is higher than other organisms due to their hyphae penetrating cellulose compound mechanically to degrade them producing a high amount of cellulase [9].
Fungi especially marine fungi become a well-known desirable source to obtain products [10]. Fungal genera as Chaetomium spp., Alternaria spp., Rhizopus spp. Penicillium spp. and Aspergillus spp., can degrade and deteriorate the cellulose chains. G. candidum was classified as a subphylum of Saccharomycotina within the phylum of Ascomycota, Fungi Kingdom. Some strain of G. candidum has the ability of cellulolytic activities which can grow on filter paper, wooden cheese boxes, and cotton [11]. However, a few investigations on cellulase production from G. candidum has been done. On the other hand, the high cost of production of cellulases is the main limitation to producing it industry [12].
Due to the high cost and unfriendly environment of gas, the scientists in the last recent year working on the production of bioethanol from biomass [13]. Production of ethanol from biomass lignocellulose especially rice straw by the biological process is considered the most important application of the cellulase enzyme due to its advantage as a renewable alternative fuel resource [14]. However, the increase in biomass, especially rice straw, in some countries is abundant because the farmers burned it in the open air that leads to problems in health and the environment. Therefore, the current study, aims to explore a high potential strain of fungal strain that has the ability to produce a massive amount of cellulase in order to produce bioethanol from crop wastes like rice straw, besides, increasing cellulase activity by optimization the condition of fermentation, enzyme characterization, and lastly, Previous studies [12, 15,16,17] were used distilled water to produce cellulase, which in turn lead to high-cost production, therefore, the current study depended on the seawater in the production process to benefit from the low cost of seawater and as a substitute to freshwater.
2 Materials and Methods
Chemicals and reagents were procured from Sigma-Aldrich, Egypt, except for AccuPrep PCR DNA Purification Kit (K-3034-1, Bioneer Corporation, South Korea) and DNA STAR SeqMan (DNA STAR Incorporation, Wisconsin, USA).
2.1 Screening of Cellulase-Producing Fungal Isolates
Sixty-one fungal isolates were isolated from different marine sources (sediments, water, algae, and wood) from the Egyptian Mediterranean Sea (Alexandria coastline). All isolates were investigated to check their abilities to digest cellulose, according to [18], which estimate the capability of fungal isolates production cellulase qualitatively. The 61 fungal isolates were inoculated on agar plates containing modified seawater Czapek’s agar medium (6 g/l Na2HPO4, 2 g/l KH2PO4, 0.5 g/l NaCl, 0.5 g/l MgSO4 7H2O, 0.3 g/l CaCl2, 18 g/l agar) plus 0.1% (CMC). pH 5 of the medium solution was attuned. Plugs of about 5 mm diameter were mounted in the center of Petri dishes. A plate-free specimen was used as a negative control. Post-incubation of 5 days at 30 °C, all Petri dishes were further kept at 50 °C for about 1 h. Later, the plates were stained by Congo red (0.1% Congo red in distilled water) and agitated at 100 rpm for 20 min on an orbital shaker at 30 °C. Congo red solution was removed, and all plates were rinsed with 1 Molar NaCl to reveal the contrast-colored hole of degradation. The degradative halo-zone and the colony diameter were then determined. The clear zone diameter around the fungal colony on CMC agar was measured with three replicates. The cellulose degradation coefficient (CDC) was calculated as follows: CDC = (degradative halo / colony diameter) [19].
2.2 Quantitative Analysis of Cellulase Activity
All fungal isolates of positive response against cellulose substrate were selected for quantitative screen regarding the enzymatic activity. Sterilized 50 mL of modified seawater Czapek–Dox broth medium (6 g/l Na2HPO4, 2 g/l KH2PO4, 0.5 g/l NaCl, 0.5 g/l MgSO4 7H2O, 0.3 g/l CaCl2, 18 g/l agar) amended with 0.1% (CMC) at pH 5 was distributed into separate 250-mL conical flasks. The flasks were injected with fungal spores and incubated at 30 °C on a rotary shaker at 180 rpm for 5 days. After 5 days, culture filtrate was collected, centrifuged at 5000 rpm for 20 min and mycelial dry weight was estimated after drying at 60 °C overnight in an oven until constant weight. The supernatant was used to determine enzyme production and estimation extracellular protein content.
2.3 Estimation of Total Protein
Secreted enzyme protein was measured by Lowry steps protocol [20]. Post the incubation of 6 days, the medium supernatant was separated by centrifugation (5000×g for 15 min) to detach the fungal mycelial mat from the liquid culture. An aliquot of this culture was used for determining the protein content. Protein standard was tested by bovine serum albumin. 200 u of separate samples were mixed with 1 mL of alkaline solution. After 10 min, 100 u of appropriately diluted Folin–Ciocalteu reagent was added. After 20 min, the color developed was read at 750 nm by using the spectrophotometer.
2.4 Enzyme Assay
The cellulase activity measurement was guided by the dinitrosalicylic acid (DNS) steps protocol, where the liberated reducing sugar was measured upon the enzyme hydrolysis activity in CMC 0.2% substrate. The volume of 0.5 mL supernatant and 0.5 mL of 0.2% substrate were incubated at 50 °C for 30 min. After incubation, 1 mL of 3,5 dinitrosalicylate (DNS) acid was added into the solution and boiled for 10 min and cooled for 10 min at room temperature to measure with a spectrophotometer at λ 570 nm. One unit (IU) of cellulase activity was defined as the amount of enzyme releasing 1 uM of reducing sugar per min.
