Abstract
We describe a hybrid MALDI/C60-SIMS Q-TOF mass spectrometer and corresponding sample preparation protocols to image intact biomolecules and their fragments in mammalian spinal cord, individual invertebrate neurons, and cultured neuronal networks. A lateral spatial resolution of 10 μm was demonstrated, with further improvement feasible to 1 μm, sufficient to resolve cell outgrowth and interconnections in neuronal networks. The high mass resolution (>13,000 FWHM) and tandem mass spectrometry capability of this hybrid instrument enabled the confident identification of cellular metabolites. Sublimation of a suitable matrix, 2,5-dihydroxybenzoic acid, significantly enhanced the ion signal intensity for intact glycerophospholipid ions from mammalian nervous tissue, facilitating the acquisition of high-quality ion images for low-abundance biomolecules. These results illustrate that the combination of C60-SIMS and MALDI mass spectrometry offers particular benefits for studies that require the imaging of intact biomolecules with high spatial and mass resolution, such as investigations of single cells, subcellular organelles, and communities of cells.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Mass spectrometry imaging (MSI) is a versatile analytical tool that enables multiplexed, non-targeted, and label-free molecular imaging of biological specimens [1–3]. MSI is commonly performed with a microprobe that is scanned across the surface of a sample of interest to acquire chemical composition details from multiple coordinates in a predefined array; this data is then used to generate ion images in which each image pixel represents a position in the interrogated array, and the relative intensity of a selected ion is represented by the (false) color of each pixel.
Many microprobe types have now been developed for MSI, each offering unique advantages for particular studies. Focused laser beams are routinely used as microprobes in biological and biomedical tissue imaging experiments. Matrix-assisted laser desorption/ ionization (MALDI) mass spectrometry (MS) provides excellent detection limits, high spatial resolution (~50 μm typical), and an extended mass range—up to hundreds of kDa for MSI— allowing detection and imaging of protein distributions [3–5]. With advanced protocols in matrix deposition, MALDI also enables the measurement of peptides in single cells and organelles [6–8]. Nanostructure initiator MS is another laser microprobe-based approach and is capable of imaging with ~20 μm resolution for smaller biomolecules (m/z <1500) in single cells, and can achieve yoctomole-range detection limits under ideal conditions [9]. Furthermore, the advent of ambient ion sources such as desorption electrospray ionization [10, 11], laser ablation electrospray ionization [12–14], and liquid microjunctions [15, 16] have recently extended microprobe MSI to ordinary environmental conditions, enabling in situ analysis of biological samples, including live organisms [16–19].
MSI with secondary ion mass spectrometry (SIMS) [20, 21] is well-suited for single-cell and subcellular investigations. In state-of-the-art systems, SIMS utilizes ion microprobes that can be focused to submicron spots, achieving <100 nm lateral resolution [22, 23], currently unmatched by other MSI probe types. Ion probes also allow etching of sample surfaces with nanometer-scale precision, offering depth profiling and three-dimensional imaging with unsurpassed spatial resolution [21, 24, 25].
Innovations in instrumentation and methodology can further extend the imaging capability of SIMS to a broader range of biomolecules and biological specimens, especially if several main challenges are met: improving molecular ionization efficiencies, providing softer ionization (reducing source fragmentation), reducing analyte suppression due to matrix effects [26], and resolving (near-)isobaric compounds. Indeed, several recent reports have described new SIMS instrumentation that combines high mass resolution, mass accuracy, tandem MS capability, and the use of polyatomic or “cluster” primary ion sources, such as Bi3 + [27], C60 + [28], and Ar2000 + [29], to extend SIMS investigations to bioanalytical analyses [30–32]. Notably, an early report by Carado et al. [30] described modification of a commercial MALDI quadrupole time-of-flight (QTOF) MS instrument with the addition of a C60 primary ion beam, thus enabling cluster SIMS imaging with high mass resolution (R >10,000) and tandem MS capability. The potential for these combined modalities was subsequently demonstrated with several biological profiling and imaging studies, including lipidomic investigations of cell populations [33] and single cells [34].
In addition to advances in instrumentation, ion yields for intact molecules have been improved via laser post-ionization of sputtered neutrals [35, 36], gas cluster ion beam doping [37], sample surface oxygen co-sputtering [38], and chemical pretreatment of the sample with metal overlays [39, 40], or organic matrixes (matrix-enhanced SIMS [ME-SIMS]) [41–44]. Interestingly, experimental methods that have been developed for “traditional” monatomic ion probes do not necessarily translate to cluster ion beams; for example, metal coatings enhanced the ionization of alkanes using Ga+ and In+ projectiles, but not with Bi3 + and C60 + probes [45]. Similar results were observed for C60-SIMS of gold-coated peptides, other polymers, and small organic molecules [46]. On the other hand, 2,5-dihydroxybenzoic acid (DHB) matrix, mixed precisely in a 10:1 ratio with the peptide gramicidin S, yielded 8-fold molecular ion signal enhancement when profiled by C60-SIMS [47], and signal enhancement was reported for intact lipids in brain tissue when coated with DHB in conjunction with a Bi3 + probe [48], though these studies did not extend the work to ME-SIMS imaging. Thus, a targeted combination of instrumentation and methodology offers the potential to further aid biomolecular investigations using SIMS imaging.
Encouraged by these successes in SIMS, we developed a hybrid MALDI/C60-SIMS quadrupole time-of-flight (Q-TOF) mass spectrometer with accompanying biological sample preparation methodologies to extend SIMS to tissue- and cell-scale molecular imaging experiments. We modified our instrument design based on the pioneering work by the Winograd research group [30], but incorporated significant changes to the incident angle of the primary ion beam and the overall design of the ion optics for the extraction and transmission of secondary ions. After evaluating the effect of these modifications on analytical performance, we utilized the combined features of this customized instrument—improved spatial resolution, high mass resolution, tandem MS, and softer secondary ion generation by cluster primary ions—to extract molecular information from chemically and structurally complex biological samples. To this end, we also devised sample preparation protocols based on matrix sublimation to enhance the signal intensity of lipid ions from biological tissues. As a result of the revised instrument design and targeted protocol development, we demonstrate that C60-SIMS/MALDI dual ion source MS can be used to uncover the spatial distribution of intact biomolecules in mammalian spinal cord samples as well as networks of cultured neurons from Aplysia californica.
2 Experimental
Mass spectrometry was performed using a customized dual-source QSTAR MALDI/SIMS mass spectrometer (QSTAR XL; AB SCIEX, Framingham, MA, USA), described in detail in the Results and Discussion section.
2.1 Materials and Chemicals
HPLC-grade methanol and acetonitrile, glycerol, formic acid (~98%), renin substrate (porcine, >97%), poly(propylene glycol) standards (Mn ~425, 1000, and 2000), DHB (98%), methionine, and α-cyano-4-hydroxycinnamic acid 98% (CHCA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ammonium acetate (98%) was from Fisher Scientific (Waltham, MA, USA). Copper mesh grids were obtained from Ted Pella, Inc. (Redding, CA, USA). For cell culture and tissue section substrates, 4-in diameter circular wafers of silicon (100) (Silicon, Inc., Boise, ID, USA) were scored and snapped to create tiles 1 × 1 cm2 in area. To estimate the C60 beam sample currents, a Faraday cup was improvised by drilling a 2 mm-diameter hole in the center of a standard 100-well stainless steel MALDI target (AB SCIEX). The sample plate was disconnected from the accelerating voltage and connected to a picoammeter via a shielded coaxial cable for measurements.
