Abstract
Peptidoglycan recognition proteins (PGRPs) contribute to the immune response of vertebrates and higher invertebrates. Animals have a varying number of PGRP genes: 4 in human and mouse, 13 in Drosophila, and 0 in an aphid. Transcriptome analyses revealed that numerous PGRP genes (> 300 in an RNA-Seq analysis) were expressed in the green rice leafhopper Nephotettix cincticeps (Uhler) (Hemiptera, Cicadellidae). PGRP genes were mostly expressed in the bacteriomes, in which two bacterial endosymbionts, Sulcia and Nasuia, are harbored. E. coli inoculation upregulated antimicrobial peptide gene expression in the leafhopper, whereas PGRP gene expression was not affected by the bacterial challenge. High and constant expression of PGRP genes in the bacteriomes and the lack of an immune response against invading bacteria suggest that these PGRPs play a hitherto unknown role in the bacteriomes, implicating a relationship with symbiotic association in N. cincticeps.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peptidoglycan recognition proteins (PGRPs) function as pattern recognition receptors (PRRs), and were first discovered as hemolymph proteins of the silkworm Bombyx mori (Linnaeus) (Yoshida et al. 1996). PGRPs recognize pathogens and contribute to the process of their elimination in vertebrates and higher invertebrates (Boneca 2009; Dziarski 2004; Michel et al. 2001; Steiner 2004; Takehana et al. 2002). Peptidoglycan (PGN), a major structural component of the cell wall of almost all bacterial species, activates signal transduction pathways for antimicrobial defense after being recognized by PGRPs (Dziarski and Gupta 2006; Guan and Mariuzza 2007; Royet and Dziarski 2007). Some PGRPs exhibit amidase activity that enables them to work as PGN scavengers, resulting in suppressing a heightened immune response to PGN (Bischoff et al. 2006; Kim et al. 2003; Mellroth et al. 2003; Persson et al. 2007; Zaidman-Remy et al. 2006, 2011). PGRPs thus work for both activation and suppression of immune response in vertebrates and invertebrates alike (Kurata, 2014; Royet et al. 2011).
Humans and mice possess four PGRP genes (Royet and Dziarski 2007). In insects, PGRPs activate the proPO cascade (Yoshida et al. 1996), Toll and Imd pathway (Royet et al. 2005), and cellular autophagy (Yano et al. 2008). In Drosophila melanogaster Meigen, PGRP-SD and -SA function as PRRs for Gram-positive bacteria upstream of the Toll pathway (Bischoff et al. 2004; Michel et al. 2001) and PGRP-LC and -LE as PRRs for Gram-negative and some of Gram-positive bacteria in the Imd pathway (Choe et al. 2002; Gottar et al. 2002; Leulier et al. 2003; Rämet et al. 2002; Takehana et al. 2002). PGRP-LE is also an intracellular recognition molecule inducing macroautophagy in Drosophila (Yano and Kurata 2011). The number of PGRP genes varies among insect species: D. melanogaster has 13 PGRP genes (Ferrandon et al. 2007); the mosquito Anopheles gambiae Giles has 7 (Christophides et al. 2002); the honeybee Apis mellifera Linnaeus has 4 (Evans et al. 2006); the beetle Tribolium castaneum (Herbst) has 8 (Zou et al. 2007); and the silkworm B. mori has 12 (Tanaka et al. 2008) but the aphid Acrythosiphon pisum Harris lacks PGRP genes (Gerardo et al. 2010).
The green rice leafhopper Nephotettix cincticeps (Uhler) (Hemiptera, Cicadellidae) (Online Resource 1A), a common rice pest found in Asian countries with moderate climates, harbors two bacterial endosymbionts and a symbiotic Rickettsia (Mitsuhashi and Kono 1975). A pair of symbiotic organs called the bacteriome, which is located on both sides of the anterior part of the abdomen (Online Resource 1B), stores Sulcia and Nasuia, members of the phylum Bacteroidetes and the class betaproteobacteria, respectively (Online Resource 1C and D) (Noda et al. 2012). Sulcia is present in an outer region of the bacteriome, whereas Nasuia is restricted to an inner region of the bacteriome. The bacteriome symbionts are vertically transmitted via the ovaries from generation to generation (Nasu 1965). The whole-genome analyses of Sulcia and Nasuia species revealed that these endosymbiotic bacteria have very small sized genomes and provide essential nutrients to their host insects (Bennett and Moran 2013; McCutcheon 2010; McCutcheon and Moran 2007, 2012). While on the other hand, expressed host genes in the bacteriome are poorly understood.