2.5 Identification of the Promising Fungal Isolate
2.5.1 Morphological Characterization and Microscopic Examination
Fungal culture was identified based on the macroscopic features and microscopic examination by aiding [1, 21] and following the taxonomic arrangement proposed in the 2nd Edition, Identification of Pathogenic Fungi [22].
2.5.2 Molecular Identification
DNA of the promising fungal species was extracted and the PCR amplifications were carried out using the primers ITS1 (5′ TCC GTA GGT GAA CCT GCG G 3′), ITS4 (5′ TCC TCC GCT TAT TGA TAT GC 3′) [23]. Amplified DNA was purified using AccuPrep PCR DNA Purification Kit (K-3034-1, Bioneer Corporation, South Korea). The PCR products were purified with the ExoSAP method and Sanger sequencing with PCR primers was performed at Macrogen Incorporation (Seoul, South Korea). The sequences were assembled using the DNA STAR SeqMan (DNA STAR Incorporation, Wisconsin, USA). Generated sequences firstly were submitted to the GenBank database. Sequence analyses obtained by BLASTN similarity search at the website (http://www.ncbi.nlm.nih.gov/BLAST) and secondly were multiple aligned and the phylogenetic tree (Neighbor-joining) constructed by maximum-likelihood method using MEGA_X_10.1.6 software.
2.6 Optimization of Culture Conditions
The most promising cellulolytic isolate was selected for further studies to optimize the culture conditions for increasing cellulase activity. The optimum pH (5, 6, 7, 8), optimum temperature (20, 25, 30, 40 °C), shaking (120, 180) rpm, static condition, and optimum incubation time from 2 to 7 days were determined.
2.7 Cellulase Purification
Cellulase was purified by precipitation (either by ammonium sulfate, ethanol, and acetone) with different fractions.
2.7.1 Precipitation Step
Ammonium sulfate fractionation and fractional precipitation by acetone and ethanol as well were applied for such procedures. The supernatant (about 100 mL) was precipitated at 4 °C using different concentrations of ammonium sulfate to obtain different fractions at 25%, 50%, 75%, and 90% saturation. The protein deposit was obtained by centrifugation (5000×g for 25 min), dissolved in a defined volume of distilled water, and dialyzed overnight in a refrigerator with constant stirring [24]. On the other hand, cooled acetone or ethanol was added to the supernatant slowly with constant stirring. Enzyme fractions were obtained at concentrations of acetone or ethanol (as in ammonium sulfate). The precipitate was obtained by centrifugation (5000×g for 25 min) overnight at 4 °C. Protein content and cellulase activity of each fraction of ammonium sulfate, ethanol, and acetone were estimated.
2.7.2 SDS–PAGE
SDS–PAGE was performed to determine the molecular weight of the purified cellulase as was described by [25].
2.8 Cellulase Characterization
2.8.1 Effect of Different Enzyme Concentrations and Substrate Concentrations
The effect of enzyme concentrations and substrate concentrations on the activity of the enzyme was determined by incubating their action mixture at enzyme concentrations ranging from 0.1 to 1% and substrate concentration ranging from 0.1 to 5% for 30 min. The cellulase activity was reported as guided before.
2.8.2 Effect of Temperature on the Activity and Stability of Cellulase
The action mixture was incubated at temperatures ranging from 25 to 70 °C for 30 min to determine the effect of temperature on the activity of the enzyme. To determine thermal stability, cellulase was incubated solitary at different Celsius degrees ranging from 40 to 70 °C for 120 min. After that, the substrate was added and the cellulase activity was reported at 15-min intervals.
2.8.3 Effect of pH on the Activity and Stability of Cellulase
2% CMC was prepared in different buffers (pH 2.0–10.0) and incubated with the enzyme to determine the effect of pH on cellulase activity. To determine pH stability, crude cellulase was incubated without substrate at pH ranging from (pH 2.0–10.0) for 120 min at room temperature. After that, the substrate was added and the activity of cellulase was measured using standard assay procedures at 15-min intervals.
2.8.4 Effect of Salinity on the Activity of Cellulase
To determine the effects of salinity on the activity of cellulase, cellulase was incubated without substrate at different salinity concentrations ranging from 1 to 30% for 120 min at room temperature and the activity of cellulase was measured using standard assay procedures.
2.8.5 Cellulase Kinetics Determination
To calculate Km (Michaelis–Menten constant) and Vmax (maximum velocity), different CMC concentrations (0.1–2.5% (w/v)) were incubated with cellulase enzyme in pH 7 at 45 °C and plotted by Lineweaver–Burk plot.
2.8.6 Activator and Inhibitor of Cellulase Activity
The effects of metal chloride ion (Fe2+, Cr2+, Cu2+, Co2+, Ni2+, Mg2 + , Ba2+, Ca2+, Na+, Mn2+, and K+), different group-specific reagents (SDS and EDTA), and solvents (petroleum ether and n-Hexane) on cellulase metabolic pathway were examined by adding different concentrations of 1 mM and 10 mM of each item to the reaction mixture for 2 h. Later, the activity was reported as guided before.
2.9 Enzymatic Hydrolysis of Biomasses and Quantification of Released Sugars
2.9.1 Physical Pretreatment of Rice Straw
Rice straw was treated physically by cutting 10 gm of rice straw into small parts and washed with distilled water then put in microwave for 5 min.