2.2 Biological Specimens
Adult male Long-Evans rats (Harlan, Indianapolis, IN, USA) were used for the vertebrate animal experiments. After rapid decapitation and dissection, intact spinal cord tissue was isolated and then frozen promptly in aluminum foil on dry ice and stored in a sealed bag at –80°C until use. Spinal cord sections, 12-μm thick, were collected on a cryotome (3050S; Leica Biosystems, Wetzlar, Germany) and thaw-mounted to clean silicon tiles, then warmed to room temperature (22–25°C) in a nitrogen-purged dry box prior to matrix application and/or direct analysis. Animal care protocols and procedures were approved by the UIUC Laboratory Animal Care Advisory Committee in full compliance with federal guidelines for the humane care and treatment of animals.
Aplysia californica (Mollusca, Gastropoda) were obtained from the University of Miami/NIH National Resource for Aplysia (Miami, FL, USA) and maintained in an aquarium filled with chilled, circulated, and aerated sea water at 14°C until used. Sea water was prepared in the lab using a synthetic sea salt mix (Instant Ocean; Aquarium Systems Inc., Mentor, OH, USA).
Prior to dissection, Aplysia were anesthetized by injection of isotonic MgCl2 (30%–50% of body weight) into the body cavity; individual neurons were isolated from the buccal, abdominal, pleural, and pedal ganglia of the molluscan central nervous system. These neurons were cultured on silicon tiles in plastic Petri dishes according to a procedure that we have established previously [49]. Cultures were allowed to develop for 24 h at room temperature, then prepared for MSI by either freeze drying or glycerol stabilization followed by rinsing, as described below.
For freeze drying, the culturing media was first substituted stepwise with 150 mM ammonium acetate (pH 7) in deionized (DI) water (Milli-Q filtration system; Millipore, Billerica, MA, USA) to remove inorganic salts from the extracellular environment. Most of this solution was then removed, leaving cells submerged in a minimal volume, and the Petri dish was then rapidly cooled by partial immersion in liquid nitrogen. Once the culturing solution was completely frozen, the Petri dish was transferred to a vacuum chamber and allowed to freeze-dry overnight. Cells were then rinsed by dipping the entire silicon substrate vertically into DI water for 1 s, followed by rapid drying with a stream of dry nitrogen gas, repeated in triplicate. The morphology of most cells was maintained throughout this procedure.
For glycerol stabilization, the culture media was substituted stepwise with mixture of 33% glycerol and 67% artificial seawater (v/v), and then decanted in ~1 s. The tiles were stored vertically overnight at ambient conditions, allowing excess solution to drain and evaporate. Stabilized cells were then rinsed by either rapid dips in DI water as described above, or by carefully applying microdroplets of DI water onto individual cells and then promptly aspirating the water using a micropipette tip. The latter technique was preferred as it generally resulted in less cell lysis and/or loss of neurites, and was used for the cells depicted in the optical and molecular images presented here.
2.3 Matrix Sublimation
The sublimation chamber apparatus and procedure were adapted from previous work described elsewhere [50]. The chamber was modified with a 25 × 75 mm2 aluminum foil boat affixed to the inner base surface with double-sided conductive copper tape as well as a 25 × 75 × 1 mm3 thick stainless steel plate similarly affixed to the bottom face of the cold finger to improve thermal conductivity to the sample; the boat-to-plate distance was 20 mm. For each sublimation procedure, 350 mg of powdered DHB was added to the aluminum foil boat and distributed evenly. Samples on silicon tiles were first weighed on an ultra-microbalance (Cubis; Sartorius, Bohemia, NY, USA), then affixed to the stainless steel plate with conductive copper tape. The chamber was closed, placed in a heating mantle, pumped to intermediate vacuum (~10 mTorr), and the cold finger was then filled with iced water (~4°C). After 5 min of temperature equilibration, the desired matrix coating was achieved by supplying 120 V to a heating mantle for 240–285 s. The chamber was then removed from the mantle, vented to room temperature air, and the sample promptly removed from the cold finger. The sample was weighed again to determine the amount of matrix applied (density estimated as total applied matrix mass/silicon tile area) before SIMS analysis.
2.4 MALDI and SIMS Imaging Experiments
All SIMS MSI experiments were performed on the customized hybrid instrument described in the Results and Discussion section. For the matrix enhancement experiments with spinal cord sections, the SIMS images were acquired in raster imaging mode with the following settings: 50 × 50 μm2 step size, m/z 60–850 detection mass window, 0.25 s/pixel dwell time, default TOF settings, and Q1 ion guide transmission biased for intact lipids (10%/20%/70% at m/z 70/200/400, respectively). The MALDI MS spine images were acquired with the laser set to 40 Hz at 60% relative power and a 100 × 100 μm2 step size with otherwise identical parameters; smaller step sizes were found to generate lower quality images due to severe oversampling. Ion distribution images for the matrix enhancement experiments were acquired from consecutive tissue sections taken from a single spinal cord specimen and imaged in successive sessions in order to minimize systematic errors; n = 6 for untreated and 0.115 mg/cm2-coated sections, and n = 5 for 0.016 and 0.181 mg/cm2-coated sections. The C60 + probe was focused to a 40-μm diameter and produced a 190 pA sample current. Mass calibration was performed for In1-7 + ions.
For the cell imaging experiments, the C60 + probe current was reduced to 50 pA with an ~15-μm beam diameter, and SIMS images were acquired by rastering the sample at a 10 μm step size with 1 s/pixel dwell time. Secondary ions were transmitted with Q1 biased to higher mass transmission (10%/40%/50% at m/z 50/100/200, respectively) and detected between m/z 60 and 450. Mass calibration was performed using In1-7 + ions, or Cu+ and In+ ions for the small metabolite tandem MS experiments. Imaging data was converted from the wiff to img format at 20 bins/unit m/z and imported to BioMap (Novartis, Basel, Switzerland) for further processing. Mass filters were ± m/z 0.05 and ion images were represented in false color, with black corresponding to zero and red to maximum signal intensity. Tandem MS experiments were performed in SIMS mode with 10–30 eV CID in argon collision gas. Optical images of cells were acquired on a Leica MZ75 light microscope (Leica Microsystems) equipped with an AxioCam MRc camera (Carl Zeiss Microscopy, Jena, Germany) and AxioVision 4 software (Carl Zeiss Microscopy).