In this study, we report the discovery and expression of PGRP genes of the green rice leafhopper N. cincticeps (hereafter referred to as “NcPGRP”). An unexpected large number of PGRP genes was discovered from expressed sequence tag (EST) and RNA-Seq analyses of this species, and their expression was mostly restricted to the bacteriome. The results of our study indicate that there is an unusual function of these PGRP genes with regards to the symbiotic association between the leafhopper and bacteriome symbionts.
Materials and methods
Insects and antibiotic treatment
The green rice leafhopper N. cincticeps, collected at Yawara, Ibaraki, were reared on rice seedlings at 26 °C under a 16:8 h light: dark cycle in a plastic box (30 × 28 × 24 cm). The leafhoppers were individually reared in glass test tubes (130 mm length and 16 mm diameter) containing rice seedlings and their developmental stages were examined. Antibiotics (tetracycline, rifampicin, and ampicillin) were individually administered to each leafhopper to kill symbionts. Antibiotics solution (0.05%) was given to rice seedlings in the glass test tubes from 0-day-old 1st instar nymphal stage (Noda et al. 2001).
Expressed sequence tag analysis
EST analysis was performed according to the method prescribed by Noda et al. (2008). mRNA was extracted from each sample; approximately 20 insects were used for whole-body libraries, ca. 50 individuals for dissected tissue or body part libraries, and ca. 200 for egg libraries (https://ncest.dna.affrc.go.jp/yokobai_lib_name.html). cDNA was synthesized using an oligo-dT primer along with a SMART II oligonucleotide in a SMART RACE cDNA Amplification Kit (Clontech Laboratories Inc., Palo Alto, CA, USA). Amplification was performed by using SMART technology, and the nested universal primer (NUP) and the 3′-polymerase chain reaction (PCR) primer available in the kit. The PCR products were cloned into pGEM®-T cloning vectors (Promega Corp., Madison, WI, USA). The NUP primer was used for the sequencing reaction by using colony PCR products amplified with SP6 and T7 primers as the PCR template. Sequencing was performed using Big Dye Terminator v3.0 or v3.1 (Applied Biosystems, USA) and a DNA analyzer (model 3700, PE Applied Biosystems). Sequencing data were first manually processed and examined, and the following EST sequences were eliminated after blast analyses: cloning vector, Escherichia coli genome, mitochondrial genome, and ribosomal rRNA gene sequences. The EST sequences of N. cincticeps are available on https://ncest.dna.affrc.go.jp.
PCR, sequencing, and RT-PCR
Full-length cDNA sequences were determined using the SMART RACE cDNA Amplification Kit (Clontech Laboratories Inc.) or 5′ RACE System for Rapid Amplification of cDNA Ends (Invitrogen). PCR products were cloned into pGEM®-T cloning vectors (Promega Corp.) and the sequences were determined according to the abovementioned method using SP6, T7, and sequence-specific primers.
The expression of 18 NcPGRP genes was determined by reverse transcription PCR (RT-PCR) using specific primers (Online Resource 2). The head, thorax, abdomen, midgut, and ovary were dissected from five 3-day-old adult females and the testis was from five 3-day-old adult males. Bacteriomes were dissected from 8 adults each of both the sexes. Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and cDNA was synthesized with SuperScript II-Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). The conditions for RT-PCR were as follows: 1 cycle of 95 °C for 1 min; 30–35 cycles of 95 °C for 30 s, 54–60 °C for 30 s, and 72 °C for 90 s; and final extension of 72 °C for 5 min. Ribosomal protein L10 (NcRpL10) gene was also amplified for validating the cDNA samples.