2.9.2 Bioethanol Production
Characterized cellulase enzyme was added separately to two flasks, one contained 10% (v/v) pretreated rice straw and another flask contained 2% (v/v) CMC. The two flasks were incubated in the water bath for 250-min intervals at 45 °C. After the incubation period, the total reducing sugars (TRS) were determined in the two flasks by DNS as mentioned before. The hydrolysate reaction mixture of incubated (rice straw with cellulase enzyme) and (2% CMC with cellulase enzyme) were transferred to bioethanol production medium (peptone (20.0 g/L), Yeast extract (10.0 g/L) and adjusted pH to 5.5 according to [26] followed by autoclaving at 121 °C for 20 min. 3 mL of prepared baker’s yeast (5 gm commercial (baker’s yeast) inoculated with 0.1% peptone water and kept at 35 °C for 20 min) was inoculated and incubated flasks at 30 °C for 48 h. After incubation, the fermented medium was evaporated by an oven evaporator at 50 °C. The ethanol yield was calculated and estimated by the dichromate reduction method described by [27]. At 660 nm, the standard curve was plotted with different ethanol concentrations from 2 to 20% which ethanol concentration in the sample was calculated from it. TRS and ethanol concentration were estimated at 48 h intervals.
The concentration of bioethanol was analyzed by the GC–MS (Thermo Scientific TSQ 9000–1 triple quadrupole GC–MS, UK). The Column (DB-5MS 30 m × 0.250 mm × 0.25 μm, Agilent, USA) was employed as a separation column, and the Chromeleon 7, Version 7.2.10., Thermo Fisher Scientific mass spectrometry software was worked according to the procedure of [28, 29]. The standard curve was plotted with different ethanol concentrations from 10 to 30% and the concentration of ethanol was estimated with the internal standard value of ethanol analysis by GC–Mass. The ethanol was identified with retention times (Rt) and peak range.
2.9.3 Statistical Analyses
The results are presented as the mean value ± one standard deviation (SD) of triplicate readings by using the Prism7 program with required controls of each experiment.
3 Results and Discussion
3.1 Fungal Screening for Cellulase Production
In nature, fungi associated with marine plant debris have many enzymes that are important for their survival which secret those enzymes such as cellulase for hydrolyzing the polysaccharide of plants producing oligosaccharides and sugars for metabolic activities. However, a few fungal communities have such ability adequately. Accordingly, the study focused on screening and finding the most potent cellulase degrading CMC for purification and industrial application.
Out of the total 61 marine fungal isolates, 46 isolates generated clear zones with varied (CDC) values within the range 1.29–2.61 on Congo red CMC agar plate, and AG1 isolate had the highest CDC value (2.61). Closely related, previous studies [19, 30, 31] have been determined CDC from different bacterial species (Bacillus licheniformis, Bacillus mojavensis, and Bacillus sp.) with radius ranges between 2.33 and 2.5. However, Goel et al. [32] have reported a CDC value (2.8) slightly higher than that achieved in the current study, while Saroj et al. [33] have reported lower values than the achieved one (1.50 and 1.24) from A. fumigatus JCM 10,253 and Aspergillus terreus, respectively. As a preliminary detection test for cellulase activity according to [34], the CDC value for isolate AG1 was selected as the most active one with the highest clear zone radius.
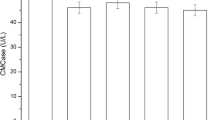
As well, protein content and cellulase activity determined for each isolate revealed that protein content ranged from 0.21 to 0.51 mg/mL and cellulase activity ranged from 6.85 to 58.07 U/mL, and AG1 isolate had the highest activity 58.07 U/mL, and protein content 0.5 mg/mL (Fig. 1) as well. Reported cellulase activity in current work using DNS achieved a higher value than other findings; 0.01 U/mL [19], 0.079 U/mL [8] but close to 34.740 U/mL [32] from Pseudomonas sp. and, 26.2 IU/mL [33] from A. fumigatus JCM 10,253. Accordingly, isolate AG1 was selected for proceeding.
3.2 Identification of the Most Promising Marine Fungal Isolate
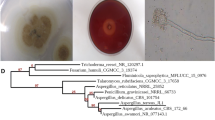
AG1 was identified based on morphological and molecular identification as G. candidum GAD1. The sequence was submitted to GenBank and the accession number was provided as MN638741. The obtained sequence showed 100% similarity with G. candidum (Fig. 2).
[35, 36] have reported G. candidum from different marine resources but without any characterization. So far, a few investigations on cellulase from G. candidum has been done.
3.3 Optimization of Culture Conditions
As shown in Fig. 3, optimization of culture conditions revealed that incubation period 6 days in shaken condition (180 rpm) at pH 7 and 30 °C were the optimum conditions giving maximum cellulase activity for each factor 57.27 U/mL, 57.27 U/mL, 56.9 U/mL, and 58.56 U/mL, respectively.
However, it has been noted that at optimum temperature the movement of the carbon source and the microorganisms increases the kinetic energy and enzyme productivity [37], but also, the higher temperature has been impressed a negative effect on the cellulase production [38], who has reported temperature 40 °C as the optimum one for cellulase production and activity from E. coli EgRK2. However, Goel et al. [32] have succeeded in producing cellulase from filaments fungus at 50 °C. The maximum cellulase production by G. candidum strain Gad1 at initial pH was reported at pH 7. This result was in agreement with [32] that produced cellulase from Sporothrix carnis at initial optimum pH 7.0. Also, [39] has produced cellulase from filamentous fungus Acremonium cellulolyticus after 48 h at pH 6. Previously reported by [5] that the activity of cellulase was reached the optimum activity at 6 days and shaken condition at 180 rpm that most likely provided an increase in the distribution of oxygen level and fungal mycelia in the medium.
The current study succeeded in the production of cellulase from seawater in the production process benefit from the low cost of seawater and as a substitute for freshwater. On the contrary, previous studies [12, 15,16,17] were used distilled water to produce cellulase, which in turn led to high-cost production.