3 Results and Discussion
3.1 Hybrid MALDI/C60-SIMS Instrument Design
A commercial QSTAR Q-TOF mass spectrometer was modified to construct the MALDI/SIMS hybrid mass spectrometer used in this work. Based conceptually on a prototype developed by the Winograd laboratory [30], design modifications were made to our instrument to facilitate the conversion and improve system performance. These adaptations included decreasing the incident angle of the primary ion beam from 45° to 30° relative to the sample surface normal to enhance the secondary ion yield, incorporating a rectilinear quadrupole (RLQ) ion guide for the extraction and transmission of secondary ions to improve performance, adding removable RLQ tips to facilitate frequent electrode cleaning for robust operation, and achieving differential vacuum pumping to aid ionization and transfer of secondary ions. We kept other aspects of the prototype C60-SIMS/MALDI system that proved successful. Specifically, we adopted the commercial oMALDI 2 ion source as it already integrated an ultraviolet (UV) MALDI source, an x/y translation stage, and a camera for optical inspection of the sample, as well as the quadrupole time-of-flight system and data analysis platform of the QSTAR, which provided tandem MS capability via collision-induced dissociation (CID) and a high mass resolving power of >10,000 FWHM. In what follows, we describe the specific modifications and discuss the rationale that inspired them.
The original QSTAR MALDI source enclosure was precision-milled to accommodate a 20 kV C60 ion gun (Ionoptika, Manchester, UK) with an ~1 μm ultimate ion probe diameter, while preserving the original MALDI functionality. A partial schematic of the hybrid instrument is shown in Figure 1a. The ion gun was supplemented with a differentially pumped and enclosed beam column that obtained intermediate vacuum (<10 mTorr) to better preserve the primary C60 + cluster ions and to achieve collisional focusing of the secondary ions [30]. In our design, the C60 ion gun was mounted on the MALDI source to bombard the sample with the primary ion beam at a 30° incidence angle relative to the surface normal, rather than 45° as in the previous work [30], because molecular dynamics simulations have indicated that impact energy and secondary ion sputter yield are maximized at shallower incidence angles [51]. The end of the ion gun was outfitted with a custom-designed electrically floating tip to improve sampling efficiency, and a blanking circuit was constructed using a pulse generator (model 4001; Global Specialties Instruments, Yorba Linda, CA, USA) to control the ion beam using the transistor-transistor logic (or TTL) signal that ordinarily triggers the nitrogen laser of the MALDI source. The original vacuum chamber of the QSTAR was extended with a set of two vacuum adapter blocks bridged with a flange adapter to fit and mechanically support the ion gun.
Schematic of the hybrid C60-SIMS/MALDI mass spectrometer and RLQ transmission profiles. (a) Custom-designed adapter blocks (outlined in red) were installed to connect the C60 + gun to the vacuum chamber, the standard UV-MALDI source, and the Q-TOF mass spectrometer. A rectilinear quadrupole (RLQ) ion guide was incorporated to transmit ions through the adapter into the ion guide Q0. The enlarged source chamber was operated at high vacuum pressure (<0.1 mTorr) by an additional turbomolecular pump (TM Pump) that was mounted at the bottom of the adapter, while nitrogen gas (represented with blue) was introduced to the chamber surrounding the Q0 for collisional focusing of ions. (b) Mass transmission profiles measured for the RLQ ion guide (2.8 MHz) confirmed the efficient transmission of ions across a broad range, approximately m/z 100–2000, at 1100 V p-p rf amplitude. Squares indicate measured data with connecting lines added as a visual guide
Ions generated from samples in the C60-SIMS and MALDI operational modalities were extracted into the ion guide Q0 of the QSTAR by using a custom-designed, additional ion guide. We chose to supplement the existing Q0 with a rectilinear quadrupole (RLQ) ion guide (Ardara Technologies, Ardara, PA, USA), rather than modifying the original Q0 as was done in the prototype, for several reasons. First, by preserving the original design of the QSTAR, the complexity of modifications was substantially minimized and the user-friendliness of the system was retained in large part by taking advantage of the commercial control software of the QSTAR to operate the ion optics downstream to Q0. Second, RLQs are characterized with high simultaneous efficiency of ion transmission across a broader mass (m/z) range compared with round-rod quadrupoles [52]. Third, both RLQs and round-rod quadrupoles are known to perform well for collisional focusing and they are tolerant to ions with lower kinetic energy (as is anticipated from C60-SIMS). The RLQ was installed between the sample plate and the front end of the Q0, without any modification to the latter; it had an 8-mm inscribed diameter and 4.7 mm square rod electrodes, and was driven by a 2.8 MHz radiofrequency (rf) and DC power supplies (Ardara Technologies). The RLQ was further modified with removable front segments (shown in Supplementary Figure S1) to facilitate removal and cleaning with fine abrasive (aluminum oxide) slurry polish; as the transmission efficiency in the C60-SIMS mode declined with extended use, frequent cleaning was important in maintaining good performance. As a result, this modification improved the overall robustness of the hybrid mass spectrometer.
The adapter blocks also incorporated vacuum gauge sensors and an additional turbomolecular pump that provided high vacuum (<0.1 mTorr) in the source chamber, whereas electrical and gas feed-through ports for the RLQ were located on the flange adapter. The adapter blocks were designed in-lab by computer-aided design freeware (DraftSight v1; Dassault Systémes, Vélizy-Villacoublay, France) and precision milled from 6061 aluminum, and the flange adapter was designed and provided by Ardara Technologies. Adapter blocks were sealed with aluminum Conflat flanges (note that for this application, soft aluminum gaskets must be used in place of standard copper gaskets). Operational settings for the hybrid instrument in the positive ion mode for both MALDI and SIMS were optimized as follows: sample plate/orifice +34 V, C60 tip +30 V, RLQ +24 V DC offset with 1100 V rf p-p amplitude, and Q0 + 20 V with 8 mTorr collisional focusing gas pressure.
To evaluate the performance effect of this change in geometry, the ionization efficiency of indium ions from pure indium foil standard was determined using the new system in C60-SIMS mode. A secondary ion yield of 1.3 × 10–3 (115In+ counts per C60 + projectile) was obtained; this result is comparable to that reported by Carado and colleagues [30] for their C60-QTOF mass spectrometer, suggesting that our hybrid instrument is equally as efficient in the low-mass range as their prototype.
3.2 Characterization of the Hybrid Mass Spectrometer
Initially, the analytical performance of the hybrid mass spectrometer was systematically evaluated; secondary ion transmission efficiency, mass resolution, capability for tandem MS, and spatial resolution for imaging were characterized for the MALDI-MS and C60-SIMS operational modes. One advantage of adding an additional ion guide rather than modifying the existing quadrupole was that downstream electrode settings required no optimization following modification. As also pointed out earlier, the rf-only RLQ was used as an ion guide through the vacuum chamber adapter because RLQ guides provide a broad mass transmission window relative to round-rod counterparts [52], thus further simplifying modification by obviating the need to ramp rf amplitude synchronously with the subsequent Q0 ion guide. The performance of the RLQ is characterized by the relative mass transmission profiles shown in Figure 1b, acquired by varying the rf voltage while measuring the ion signal for ions generated from a poly(propylene glycol) standard with a continuously-rastered laser probe. Transmission of larger ions (1–2 kDa) was greatest at the maximum accessible rf amplitude, 1100 V peak-to-peak (V p-p ), which still provided efficient (>90% relative) transmission for the smaller ions. Thus, by operating the guide at this fixed rf amplitude and 2.8 MHz frequency, our hybrid mass spectrometer ensured high transmission efficiency for both large and small ions in the broad, m/z 100–2000 mass range, opening the possibility of measuring intact metabolites, lipids, and small peptides.