The expression levels of NcPGRP1 and NcPGRP12 genes were determined based on each developmental stage of the leafhopper by real-time reverse transcription PCR (RRT-PCR). The stages tested were 5- to 7-day-old eggs (50 eggs), 1st instar nymph (30 nymphs), 2nd instar nymph (20 nymphs), 3rd–5th instar nymph (10 nymphs), and 0-, 3-, 7-day-old males and females (5 adults each from both the sexes). cDNA templates were prepared as described above and RRT-PCR was performed using LightCycler 480 SYBR Green I Master (Roche Diagnostics, Indianapolis, IN, USA) at the thermal conditions of 1 cycle of 95 °C for 5 min; 50 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 10 s. Absence of undesirable byproducts was confirmed by automated melting curve analysis. The details of the primers used for RRT-PCR are provided in Online Resource 2.
Computing data and phylogenetic analyses
Clustering analysis of the EST sequences was performed using CLOBB and CLOBB2 algorithms (Parkinson et al. 2002). The parameters used for clustering were 95% identity and 50-bp coverage. Blast analysis was performed using NCBI blast 2.2.26 (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). Signal peptides were predicted using SignalP 4.1 (https://www.cbs.dtu.dk/services/SignalP-4.1/), SignalP 5.0 (https://www.cbs.dtu.dk/services/SignalP/), and Phobius (https://phobius.sbc.su.se), and transmembrane regions were examined using TMHMM ver. 2.0 (https://www.cbs.dtu.dk/services/TMHMM/) and Phobius. Multiple alignments were performed using Clustal X ver. 1.83 and the alignment was manually corrected. Phylogenetic analysis was performed by the Neighbor-Joining method using MEGA ver. 4.1. Bootstrap analysis was performed with 1000 replicates.
Illumina sequencing and de novo assembly
Total RNAs were isolated from the whole bodies of 10 adult males, adult females, and mature (4th–5th instar) nymphs each using an RNeasy Mini kit (Qiagen). mRNA was purified by passing the total RNA through a chromatographic column of beads with oligo (dT)-cellulose, and was subsequently fragmented into short sequences of ca. 200 bp. The fragmented mRNA was ligated with the adaptor mix and was transcribed to cDNA by reverse transcriptase before sequencing (Hokkaido System Science Co., Ltd., Japan). Sequencing was performed on an Illumina HiSeq-2000 sequencing system, which generated 40.3 million pairs of 100 base length paired-end reads (8.07 Gb). The raw sequence data were submitted to the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) database and is accessible through the BioProject [DRA008970]. Prior to assembly, the sequenced raw data were filtered with ea-utils (version 1.1.2) to obtain clean reads by removing adaptors, and this process resulted in a total of 39,951,943 pairs of high-quality reads (99.05% of total raw reads). De novo assembly of the quality-processed reads was performed using the Trinity assembly program (version r2013-02-25).
Calculation of bacterial density
The number of bacteria in the leafhoppers was counted by calculating the copy number of the rRNA gene of Nasuia and Sulcia by quantitative PCR (qPCR). DNA was individually isolated from antibiotic-treated 0-day-old 5th instar nymphs by using the DNeasy Blood and Tissue Kit (Qiagen). qPCR was performed as mentioned earlier using LightCycler 480 (Roche Diagnostics). The details of the primers used for the qPCR are provided in Online Resource 2. Since Nasuia and Sulcia possess one copy of the rRNA gene in the genome (Bennett and Moran 2013; McCutcheon and Moran 2007), the copy number of the gene calculated in qPCR was regarded as the number of bacteria in one leafhopper individual.
Western blotting
Mouse anti-NcPGRP12 antibody was prepared using a recombinant protein expressed in E. coli. The sequence corresponding to the open reading frame of the NcPGRP12 gene was cloned into Novagen’s pET42a plasmid (Merck-Millipore), and expressed in the E. coli BL21. Polyclonal antibody was obtained by injecting the recombinant protein into a mouse after His-tag purification. Protein samples for Western blotting were prepared from the head, thorax, 1st–4th segments of the abdomen, the rest of the segments of the abdomen, and the bacteriome obtained from 3-day-old females. The bacteriomes are found in the 2nd–4th segments of the abdomen (Online Resource 1B). Samples were homogenized with the phosphate buffer with 1% triton X-100 and sonicated using the ultrasonic homogenizer ML-2000 (Misonix Inc, Farmingdale, NY, USA). Water-soluble fraction was electrophoresed in a 12.5% polyacrylamide gel (e-PAGEL E-T 12.5L, ATTO Corporation, Tokyo, Japan). Proteins were transferred from the polyacrylamide gel to the PVDF sheet (Hybond-P, GE Healthcare, Buckinghamshire, UK) using the transfer blotting apparatus Trans-Blot SD (Bio-Rad, Hercules, CA, USA) and made to react with the antiserum to NcPGRP12 (1000-fold dilution). After treatment with anti-mouse IgG-HRP (MBL Co., Ltd, Nagoya, Japan) (2000-fold dilution), the proteins were detected with the HistoMark TrueBlue Peroxidase System (KPL, Gaithersburg, MD, USA).