3.4 Cellulase Purification from G. candidum Strain Gad1
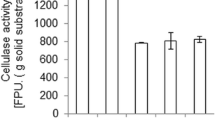
The main purpose of using a method of precipitation is to concentrate the target molecule. With ammonium sulfate precipitation, more than 55.3% of total activity was recovered with maximum specific activity reach to 209.14 U/mg in the 50% saturation fraction. The cellulase activity recovery increased up to 27.5% and 28.93% for acetone and ethanol, respectively, with maximum specific activities for acetone (138.15 U/mg) and ethanol (192.8 U/mg) at a ratio of saturation fraction 70% and 50%, respectively (Fig. 4). Accordingly, the yield of partially purified cellulase produced by 50% ammonium sulfate fraction was subjected to SDS–PAGE examination.
3.5 SDS–PAGE
Figure 5 shows that ammonium sulfate fraction 50% gave a one definite protein band at 72 KDa indicating that cellulase enzyme was purified and ready for further characterization.
This result has been found matching with findings obtained by Goyari et al. [40, 41] who produced cellulase from Talaromyces verruculosus SGMNPf3 and Escherichia coli, respectively, but close to other findings (48, 55, 58, and 60 KDa) produced from Bacillus sp., Aspergillus flavus (TISTR 3637), E. coli BPPTCC-EGRK2 and Pseudomonas sp. by [6, 31, 32, 40], respectively. Unlikely, [42, 43] have reported cellulase molecular weight at 20 and 32 KDa from Bacillus licheniformis 380 and Bacillus subtilis YJ1, respectively.
3.6 Cellulase Characterization
3.6.1 Effect of Different Enzyme Concentrations and Substrate Concentrations
Figure 6a indicates that the cellulase activity was increased proportionally with increasing cellulase concentration. Accordingly, 1 mL of cellulase activity equivalent to 0.17 mg/mL was selected for proceeding. As shown in Fig. 6b, the highest enzyme activity (33.46 U/mL) peak for the purified cellulase was obtained at 2% CMC concentration, afterward, followed by a gradual decline pattern, which may refer to elevated viscosity of the substrate. This result of the substrate concentration was in agreement with the previous record by [6], but higher substrate concentration than achieved by [7], who reported 1% concentration.
3.6.2 Cellulase Kinetic Estimation
The kinetic Km and Vmax of cellulase were calculated with different CMC concentrations (0.1–2.5% (w/v)) in pH 7 at 45 °C and plotted by Lineweaver–Burk plot. Figure 7 shows that the Vmax for CMC degradation by cellulase of G. candidum strain Gad1 was 238.09 μM/min, while Km was 0.42 mg/mL that has appeared lower than other fungi such as Aspergillus flavus (Km value of 3.0 mg/mL), which reported by [44]. Pachauri et al. [45] have suggested that the genetic variability of fungi could be the reason for different km with different fungi. The decrease in Km value suggested that the cellulase produced by G. candidum strain Gad1 has a high affinity with CMC substrate [31].
3.6.3 Optimum Enzyme Reaction pH, Temperature, and Salinity for Cellulase Activity
As shown in Fig. 8, the optimum temperature, pH, and salinity reported (pH 7, 45 °C and 10%) with maximum cellulase activity value (36.46 U/mL, 45.78 U/mL, and 49.78 U/mL), respectively, giving by the end 1.39-fold the value (35.75 U/mL) of starting basic conditions (pH 6.0, 50 °C and 0% salinity). The optimal temperature for the purified cellulase was 45 °C. this result has been matching with those obtained by [45], but higher than (20 °C) that obtained by [14] from Chaetomium sp. and lower than 60 °C and 50 °C that obtained by [31, 32, 38, 43, 44, 46] from Bacillus subtilis YJ1, Trichoderma sp. strain IS-05, E. coli EgRK2, Aspergillus flavus (TISTR 3637), Bacillus sp. and Pseudomonas sp., respectively. Maximum cellulase activity has been found at pH 7.0 and matches with the results obtained by [6, 7, 32] from Paenibacillus sp., E. coli, and Pseudomonas sp., but higher than pH 4.8 and pH 5.0 which reported by [17, 45] using Trichoderma longibrachiatum and Chaetomium sp., respectively.
While the achieved pH has been lower than pH 9 and pH 10.0 reported by [42, 44] using Aspergillus flavus (TISTR 3637) and Bacillus licheniformis 380, respectively. The addition of NaCl concentration (more than 10%) did cause a decrease in enzyme activity. This finding is in agreement with [44] finding who indicates that more than 10% NaCl concentration strongly inhibited the cellulase activity produced by the Aspergillus flavus strain (TISTR 3637).
3.6.4 Thermal Stability and pH Stability of Cellulase
Heat-resistant enzymes production permits enzymatic reactions at elevated temperatures. Consequently, increasing substrates solubility, conversion rates, minimizing the risk of microbial contamination, and the viscosity of the medium reaction would be achieved [47].
As depicted in Fig. 9a, the thermal stability of cellulase ranged from 40 to 60 °C approximately was steady for 2 h, however, a slight decrease in cellulase activity with each elevation of temperature set, while at 70 °C the cellulase was lost more than half of the activity after 2 h. Cellulase was stable between pH 4.0 and 10.0 (Fig. 9b) for 2 h exposure. However, the enzyme activity had no resistance for environment pH less than 4. This result was almost similar to the previous research done by [8]. This result indicates that cellulase from G. candidum strain Gad1 could be used in acidic, neutral, and alkaline environments, which in turn could be available for large-scale industrial processes. Cellulase activity in the acidic pH range was reported in a few works of literature [46]. The activity and stability of cellulase enzyme depend strongly on the pH value of the solution reaction mixture. As indicated by [48], the increase in enzyme activity at the optimum pH range was due to a reaction that involved deionization or ionization of basic or acidic groups in the active center in the protein of the enzyme.