A differentially pumped interface was designed to suit dissimilar vacuum requirements imposed by the C60 + gun and the mass spectrometer. The commercial QSTAR originally called for ~1 Torr in the MALDI source chamber to collisionally cool ions, and ~10 mTorr in the ion guide Q0 to collisionally focus them for transmission to the analyzer [53]. However, SIMS is incompatible with low and medium vacuum as the primary ion beam is defocused by collisions with gas [30]. At 10 mTorr, the mean free path of a C60 + ion is ~1 mm; thus, few intact projectiles reach the sample from a practical (>5 mm) working distance. To resolve this conflict, the adapter block was differentially pumped to maintain the sample chamber at high vacuum while collisional focusing was accomplished downstream with ~10 mTorr in the back half of the RLQ and throughout Q0. Operation with intermediate source pressure (1–10 mTorr) was found to have no effect on C60-SIMS performance until ~5 mTorr (data not shown), beyond which the ion signal intensity decreased, likely as a result of beam destruction [30].
To evaluate the effective spatial resolution in the SIMS mode, a copper mesh grid was imaged at a 10-μm step size, the highest spatial resolution offered by the MALDI sample stage. The reconstructed Cu+ ion map showed a matching 10 μm effective lateral spatial resolution (Supplementary Figure S1). As the ion gun is capable of smaller (~1 μm ultimate diameter) probe dimensions, we are currently implementing synchronized beam control using built-in raster plates to improve the lateral resolution by approximately an order of magnitude; single-micron (and better) spatial resolution importantly enables adequate resolution for visualization of organelles and other subcellular structures within ion images [54]. A high mass resolution of >13,000 by FWHM is also demonstrated on the adapted system with MALDI-MS analysis of renin substrate tetradecapeptide standard (shown in Supplementary Figure S1).
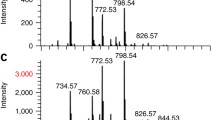
The hybrid mass spectrometer provided complementary imaging capabilities in the C60-SIMS and MALDI MSI modes. As the laser optics on the oMALDI source were unaffected by the instrumental modifications undertaken here, a ~100 × 200 μm2 laser microprobe area was obtainable as on the unmodified commercial QSTAR. To evaluate the complementarity of SIMS and MALDI MSI by this hybrid instrument, tissue sections of rat spinal cord were imaged in each mode with DHB sublimed on the sample for MALDI MSI. The results are compared in Figure 2. The MS lipid profile spectra acquired from the gray matter show that MALDI generated a several times higher ion abundance for intact lipid ions (pseudomolecular and salt adduct) over SIMS for a comparable analysis area. The smaller probe size of SIMS enabled imaging with higher spatial resolution, improving the definition of small anatomical features such as the central canal and dorsal horns, which both show as intense localizations of PC(32:0) with m/z 772.53. Also notable is that MALDI generated relatively fewer lipid fragments at m/z 713.46, 723.49, and 739.47, corresponding to the loss of trimethylamine observed in earlier investigations on mouse brain using a similar C60-SIMS system [55].
Imaging of a mammalian spinal cord in MALDI and C60-SIMS modes using the hybrid Q-TOF mass spectrometer. (a) MS lipid profiles of central gray matter acquired by MALDI (top) and SIMS (bottom, inverted and intensity scaled × 10) show similar overall ion composition from comparable tissue areas, with certain differences. MALDI yielded several-fold more intact lipid ions, including m/z 734.56 – PC(32:0) [M + H]+, m/z 756.55 – PC(32:0) [M + Na]+, m/z 760.58 – PC(34:1) [M + H]+, 772.53 – PC(32:0) [M + K]+, m/z 782.56 – PC(34:1) [M + Na]+, and m/z 798.55 – PC(32:0) [M + K]+. In comparison, SIMS generated relatively more fragments by trimethylamine loss including at m/z 713.46 – PC(32:0) [M – N(CH3)3 + K]+, m/z 723.49 – PC(34:1) [M – N(CH3)3 + Na]+, and m/z 739.47 – PC(34:1) [M – N(CH3)3 + K]+. Ions were assigned tentatively by mass match to previous work with similar tissue [55]. (b) MALDI-mode ion image of PC(32:0) [M + K]+ at m/z 772.53 provided higher signal intensity and image contrast compared with (c) SIMS-mode image for the same ion. SIMS provided imaging higher spatial resolution, allowing improved definition of small anatomical features in the spinal cord such as the central canal and dorsal horns, which show higher relative PC(32:0) abundance. Decreasing MALDI step size resulted in significant oversampling and did not improve image quality. Scale bar = 500 μm for both ion images
3.3 Imaging of Single Cells and Neuronal Networks
The hybrid mass spectrometer was utilized to uncover chemical differences between the soma and processes of individual cultured A. californica neurons. Buccal neurons were placed on a silicon substrate in culturing solution for 1–2 d, where they formed processes (Figure 3). Each step of the sample preparation workflow was judiciously assessed to ensure minimal chemical changes and avoid potential morphologic alterations to the neurons before removing them from the culture solution and introducing the samples into vacuum for C60-SIMS imaging. We previously found that fixation with formaldehyde and paraformaldehyde preserved cell morphology for SIMS imaging [56]. However, because this treatment chemically modifies the specimen, we explored alternative treatment options. Glycerol cell stabilization [6] has been used successfully and has also been found to preserve cell function [57]. We also tried freeze-drying, common to the typical SIMS workflow [58, 59], in an effort to avoid chemical modification of analytes. While freeze-drying preserved cell somas, it destroyed their processes (data not shown), whereas the glycerol treatment preserved both. As alternatives, cryogenic techniques, such as frozen hydration [60] and critical-point drying [61], may also preserve the fragile cell outgrowth; these preparation techniques have recently been evaluated for SIMS imaging by Robinson and Castner [62]. Here, glycerol stabilization was found to yield robust results, and was therefore adopted in our protocol for the preparation of the cultured neurons for MSI.