Microarray analysis
The leafhopper 15 K oligo-microarray slide (60-mer oligonucleotides on 15,118 spots for Nephotettix genes, Agilent Technologies, Palo Alto, CA, USA; #2527323) was constructed using EST sequences. One or 2 oligo-probe sequences were designed from the 9180 gene clusters from the EST sequences. cDNA and labeled cRNA were synthesized using the Quick Amp Labeling Kit according to the manufacturer’s instruction (Agilent Technologies). Total RNA 400 ng each for Cyanine 3-CTP (Cy3) and Cyanine 5-CTP (Cy5) was converted to cDNAs with the T7 promoter oligo-dT primer and Moloney Murine Leukemia Virus reverse transcriptase (MMLV-RT). Second-strand cDNA was transcribed to cRNA using T7 RNA polymerase together with Cy3 or Cy5. The labeled cRNA was purified using the RNeasy Mini Kit (Qiagen). The labeled cRNA (825 ng) was hybridized to a microarray slide for 17 h at 65 °C. The arrays were then washed using the Wash Buffer Kit (Agilent Technologies) and scanned at 5 µm resolutions using Agilent Microarray Scanner. The scanned signals were calculated with Feature Extraction Software v. 9.5 (Agilent Technologies). Signal data were analyzed using GeneSpring software v. 9.0 (Agilent Technologies). Statistical analysis was performed using t test in the GeneSpring.
Two-color dye-swap microarray analyses were performed for the bacteria (E. coli)-injected female adults and distilled water-injected female adults to elucidate the effect of bacterial challenge on PGRP gene expression. E. coli-suspended water was infused into a glass capillary tube and set on a manipulator model MYN-1 (Narishige, Tokyo, Japan). Leafhoppers were anesthetized by putting them on ice before injection. The capillary was inserted at the intersegmental region between thorax and abdomen on the ventral side under a microscope, and the bacteria-suspended water (0.03 µl) was injected into the body cavity using Transjector 5246 (Eppendorf, Hamburg, Germany). Three or 12 h after injection, total RNA was extracted from the abdomen of 5 leafhoppers using RNeasy Mini Kit (Qiagen).
Another set of the same analyses was also performed between Rickettsia-infected and Rickettsia-eliminated leafhopper adults (Watanabe et al. 2014) for comparing the PGRP gene expression level. The microarray analysis data were submitted to NCBI (GEO accession numbers, GSE135974 and GSE135973, respectively).
Results
PGRP genes in the ESTs of N. cincticeps
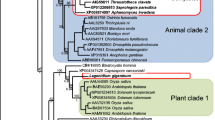
EST analyses were performed for each tissue of the leafhopper (https://ncest.dna.affrc.go.jp). Genes dominantly- and specifically-expressed in the bacteriome were selected from Nephotettix ESTs for obtaining insect genes related to symbiosis. Genes were first surveyed in 22,097 EST sequences that were generated from the bacteriome, ovary, testis, salivary glands, and a cell line (Nc-24, Kimura and Omura 1988). The total EST sequences from the different tissues were grouped together using the clustering script CLOBB (Parkinson et al. 2002) under the parameters of 95% identity and 50-bp coverage, resulting in the identification of 6773 clusters including 3895 singlets. The gene clusters that exclusively contained clones from the bacteriome library were then manually surveyed. Together with many undefined genes and housekeeping genes, PGRP genes were noticeably expressed in the bacteriome. Eighteen gene clusters, which include 5 or more clones whose sequences highly showed homology to the PGRP genes, were selected. PGRP gene clusters that contained 4 or less clones were discarded for further analyses. Complete sequences of the 18 PGRP genes were determined by 5′ rapid amplification of cDNA ends (5′ RACE) and 3′ RACE. The genes were labeled as NcPGRP1 to NcPGRP18 in the order of descending prevalence of the number of EST clones in each cluster and then in the order of the descending prevalence of the size of coding sequence (accession numbers LC484294–LC484311). The size of an amino acid sequence of the NcPGRPs ranged from 148 to 300 with an average size of 195, which is equivalent to or smaller than those of Drosophila PGRPs-S (~ 203) and PGRPs-L (~ 520). The evolutionary relationship between NcPGRPs and PGRPs of other insects was elucidated by the phylogenetic analysis of the amino acid sequences; the phylogenetic tree indicated that NcPGRPs formed a monophyletic group together with one PGRP from T. castaneum (PGRP-LF) (Fig. 1).