3.6.5 Effect of Activators and Inhibitors on Cellulase Activity
Metal ions are the key elements of the most enzymatic reaction, where the catalytic process was proceeded through donating or attracting electrons. Some metal ions bind the substrate by links called coordination. Others maintain the quaternary and tertiary structures of the enzyme molecule [44]. As shown in Fig. 10, most tested metal ions (Fe2+, Co2+, Mg2+, Ca2+, Na+, Mn2+, and K+) increased the cellulase activity with both concentrations especially Fe2+ which increased the enzyme activity 200% (90.2 u/mL) at 10 mM. This suggests that (Fe2+, Co2+, Mg2+, Ca2+, Na+, Mn2+, and K+) conjugate with certain amino acid residues in the active site of the halo cellulase enzyme, lead to a conformational change which promotes increasing the activity of the cellulase [49].
While Cu2+, Ni2+, and Ba2+ decreased the cellulase activity with all concentrations. Cr2+ as metal ion and SDS as biosurfactant had a neutral effect on the activity of cellulase. However, the EDTA and two solvents (petroleum ether and n-Hexane) deactivated the cellulase activity suggesting that EDTA and two solvents (petroleum ether and n-Hexane) worked as a chelating agent therefore, must be avoided in future cultivations for increasing the cellulase production.
3.7 Bioethanol Production
3.7.1 Reaction Mixture Yield Course Time
Reaction mixture yield course time is a key factor for evaluating the cellulase activity reciprocal to sugar yield in a sense of converting insoluble glucose polymer in a substrate to soluble forms of glucose oligosaccharides that fermented by yeast to bioethanol production. Upon the outcome of time course results of cellulase activity with CMC salt by G. candidum strain Gad1 presented in Fig. 11, after 105-min incubation, the maximum cellulase activity was achieved (55.54 u/mL). This result encouraging to compare this result with that found by [50] who showed that maximum rice straw hydrolysis by cellulase of A. niger strain BK01 occurred after 2.5 h.
3.7.2 Reducing Sugars Before and After Fermentation
In general, the ethanol produced by yeasts such as Saccharomyces and Kluyveromyces lack cellulase expression, therefore, fungi that have the cellulolytic ability as Chaetomium, Aspergillus, and Trichoderma are used for the production of cellulase to bioethanol production. Hence, in the present study purified cellulase from G. candidum strain, Gad1 was directly administered to degrade glucose backbone present in CMC time and from pretreated rice straw another time to produce soluble sugar that used in bioethanol fermentation media, which in turn would decrease the time of bioethanol production. Pretreatment of rice straw is essential to remove lignin and cellulose become ready to degradation by an enzyme to glucose which is fermented by yeast to ethanol [51]. Maximum rice straw and CMC hydrolysis by cellulase of G. candidum strain Gad1 were 30.2 g/L and 26.6 g/L, respectively. However, in the present study, the results of rice straw hydrolysis by cellulase from G. candidum strain Gad1 don’t support the previous study [11] that showed three strains of G. candidum lack of growth on wheat straw. The belated onset of CMC and rice straw utilization could be referred to the higher affinity of the cellulose transporter to glucose [52].
3.7.3 Determination of Bioethanol by Dichromate Reduction Method
As shown in Fig. 12, the amount of reducing sugar decreased significantly at the end of the fermentation process with maximum bioethanol production pointing to a direct relationship between the reducing sugar and the amount of bioethanol produced during the fermentation process. Consequently, by using the dichromate reduction method and a calibration curve of ethanol standards. Straw and CMC were hydrolyzed producing 17.5% and 16.3% ethanol, respectively. As depicted in Table 1, ethanol yield (g/g of sugar/L) by fermentation of pretreated rice straw and CMC using cellulase enzyme produced by G. candidum strain Gad1 were 11.5 and 12.2 g/g of sugar/L, respectively. This result of Ethanol yield, higher than 0.42, 0.125, and 0.137 g/L reported by [51, 53, 54] from fermentation rice straw, rice bran, and banana peel, respectively, using Trichoderma reesei F-418 B., licheniformis strain AMF-07 and Klebsiella sp. SWET4, respectively.
3.7.4 Evaluation of Bioethanol Production by GC–MS Analysis
The bioethanol produced by the degradation of CMC and rice straw from the fermented medium was collected from the oven evaporator and injected in GC–MS for determination of bioethanol concentration (Fig. 13). The chromatogram peak of the produced bioethanol by the degradation of CMC and rice straw compatible with standard one was observed at RT of 1.73.1 min giving the concentration 15.7% and 20%, respectively, and ethanol yield 13.2 and 11.9 g/g of sugar/L, respectively. Similarly [15] have been reported ethanol concentration from rice straw using Aspergillus terreus F98 and Trichoderma viride F94 within a range between 15.36 and 12.85%, respectively. While the achieved ethanol concentration has been higher than 2.67% and 0.56% reported by [55] from corncob using A. niger and P. decumbens, respectively. Finally, no significant differentiation between the determination of bioethanol by GC–MS analysis and the dichromate reduction method indicating assured the potentiality of such enzyme.