C60-SIMS ion images of cultured A. californica neurons. Cells were cultured on silicon tiles and stabilized with glycerol substitution. Cell bodies and processes are apparent in the phosphocholine (PC) image (m/z 184.08, 0–200 counts), while α-tocopherol accumulated on the cell bodies (m/z 430.39, 0–50 counts). The isobaric ions m/z 103.98, 104.05 (0–60 counts), and 104.11 (bottom row, 0–60 counts) were detected with different distributions, assigned to background contaminant, nonspecific organic fragment, and choline, respectively. Scale bar = 200 μm for ion images; the optical image was scaled manually to match ion images
C60-SIMS imaging produced characteristic ion images of the distribution of several biomolecules in the cultured neurons. SIMS-generated ions were identified based on mass measurements and fragmentation behavior under CID. Representative identifications are presented for phosphocholine and α-tocopherol in Supplementary Figure S2. The tandem mass spectra recorded on these ions matched well those published in the METLIN mass spectrometric metabolite database [63]. Phosphocholine was detected at m/z 184.08 as a fragment from the phospholipid head group of lipids, in agreement with other studies using SIMS [49, 56, 64]. This signal was particularly useful in highlighting localization of the cell membrane in our C60-SIMS images. The ion of α-tocopherol, one form of vitamin E, was observed as M•+ at m/z 430.38. This membrane-localized, multi-functional nutrient fulfills roles in oxygen radical scavenging as well as modulation of signal transmission and gene expression [65]; thus, it is an analyte of particular interest in this work. The C60-SIMS ion images of phosphocholine and α-tocopherol were acquired from two cultured and glycerol-stabilized neurons (Figure 3). Neural processes are readily visible in the phosphocholine ion image, whereas α-tocopherol accumulated almost exclusively within the cell bodies. We previously reported localization of α-tocopherol at the process-soma junction in similar A. californica neurons using a commercial TOF-SIMS instrument that was equipped with a Au+ ion probe [49]; although distribution in the present work appears to differ, these cells were also cultured in close proximity and, thus, may be influenced by cell signaling molecules that affect chemical composition. These combined results demonstrate that the hybrid mass spectrometer provided biomolecular images with high spatial resolution, serving as a powerful bioanalytical platform to help decipher the molecular architecture of single cells.
Attaining high mass resolution, the hybrid mass spectrometer resolved multiple isobaric ions by their nominal molecular mass throughout the m/z 100–300 range. This observation highlights the importance of the high mass resolution for SIMS and also suggests that some of these ions may correspond to biomolecules rather than nonspecific organic fragments. Moreover, useful biological information might be gleaned from them, provided they can be confidently identified. That task is made challenging by several factors: many metabolites match a given nominal mass, some may be structural isomers (i.e., share an exact mass) and spectral complexity in this range is also increased by fragments generated from larger biomolecules during the SIMS sputtering process. Approaches that offer high mass resolution, mass accuracy, and tandem MS capability, used in combination with a well-characterized biological model and existing metabolite databases such as METLIN [63], have become valuable and provide a useful starting point for ion signal identification in SIMS experiments [66].
We further stress the importance of distinguishing isobaric ions in high-fidelity SIMS imaging data to aid in bioanalytical discovery. This point is demonstrated for three isobars detected at a nominal mass of m/z 104. As the ion map at m/z 103.98 homogeneously distributed the entire sample area (Figure 3) and did not match any metabolites in METLIN, this signal was assigned as background and disregarded in further analysis. The other isobars at 104.11 and 104.05 exhibited characteristic images that correlated with the neuron structure, suggesting that they may be biologically relevant. An accurate-mass search in METLIN resulted in three possible metabolites for 104.11 Da with 10 mDa accuracy: choline, neurine, and valinol (2-amino-3-methyl-1-butanol). Likewise, seven matches were found as potential intact biomolecules for 104.05 Da, though none of these compounds seemed biologically feasible given the existing knowledge on A. californica neurons. Thus, we propose that this ion may instead be an organic fragment produced during sputtering. In agreement, this mass was recently assigned as a unique methionine fragment amongst 20 common metabolites identified by C60-SIMS tandem MS [66]. To confirm these assignments, tandem MS was performed on the cultured neurons, with the resulting spectrum shown in Figure 4. Importantly, these experiments were conducted in SIMS mode since MALDI may generate different ions, and also because the addition of a matrix would complicate the low mass spectrum with additional matrix-related cluster, adduct, and fragment ions.
C60-SIMS tandem mass spectrum of m/z 104 acquired from A. californica ganglion tissue. Several precursor ions are included in the isolation window (see the inset). The ion at m/z 104.11 was assigned to choline based on the precursor mass and characteristic fragment ions at m/z 45.03, 58.07, and 60.08 (red text)
The tandem MS capability of the hybrid instrument facilitated the identification of ions. As the isobars at m/z 104 could not be isolated individually, the tandem MS spectrum on this mass represented a convolution of fragments from the three precursors. The abundant fragments observed at m/z 45.03, 58.07, and 60.08 closely match the tandem MS spectrum of choline reported in METLIN, supporting this assignment. Choline is also biologically the most likely of the three candidate compounds matching m/z 104.11 to be present in the cells as a metabolite or generated as a membrane lipid fragment. On the other hand, the expected smaller fragments of methionine—m/z 61.01 and 56.05—were not present. Furthermore, C60-SIMS analysis of methionine standard yielded the MH+ ion at m/z 150.05, along with multiple characteristic fragments including m/z 104.06, 61.01, and 56.05 (see Figure S3). Isolation and tandem MS of the m/z 104.06 fragment itself—essentially a pseudo MS3 experiment in which SIMS source fragmentation provides MS2—also yielded the expected smaller fragments at m/z 61.01 and 56.05 from the standard. The absence of these fragments in the tandem MS spectrum of m/z 104 acquired from the cells indicates that this ion was either not a methionine fragment, or that the abundance was too low to generate detectable fragments during pseudo-MS3. Fragments observed at m/z 77.04 and 78.05 could not be assigned to either choline or the methionine fragment; these may be fragments of the third precursor ion m/z 103.98, which is likely an inorganic cluster ion based on elemental permutation calculations, and may originate from the cell culture solution (artificial seawater containing numerous inorganic salts). Although the analysis of these ions did not yield complete identification here, it demonstrates how high mass resolution and tandem MS capability may be combined with existing databases to decipher convoluted chemical images.
The C60-SIMS instrument offers an opportunity to evaluate cultured neural networks. When cultured in close proximity, A. californica neurons reconnect with processes to form a neural network, and by co-culturing specific cells such as sensory and motor neurons, simple neural circuits can be assembled and studied as a model of fundamental nervous system function [67]. Towards this goal, we cultured neural networks and imaged them using the C60-SIMS modality of the platform, with results shown in Figure 5. The optical image revealed extensive neurite outgrowth, including visible intercellular connections between the neurons, and these connections were resolved in the SIMS ion images as well. Phosphocholine and choline were detected throughout the network, corresponding to the exposed cell membranes. In contrast, α-tocopherol accumulated at the cell somata at specific subcellular locations, in general agreement with recent SIMS data [34]. Although additional work, including image normalization, is necessary to establish the biological significance of the molecular distributions observed here (e.g., discern actual subcellular distributions from artifactual signal bias), these results demonstrate the capability of our new instrumentation, combined with existing biological knowledge, to interrogate the chemical nature of cultured neural networks.
C60-SIMS ion images of an A. californica neuronal network. Cells were cultured on a silicon tile and stabilized with glycerol. Outgrowth and interconnecting processes are visible in the optical image and correlate with the total ion and phosphocholine (PC, 0–400 counts) ion images, while α-tocopherol (0–150 counts) is concentrated to cell bodies and also shows subcellular localization. Scale bar = 200 μm for ion images; the optical image was scaled manually to match ion images
3.4 ME-SIMS of Biological Tissue
To evaluate matrix-enhancement for C60-SIMS imaging, DHB was applied via sublimation to mammalian spinal cord tissue sections. The spinal cord was selected as a specimen for this study since it has been well-characterized in previous MSI studies [64, 68], has a distinctive anatomy consisting primarily of central gray matter mostly containing cell bodies and peripheral white matter populated with myelinated axons, and has a similar chemical composition axially, which allows comparisons between consecutive tissue sections. Vacuum sublimation was chosen for matrix deposition as this dry process avoids lateral analyte delocalization and also generates small crystals that do not preclude micron-scale lateral resolution [50]. The applied matrix amount was controlled by sublimation time and quantified as matrix density on the surface following an approach that was established elsewhere [69]. Three matrix densities were evaluated by C60-SIMS imaging along with untreated sections. The overall results are shown in Figure 6.
Results of matrix-enhanced C60-SIMS imaging experiments using sublimed DHB on spinal cord tissue sections. The chart compares average peak counts detected in gray matter for a lipid phosphocholine (PC) fragment as well as putative PC(32:0) and PC(34:1), two intact lipids detected as [M + K]+ adduct ions, from samples that were untreated or coated with one of three different matrix densities. Statistically significant (P < 0.05) intact lipid enhancement was achieved with a 0.12 mg/cm2 sublimed matrix coating, indicated with asterisks. Inset shows representative ion images from uncoated and matrix-coated specimens; the identical color scale is used for each pair of images for accurate comparison. Scale bar = 500 μm for all ion images
Statistically significant enhancement of intact lipid signal amplitudes was achieved with a 0.12 mg/cm2 sublimed matrix coating (P < 0.05 by one-tailed, heteroscedastic Student’s t-test), yielding an approximately threefold increase in signal for intact lipid ions such as [M + K]+ ions of PC(32:0) and PC(34:1), shown here (Figure 6). The phosphocholine fragment appeared suppressed with all of the tested matrix treatments, possibly indicating reduced fragmentation of lipids by the primary beam in the presence of matrix. Heavier and lighter matrix coats (0.18 mg/cm2 and 0.02 mg/cm2, respectively) resulted in reduced signal relative to uncoated sections. The observation that this treatment appeared to enhance non-protonated molecular lipid ions suggests that signal enhancement arises primarily from concentration of analytes at the sample surface rather than by proton donation in solid or gas phases. Although sublimation is a dry matrix application method, which is not expected to solvate analytes from a tissue section, some analyte migration may nevertheless occur. This behavior may be enabled by residual tissue moisture and/or brief water condensation on the sample prior to removal from the sublimation cold finger, similar to recrystallization steps employed elsewhere [69]. While the details of this enhancement mechanism are the subject of ongoing investigation, these results indicate that the combination of a C60 ion probe and matrix application provides significant signal enhancement.
4 Conclusions
In the broader context, this study adds to a growing body of work where the spatial distribution of intact biomolecules is interrogated in high resolution using multiple MSI modalities. The complementary nature of accurate-mass SIMS and MALDI is apparent from the type of biochemical data collected using the MALDI/C60-SIMS hybrid ion source Q-TOF mass spectrometer described here, as well as its prototype from the Winograd research group (Penn State University). By integrating these fundamentally dissimilar MS technologies into a single analytical platform, it becomes possible to query the molecular architecture of the same specimen, such as a section of tissue, for small- to mid-sized (m/z 100–2000) biomolecules in high lateral resolution (below 1 μm by SIMS), as well as for large biomolecules (m/z >1000) in moderate to relatively low imaging resolution (~5–250 μm by MALDI). The MSI of the model neuronal networks described here demonstrates that hybridization of MALDI and C60-SIMS, in combination with targeted sample preparation protocols, can aid basic research in cell biology and neurobiology.
We have made a number of refinements to the instrument design of the MALDI/C60-SIMS hybrid ion source Q-TOF mass spectrometer as well as sample preparation in an effort to improve performance, robustness, and user experience; these include the reduction of the primary ion beam incidence angle, utilization of a rectilinear ion guide, use of removable tips for the rectilinear ion guide, utilization of a dual-step vacuum chamber, and enhancement by matrix application to the sample surface. Despite these advances, to capture a broader range of biomolecules in a large network of neurons, further developments are needed in MALDI/SIMS hybrid MS. Continuing work should also be focused on expanding the chemical information that can be obtained from single neurons and cultured neural networks. To this end, we have begun systematically evaluating different matrixes and matrix-application protocols to enhance signals and optimize profiling and imaging using dual SIMS/MALDI. We are also working to improve the identification of endogenous metabolites using tandem MS to a higher degree of confidence, including the characterization of nonspecific chemical backgrounds. In order to capture the subcellular localization of intact biomolecules, the spatial resolution also needs to be increased. As our current design has a C60 + beam focus limit of ~1 μm, integration of precise electronic control of microprobe and/or sample position would be highly advantageous. Furthermore, better characterization of sample morphology would be equally important as it would help uncover physical connections such as neurites among a network of communicating neurons. At present, we are incorporating a secondary electron detector into our hybrid MALDI/SIMS instrument to add electron microscope functionality. We envision this “tribrid analyzer” will contribute to ongoing advancements in mass spectrometric technologies, which we believe will play an ever-increasing role in biological investigations conducted at cellular and molecular levels.