4 Conclusion
The fungal world still provides the universe with interesting benefits. During this study, a promising marine yeast isolate G. candidum strain Gad1 approached a perceptible capability to produce cellulase from the agricultural waste (rice straw) as a model for sustainable production of an interesting enzyme as an efficient versatile tool for having different industrial applications. Medium optimization and cellulase characterization have succeeded in maximizing the cellulase biosynthesis with cellulolytic activity of CMC and rice straw to sugar that is subsequently fermented to bioethanol as an alternative source of the renewable energy. Eventually, the study succeeded to achieve two important goals serving the idea of sustainable development; production of bioethanol as renewable fuel from the agricultural waste as an inspiring waste management tool.
References
Suleiman, W.B.; El Bous, M.M.; El Said, M.; El Baz, H.: In vitro evaluation of syzygium aromaticum l. Ethanol extract as biocontrol agent against postharvest tomato and potato diseases. Egypt. J. Bot. 59, 81–94 (2019). https://doi.org/10.21608/ejbo.2018.3838.1180
Hashem, A.H.; Hasanin, M.S.; Khalil, A.M.A.; Suleiman, W.B.: Eco-green conversion of watermelon peels to single cell oils using a unique oleaginous fungus: lichtheimia corymbifera AH13. Waste Biomass Valoriz. 11, 5721–5732 (2019). https://doi.org/10.1007/s12649-019-00850-3
Hashem, A.H.; Suleiman, W.B.; Abu-elreesh, G.; Shehabeldine, A.M.; Khalil, A.M.A.: Sustainable lipid production from oleaginous fungus Syncephalastrum racemosum using synthetic and watermelon peel waste media. Bioresour. Technol. Rep. 12, 100569 (2020). https://doi.org/10.1016/j.biteb.2020.100569
Hashem, A.H.; Suleiman, W.B.; Abu-Elrish, G.M.; El-Sheikh, H.H.: Consolidated bioprocessing of sugarcane bagasse to microbial oil by newly isolated oleaginous fungus: Mortierella wolfii. Arab. J. Sci. Eng. 46, 199–211 (2021). https://doi.org/10.1007/s13369-020-05076-3
Abdel-Razek, A.S.; El-Sheikh, H.H.; Suleiman, W.B.; Taha, T.H.; Mohamed, M.K.: Bioelimination of phenanthrene using degrading bacteria isolated from petroleum soil: safe approach. Desalin. Water Treat. 181, 131–140 (2020). https://doi.org/10.5004/dwt.2020.25109
Haryanto, M.G.; Setyahadi, S.; Sahlan, M.; Yohda, M.: Characterization of cellulase from E. coli BPPTCC-EGRK2. E3S Web Conf 52, 00024 (2018). https://doi.org/10.1051/e3sconf/20185200024
Islam, F.; Roy, N.: Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res. Notes. (2018). https://doi.org/10.1186/s13104-018-3558-4
Ariffin, H.; Abdullah, N.; Shirai, Y.: Production and characterization of cellulase by Bacillus pumilus EB3. Int. J. Eng. Technol. 3(1), 47–55 (2006)
Imran, M.; Anwar, Z.; Irshad, M.; Asad, M.J.; Ashfaq, H.: Cellulase production from species of fungi and bacteria from agricultural wastes and its utilization in industry: a review. Adv. Enzym. Res. 04, 44–55 (2016). https://doi.org/10.4236/aer.2016.42005
Amer, M.S.; Ellatif, H.H.A.; Hassan, S.W.M.; Aboelela, G.M.; Gad, A.M.: Characterization of some fungal strains isolated from the Eastern coast of Alexandria, Egypt, and some applications of Penicillium crustosum. Egypt. J. Aquat. Res. 45, 211–217 (2019). https://doi.org/10.1016/j.ejar.2019.06.006
Ladevèze, S.; Haon, M.; Villares, A.; Cathala, B.; Grisel, S.; Herpoël-Gimbert, I.; Henrissat, B.; Berrin, J.G.: The yeast Geotrichum candidum encodes functional lytic polysaccharide monooxygenases. Biotechnol. Biofuels. 10, 1–11 (2017). https://doi.org/10.1186/s13068-017-0903-0
Sulyman, A.O.; Igunnu, A.; Malomo, S.O.: Isolation, purification and characterization of cellulase produced by Aspergillus niger cultured on Arachis hypogaea shells. Heliyon. 6, e05668 (2020). https://doi.org/10.1016/j.heliyon.2020.e05668
Abo-State, M.A.; Ragab, A.M.E.; El-Gendy, N.S.; Farahat, L.A.; Madian, H.R.: Bioethanol production from rice straw enzymatically saccharified by fungal isolates, Trichoderma viride F94 and Aspergillus terreus F98. Soft 03, 19–29 (2014). https://doi.org/10.4236/soft.2014.32003
Al-kharousi, M.M.; Sivakumar, N.; Elshafie, A.: from books and archives characterization of cellulase enzyme produced by Chaetomium sp. isolated from books and archives. EurAsian J. Biosci. 9, 52–60 (2015). https://doi.org/10.5053/ejobios.2015.9.0.7
Abo-State, M.A.M.; Ghaly, M.F.; Abdellah, E.M.: Optimization of cellulase (s) and xylanase production by thermophilic and alkaliphilic bacillus isolates. Am. J. Agri. Environ. Sci. 13, 553–564 (2013). https://doi.org/10.5829/idosi.aejaes.2013.13.04.15513
Marín, M.; Artola, A.; Sánchez, A.: Optimization of down-stream for cellulases produced under solid-state fermentation of coffee husk. Waste Biomass Valoriz. 10, 2761–2772 (2019). https://doi.org/10.1007/s12649-018-0327-5