References
Rubakhin, S.S., Sweedler, J.V. Mass Spectrometry Imaging. In: Rubakhin S.S., Sweedler J.V., (Eds.), pp. 21–49. Humana Press: Totowa (2010)
Setou, M.: Imaging Mass Spectrometry: Protocols for Mass Microscopy, pp. 1–257. Springer, Tokyo (2010)
Chughtai, K., Heeren, R.M.A.: Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110, 3237–3277 (2010)
Schwamborn, K., Caprioli, R.M.: Innovation molecular imaging by mass spectrometry - looking beyond classical histology. Nat. Rev. Cancer 10, 639–646 (2010)
Cornett, D.S., Reyzer, M.L., Chaurand, P., Caprioli, R.M.: MALDI imaging mass spectrometry: Molecular snapshots of biochemical systems. Nat. Methods 4, 828–833 (2007)
Rubakhin, S.S., Greenough, W.T., Sweedler, J.V.: Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal. Chem. 75, 5374–5380 (2003)
Rubakhin, S.S., Garden, R.W., Fuller, R.R., Sweedler, J.V.: Measuring the peptides in individual organelles with mass spectrometry. Nat. Biotechnol. 18, 172–175 (2000)
Rubakhin, S.S., Sweedler, J.V.: Quantitative measurements of cell−cell signaling peptides with single-cell MALDI MS. Anal. Chem. 80, 7128–7136 (2008)
Greving, M.P., Patti, G.J., Siuzdak, G.: Nanostructure-initiator mass spectrometry metabolite analysis and imaging. Anal. Chem. 83, 2–7 (2011)
Eberlin, L.S., Ferreira, C.R., Dill, A.L., Ifa, D.R., Cooks, R.G.: Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim. Biophys. Acta 1811, 946–960 (2011)
Eberlin, L.S., Ifa, D.R., Wu, C., Cooks, R.G.: Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew. Chem. Int. Ed. 49, 873–876 (2010)
Nemes, P., Woods, A.S., Vertes, A.: Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 82, 982–988 (2010)
Nemes, P., Barton, A.A., Vertes, A.: Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 81, 6668–6675 (2009)
Shrestha, B., Patt, J.M., Vertes, A.: In situ cell-by-cell imaging and analysis of small cell populations by mass spectrometry. Anal. Chem. 83, 2947–2955 (2011)
Laskin, J., Heath, B.S., Roach, P.J., Cazares, L., Semmes, O.J.: Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 84, 141–148 (2011)
Watrous, J., Roach, P., Heath, B., Alexandrov, T., Laskin, J., Dorrestein, P.C.: Metabolic profiling directly from the petri dish using nanospray desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 85, 10385–10391 (2013)
Nemes, P., Vertes, A.: Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. TraC Trends Anal. Chem 34, 22–34 (2012)
Harris, G.A., Galhena, A.S., Fernandez, F.M.: Ambient sampling/ionization mass spectrometry: applications and current trends. Anal. Chem. 83, 4508–4538 (2011)
Ifa, D.R., Wu, C.P., Ouyang, Z., Cooks, R.G.: Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst 135, 669–681 (2010)
Boxer, S.G., Kraft, M.L., Weber, P.K.: Advances in imaging secondary ion mass spectrometry for biological samples. Annu. Rev. Biophys. 38, 53–74 (2009)
Lanni, E.J., Rubakhin, S.S., Sweedler, J.V.: Mass spectrometry imaging and profiling of single cells. J. Proteome 75, 5036–5051 (2012)
CAMECA (2010) NanoSIMS 50/50 l. SIMS microprobe for ultra fine feature analysis. Available at: http://www.cameca.fr/instruments-for-research/nanosims.aspx. Accessed 4/3/2014.
Kollmer, F., Paul, W., Krehl, M., Niehuis, E.: Ultra high spatial resolution SIMS with cluster ions—approaching the physical limits. Surf. Interface Anal. 45, 312–314 (2012)
Fletcher, J.S., Lockyer, N.P., Vaidyanathan, S., Vickerman, J.C.: TOF-SIMS 3D biomolecular imaging of xenopus laevis oocytes using buckminsterfullerene (C60) primary ions. Anal. Chem. 29, 2199–2206 (2007)
Fletcher, J.S., Vickerman, J.C., Winograd, N.: Label free biochemical 2D and 3D imaging using secondary ion mass spectrometry. Curr. Opin. Chem. Biol. 15, 733–740 (2011)
Jones, E., Lockyer, N., Kordys, J., Vickerman, J.: Suppression and enhancement of secondary ion formation due to the chemical environment in static-secondary ion mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 1559–1567 (2007)
Kollmer, F.: Cluster primary ion bombardment of organic materials. Appl. Surf. Sci. 231/232, 153–158 (2004)
Boussofiane-Baudin, K., Bolbach, G., Brunelle, A., Della-Negra, S., Håkansson, P., Le Beyec, Y.: Secondary ion emission under cluster impact at low energies (5–60 kev); influence of the number of atoms in the projectile. Nucl. Instrum. Meth. Phys. Res. B 88, 160–163 (1994)
Matsuo, J., Okubo, C., Seki, T., Aoki, T., Toyoda, N., Yamada, I.: A new secondary ion mass spectrometry (SIMS) system with high-intensity cluster ion source. Nucl. Instrum. Meth. Phys. Res. B 219–220, 463–467 (2004)
Carado, A., Passarelli, M.K., Kozole, J., Wingate, J.E., Winograd, N., Loboda, A.V.: C60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal. Chem. 80, 7921–7929 (2008)
Smith, D.F., Robinson, E.W., Tolmachev, A.V., Heeren, R.M.A., Paša-Tolić, L.: C60 secondary ion Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 83, 9552–9557 (2011)
Fletcher, J.S., Rabbani, S., Henderson, A., Blenkinsopp, P., Thompson, S.P., Lockyer, N.P., Vickerman, J.C.: A new dynamic in mass spectral imaging of single biological cells. Anal. Chem. 80, 9058–9064 (2008)
Passarelli, M.K., Ewing, A.G., Winograd, N.: C(60)-SIMS studies of glycerophospholipid in a lipid maps model system: kdo(2)-lipid a stimulated raw 264.7 cells. Surf. Interface Anal. 45, 298–301 (2013)
Passarelli, M.K., Ewing, A.G., Winograd, N.: Single-cell lipidomics: characterizing and imaging lipids on the surface of individual Aplysia californica neurons with cluster secondary ion mass spectrometry. Anal. Chem. 85, 2231–2238 (2013)
Willingham, D., Brenes, D.A., Winograd, N., Wucher, A.: Investigating the fundamentals of molecular depth profiling using strong-field photoionization of sputtered neutrals. Surf. Interface Anal. 43, 45–48 (2011)
Winograd, N.: Ion beams and laser postionization for molecule-specific imaging. Anal. Chem. 65, 622A–629A (1993)
Wucher, A., Tian, H., Winograd, N.: A mixed cluster ion beam to enhance the ionization efficiency in molecular secondary ion mass spectrometry. Rapid Commun. Mass Spectrom. 28, 396–400 (2014)
Liao, H.-Y., Lin, K.-Y., Kao, W.-L., Chang, H.-Y., Huang, C.-C., Shyue, J.-J.: Enhancing the sensitivity of molecular secondary ion mass spectrometry with C60+-o2+ cosputtering. Anal. Chem. 85, 3781–3788 (2013)
Grade, H., Cooks, R.G.: Secondary ion mass spectrometry. Cationization of organic molecules with metals. J. Am. Chem. Soc. 100, 5615–5621 (1978)
Altelaar, A.F.M., Klinkert, I., Jalink, K., de Lange, R.P.J., Adan, R.A.H., Heeren, R.M.A., Piersma, S.R.: Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal. Chem. 78, 734–742 (2005)
Wu, K.J., Odom, R.W.: Matrix-enhanced secondary ion mass spectrometry: a method for molecular analysis of solid surfaces. Anal. Chem. 68, 873–882 (1996)
Nicola, A., Muddiman, D., Hercules, D.: Enhancement of ion intensity in time-of-flight secondary-ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 7, 467–472 (1996)
Altelaar, A.F.M., Luxembourg, S.L., McDonnell, L.A., Piersma, S.R., Heeren, R.M.A.: Imaging mass spectrometry at cellular length scales. Nat. Protoc. 2, 1185–1196 (2007)
Altelaar, A.F.M., Taban, I.M., McDonnell, L.A., Verhaert, P.D.E.M., de Lange, R.P.J., Adan, R.A.H., Mooi, W.J., Heeren, R.M.A., Piersma, S.R.: High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int. J. Mass Spectrom. 260, 203–211 (2007)
Wehbe, N., Heile, A., Arlinghaus, H.F., Bertrand, P., Delcorte, A.: Effects of metal nanoparticles on the secondary ion yields of a model alkane molecule upon atomic and polyatomic projectiles in secondary ion mass spectrometry. Anal. Chem. 80, 6235–6244 (2008)
Delcorte, A., Yunus, S., Wehbe, N., Nieuwjaer, N., Poleunis, C., Felten, A., Houssiau, L., Pireaux, J.J., Bertrand, P.: Metal-assisted secondary ion mass spectrometry using atomic (ga+, in+) and fullerene projectiles. Anal. Chem. 79, 3673–3689 (2007)
Locklear, J.E., Guillermier, C., Verkhoturov, S.V., Schweikert, E.A.: Matrix-enhanced cluster-SIMS. Appl. Surf. Sci. 252, 6624–6627 (2006)
McDonnell, L.A., Heeren, R.M.A., de Lange, R.P.J., Fletcher, I.W.: Higher sensitivity secondary ion mass spectrometry of biological molecules for high resolution, chemically specific imaging. J. Am. Soc. Mass Spectrom. 17, 1195–1202 (2006)
Monroe, E.B., Jurchen, J.C., Lee, J., Rubakhin, S.S., Sweedler, J.V.: Vitamin E imaging and localization in the neuronal membrane. J. Am. Chem. Soc. 127, 12152–12153 (2005)
Hankin, J., Barkley, R., Murphy, R.: Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652 (2007)
Ryan, K.E., Smiley, E.J., Winograd, N., Garrison, B.J.: Angle of incidence effects in a molecular solid. Appl. Surf. Sci. 255, 844–846 (2008)
Taormina, C.R., Nicolette, E., Pedder, R.E., Novak, T.T.: An ion guide study: quadrupoles, rectilinear quadrupoles, hexapoles, and octopoles, 2009. Available at: http://www.ardaratech.com/document-library/technical-notes/11-tn-3007-practical-quadrupole-theory-ion-guides. Accessed 8 Aug 2014
Loboda, A.V., Krutchinsky, A.N., Bromirski, M., Ens, W., Standing, K.G.: A tandem quadrupole/time-of-flight mass spectrometer with a matrix-assisted laser desorption/ionization source: design and performance. Rapid Commun. Mass Spectrom. 14, 1047–1057 (2000)
Brison, J., Robinson, M.A., Benoit, D.S., Muramoto, S., Stayton, P.S., Castner, D.G.: TOF-SIMS 3D imaging of native and non-native species within hela cells. Anal. Chem. 85, 10869–10877 (2013)
Passarelli, M.K., Winograd, N.: Characterizing in situ glycerophospholipids with SIMS and MALDI methodologies. Surf. Interface Anal. 43, 269–271 (2011)
Tucker, K.R., Li, Z., Rubakhin, S.S., Sweedler, J.V.: Secondary ion mass spectrometry imaging of molecular distributions in cultured neurons and their processes: comparative analysis of sample preparation. J. Am. Soc. Mass Spectrom. 23, 1931–1938 (2012)
Miyamoto, M.D.: Binomial analysis of quantal transmitter release at glycerol treated frog neuromuscular junctions. J. Physiol. 250, 121–142 (1975)
Galle, P.: [Sur une nouvelle methode d'analyse cellulaire utilisant le phenomene d'emission ionique secondaire.]. Ann. Phys. Biol. Med. 42, 83–94 (1970)
Ostrowski, S.G., Van Bell, C.T., Winograd, N., Ewing, A.G.: Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science 305, 71–73 (2004)
Rabbani, S., Fletcher, J.S., Lockyer, N.P., Vickerman, J.C.: Exploring subcellular imaging on the buncher-ToF j105 3D chemical imager. Surf. Interface Anal. 43, 380–384 (2011)
Tokareva, E.N., Fardim, P., Pranovich, A.V., Fagerholm, H.P., Daniel, G., Holmbom, B.: Imaging of wood tissue by ToF-SIMS: critical evaluation and development of sample preparation techniques. Appl. Surf. Sci. 253, 7569–7577 (2007)
Robinson, M.A., Castner, D.G.: Characterization of sample preparation methods of nih/3t3 fibroblasts for ToF-SIMS analysis. Biointerphases 8, 15 (2013)
Smith, C.A., O'Maille, G., Want, E.J., Qin, C., Trauger, S.A., Brandon, T.R., Custodio, D.E., Abagyan, R., Siuzdak, G.: Metlin: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 (2005)
Monroe, E.B., Annangudi, S.P., Hatcher, N.G., Gutstein, H.B., Rubakhin, S.S., Sweedler, J.V.: SIMS and MALDI MS imaging of the spinal cord. Proteomics 8, 3746–3754 (2008)
Zingg, J.-M.: Modulation of signal transduction by vitamin E. Mol. Aspects Med. 28, 481–506 (2007)
Fletcher, J., Kotze, H., Armitage, E., Lockyer, N., Vickerman, J.: Evaluating the challenges associated with time-of-fight secondary ion mass spectrometry for metabolomics using pure and mixed metabolites. Metabolomics 9, 535–544 (2013)
Glanzman, D.L., Kandel, E.R., Schacher, S.: Identified target motor neuron regulates neurite outgrowth and synapse formation of Aplysia sensory neurons in vitro. Neuron 3, 441–450 (1989)
Girod, M., Shi, Y., Cheng, J.-X., Cooks, R.G.: Desorption electrospray ionization imaging mass spectrometry of lipids in rat spinal cord. J. Am. Soc. Mass Spectrom. 21, 1177–1189 (2010)
Yang, J., Caprioli, R.M.: Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 83, 5728–5734 (2011)
Acknowledgments
The authors are very grateful for instrumental design and troubleshooting advice provided by Alexandre Loboda at AB SCIEX (presently at Fludigm Corporation), Randy Pedder at Ardara Technologies, and the Winograd research group at Penn State University, particularly with Professor Nicholas Winograd, Melissa Passarelli, Anita Durairaj, and Lauren Jackson. Design assistance by Zhen Li and Kevin Tucker, precision machining by Michael A. Harland and Roger Smith at the Machine Shop of the School of Chemical Sciences at the University of Illinois at Urbana-Champaign, and cell dissection and culture assistance by Xiying Wang and Callie (Croushore) Kindt are also acknowledged. This work was supported by the Department of Energy Office of Biological and Environmental Research through grant DE SC0006642, and The National Resource for Aplysia funded by PHS grant P40 OD010952. The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 532 kb)
Rights and permissions
About this article
Cite this article
Lanni, E.J., Dunham, S.J.B., Nemes, P. et al. Biomolecular Imaging with a C60-SIMS/MALDI Dual Ion Source Hybrid Mass Spectrometer: Instrumentation, Matrix Enhancement, and Single Cell Analysis. J. Am. Soc. Mass Spectrom. 25, 1897–1907 (2014). https://doi.org/10.1007/s13361-014-0978-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-014-0978-9