Aguiar, C.L.: Biodegradation of the cellulose from sugarcane bagasse by fungal cellulase. Cienc. y Tecnol. Aliment. 3, 117–121 (2001). https://doi.org/10.1080/11358120109487655
Coniglio, R.O.; Fonseca, M.I.; Villalba, L.L.: Screening of new secretory cellulases from different supernatants of white rot fungi from fungi from Misiones. Argentina. Mycol. 8, 1–10 (2017). https://doi.org/10.1080/21501203.2016.1267047
Science, E.: Characterization of crude cellulase enzyme produced by Bacillus licheniformis P12 isolate. IOP Conf. Ser. Earth Environ. Sci. Pap. (2020). https://doi.org/10.1088/1755-1315/475/1/012085
Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J.: Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951). https://doi.org/10.1016/s0021-9258(19)52451-6
Gad, A.; Suleiman, W.B.; Beltagy, E.A.; El-sheikh, H.; Ibrahim, H.A.H.: Characterization and screening of marine-derived fungi along the coastline of Alexandria, Mediterranean Sea. Egypt. Egypt. J. Aquat. Biol. Fish. 25, 215–239 (2021). https://doi.org/10.21608/EJABF.2021.198560
Campbell, C.K.; Johnson, E.M.: Identification of Pathogenic Fungi, 2nd edn. Wiley, Hoboken (2013)
White, T.J.; Bruns, T.; Lee, S.; Taylor, J.: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Prot. Guid. Methods Appl. 18(1), 315–322 (1990). https://doi.org/10.1016/b978-0-12-372180-8.50042-1
Farag, A.M.; Hassan, M.A.: Purification, characterization and immobilization of a keratinase from Aspergillus oryzae. Enzyme Microb. Technol. 34, 85–93 (2004). https://doi.org/10.1016/j.enzmictec.2003.09.002
Laemmli, U.K.: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (1970). https://doi.org/10.1038/227680a0
Yu, Z.; Zhang, H.: Ethanol fermentation of acid-hydrolyzed cellulosic pyrolysate with Saccharomyces cerevisiae. Bioresour. Technol. 93, 199–204 (2004). https://doi.org/10.1016/j.biortech.2003.09.016
Yoswathana: Bioethanol Production from Rice Straw. Energy Res. J. 1, 26–31 (2010). https://doi.org/10.3844/erjsp.2010.26.31
Suleiman, W.B.: In vitro estimation of superfluid critical extracts of some plants for their antimicrobial potential, phytochemistry, and GC-MS analyses. Ann. Clin. Microbiol. Antimicrob. (2020). https://doi.org/10.1186/s12941-020-00371-1
Tamil Selvan, S.; Velramar, B.; Ramamurthy, D.; Balasundaram, S.; Sivamani, K.: Pilot scale wastewater treatment, CO2 sequestration and lipid production using microalga, Neochloris aquatica RDS02. Int. J. Phytoremed. 22, 1462–1479 (2020). https://doi.org/10.1080/15226514.2020.1782828
Reddy, K.V.; Vijayalashmi, T.; Ranjit, P.: Characterization of some efficient cellulase producing bacteria isolated from pulp and paper mill effluent contaminated soil. Brazil. Arch. Biol. Technol. 60, 1–6 (2017)
Sriariyanun, M.; Tantayotai, P.; Yasurin, P.; Pornwongthong, P.; Cheenkachorn, K.: Production, purification and characterization of an ionic liquid tolerant cellulase from Bacillus sp. isolated from rice paddy field soil. EJBT 19, 23–28 (2016). https://doi.org/10.1016/j.ejbt.2015.11.002
Goel, N.; Patra, R.; Verma, S.K.; Sharma, P.C.: Purification and characterization of cellulase from Pseudomonas sp. isolated from waste dumping site soil. J. Appl. Biotechnol. Bioeng. Res. 6, 118–124 (2019). https://doi.org/10.15406/jabb.2019.06.00183
Saroj, P.; Manasa, P.; Narasimhulu, K.: Characterization of thermophilic fungi producing extracellular lignocellulolytic enzymes for lignocellulosic hydrolysis under solid-state fermentation. Bioresour. Bioprocess. (2018). https://doi.org/10.1186/s40643-018-0216-6
Legodi, L.M.; La Grange, D.; Van Rensburg, E.L.J.; Ncube, I.: Isolation of cellulose degrading fungi from decaying banana pseudostem and strelitzia alba. Enzyme Res. 2019, 10 (2019). https://doi.org/10.1155/2019/1390890
Liu, H.; de Souza, F.Z.R.; Liu, L.; Chen, B.S.: Immobilized and free cells of geotrichum candidum for asymmetric reduction of ketones: stability and recyclability. Molecules (2018). https://doi.org/10.3390/molecules23092144
Samuel, P.: Antibacterial activity of marine derived fungi collected from South East Coast of Tamilnadu, India. Screening 1, 86–94 (2011)
Merlin Christy, P.; Gopinath, L.R.; Divya, D.: A review on anaerobic decomposition and enhancement of biogas production through enzymes and microorganisms. Renew. Sustain. Energy Rev. 34, 167–173 (2014). https://doi.org/10.1016/j.rser.2014.03.010
Amraini, S.Z.; Ariyani, L.P.; Hermansyah, H.; Setyahadi, S.; Rahman, S.F.; Park, D.: Production and characterization of cellulase from E. coli EgRK2 recombinant based on oil palm empty fruit bunches. Biotechnol. Bioprocess Eng. 295, 287–295 (2017). https://doi.org/10.1007/s12257-017-0034-2
Prasetyo, J.; Sumita, S.; Okuda, N.; Park, E.Y.: Response of cellulase activity in pH-controlled cultures of the filamentous fungus acremonium cellulolyticus. Appl Biochem Biotechnol (2010). https://doi.org/10.1007/s12010-009-8826-2
Goyari, S.; Devi, S.H.; Bengyella, L.; Khan, M.; Sharma, C.K.; Kalita, M.C.; Talukdar, N.C.: Unveiling the optimal parameters for cellulolytic characteristics of Talaromyces verruculosus SGMNPf3 and its secretory enzymes. J. Appl. Microbiol. 119, 88–98 (2015). https://doi.org/10.1111/jam.12816
Jantasaeng, O.; Thaenkudrua, P.; Tan, J.S.; Phapugrangkul, P.: Expression of the endoglucanase gene in Escherichia coli for the second-generation bioethanol production. Food Appl. Biosci. 7, 27–35 (2019)
Marco, É.G.D.E.; Heck, K.; Martos, E.T.; van der Sand, S.T.: Purification and characterization of a thermostable alkaline cellulase produced by Bacillus licheniformis 380 isolated from compost. Ann. Brazil. Acad. Sci. 89, 2359–2370 (2017). https://doi.org/10.1590/0001-3765201720170408
Yin, L.; Lin, H.; Xiao, Z.: Purification and characterization of a cellulase from Bacillus subtilis YJ1. J. Mar. Sci. Technol. 18, 466–471 (2010)
Bano, A.; Chen, X.; Prasongsuk, S.; Akbar, A.: Purification and characterization of cellulase from obligate halophilic aspergillus flavus (TISTR 3637) and its prospects for bioethanol production. Appl. Biochem. Biotechnol. 189(4), 1327–1337 (2019). https://doi.org/10.1007/s12010-019-03086-y
Pachauri, P.; Aranganathan, V.; More, S.; Sullia, S.B.: Purification and characterization of cellulase from a novel isolate of Trichoderma longibrachiatum. Biofuels (2017). https://doi.org/10.1080/17597269.2017.1345357
Andrade, J.P.; Sim, A.; Bispo, R.; Arthur, P.; Marbach, S.; Pires, R.: Production and partial characterization of cellulases from Trichoderma sp. IS-05 isolated from sandy coastal plains of Northeast Brazil. Enzyme Res. 2011, 7 (2011). https://doi.org/10.4061/2011/167248
Longo, M.A.; Combes, D.: Analysis of the thermal deactivation kinetics of α-chymotrypsin modified by chemoenzymatic glycosylation. Prog. Biotechnol. 15, 135–140 (1998). https://doi.org/10.1016/S0921-0423(98)80022-1
Frankenberger, W.T.; Johanson, J.B.: Effect of pH on enzyme stability in soils. Soil Biol. Biochem. (1982). https://doi.org/10.1016/0038-0717(82)90101-8
Olajuyigbe, F.M.; Ogunyewo, O.A.: Biocatalysis and Agricultural Biotechnology Enhanced production and physicochemical properties of thermostable crude cellulase from Sporothrix carnis grown on corn cob. Biocatal. Agric. Biotechnol. 7, 110–117 (2016). https://doi.org/10.1016/j.bcab.2016.05.012
Aggarwal, N.K.; Goyal, V.; Saini, A.; Yadav, A.; Gupta, R.: Enzymatic saccharification of pretreated rice straw by cellulases from Aspergillus niger BK01. 3 Biotech. 7, 1–10 (2017). https://doi.org/10.1007/s13205-017-0755-0
Belal, E.B.: Bioethanol production from rice straw residues. Brazil. J. Microbiol. 44, 225–234 (2013). https://doi.org/10.1590/S1517-83822013000100033
Mattam, A.J.; Kuila, A.; Suralikerimath, N.; Choudary, N.; Rao, P.V.C.; Velankar, H.R.: Cellulolytic enzyme expression and simultaneous conversion of lignocellulosic sugars into ethanol and xylitol by a new Candida tropicalis strain. Biotechnol. Biofuels. 9, 1–12 (2016). https://doi.org/10.1186/s13068-016-0575-1
Access, O.; Article, O.; Azadian, F.; Badoei-dalfard, A.; Namaki-shoushtari, A.; Hassanshahian, M.: Purification and biochemical properties of a thermostable, haloalkaline cellulase from Bacillus licheniformis AMF-07 and its application for hydrolysis of different cellulosic substrates to bioethanol production. MBRC 5, 143–155 (2016)
Sarkar, D.; Prajapati, S.; Poddar, K.; Sarkar, A.: Ethanol production by Klebsiella sp SWET4 using banana peel as feasible substrate. Biomass Convers. Biorefinery 1, 13 (2020). https://doi.org/10.1007/s13399-020-00880-1
Saliu, B.K.; Sani, A.: Bioethanol potentials of corn cob hydrolysed using cellulases of Aspergillus niger and Penicillium decumbens. EXCLI J. 11, 468–479 (2012)
Acknowledgements
The authors thank the National Institute of Oceanography and Fisheries (NIOF), Alexandria-Egypt, for giving laboratory facilities to present this work in its current form.
Funding
The authors declare that this work had been performed by themselves and they did not receive any financial support from any agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest concerning this article.
Rights and permissions
About this article
Cite this article
Gad, A.M., Suleiman, W.B., El-Sheikh, H.H. et al. Characterization of Cellulase from Geotrichum candidum Strain Gad1 Approaching Bioethanol Production. Arab J Sci Eng 47, 6837–6850 (2022). https://doi.org/10.1007/s13369-021-06391-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13369-021-06391-z