Phylogenetic tree of PGRP in insects. NJ-tree of PGRP domain (164 sites) was constructed using MEGA ver. 4. Amino acid alignment was performed using ClustalW. Closed circles show the nodes whose bootstrap values were above 50%. Initial letters show the insect species: Ag Anopheles gambiae, Am Apis mellifera, Bm Bombyx mori, Dm Drosophila melanogaster, Gmm Glossina morsitans morsitans, Nc Nephotettix cincticeps (underlined), Sz Sitophilus zeamais, Tc Tribolium castaneum, Ir Ixodes ricinus (outgroup)
Bacteriome-specific expression of PGRP genes
The level of NcPGRP gene expression in the head, thorax, abdomen (without the bacteriome and gut), midgut, ovary, testis, female bacteriome, and male bacteriome was determined by RT-PCR. The thorax and abdomen included the fat body. All the 18 genes were almost exclusively expressed in the bacteriome (Fig. 2). PGRPs are generally found in immune-competent organs and hemolymph in insects, and PGRP genes are mainly expressed in the fat body, and lesser in the cuticular epidermis, midgut, and hemocytes (Dziarski and Gupta 2006). However, in the samples of our study, the 18 NcPGRP genes were not expressed in the thorax and abdomen that include the fat body and epidermis. Expression was not also observed in the midgut (Fig. 2). The expression profile throughout the leafhopper developmental stages was examined by RRT-PCR in 2 NcPGRP genes, NcPGRP1 that was highly expressed based on the number of EST (described below) and NcPGRP12 that had the longest translation size (300 amino acids) among the 18 genes. These NcPGRP genes were ubiquitously expressed in the leafhopper, showing moderate fluctuation in expression level through the developmental stages (Online Resource 3).
Expression of 18 PGRP genes in various tissues of Nephotettix cincticeps. All the tissue samples were prepared from 3-day-old adult females except the testis and male bacteriome samples, which were prepared from 3-day-old adult males. The sample from the abdomen did not include the bacteriome. The ribosomal protein L10 (NcRpL10) gene was used as a control for RT-PCR
Bacteriome-specific localization of PGRP
Prediction analyses of signal peptides and the transmembrane region of NcPGRPs suggested that NcPGRPs were located in the bacteriome rather than being secreted from the organ. The deduced amino acid sequences were analyzed using SignalP 4.0 (https://www.cbs.dtu.dk/services/SignalP/), Phobius (https://phobius.sbc.su.se/), and TMHMM Server v. 2.0 (https://www.cbs.dtu.dk/services/TMHMM-2.0/). All NcPGRPs did not possess any signal peptide sequences and transmembrane region, except NcPGRP10. NcPGRP10 was predicted to contain a signal peptide sequence from the 1–17 residues and a transmembrane region in 20–45 residues by Phobius, with their provability value 0.6–0.7. However, SinalP and TMHMM did not support the presence of signal peptide and a transmembrane region in NcPGRP10.
To detect the location of PGRP proteins, mouse antisera against PGRP1 and PGRP12 were prepared for Western blot analysis. Since the antiserum against PGRP1 did not show high antibody titre, the location of PGRP was determined using the antiserum against NcPGRP12. NcPGRP12 was detected in the bacteriome and the anterior part of the abdomen that includes the bacteriome; no signal was observed in the head, thorax, and posterior part of the abdomen (Fig. 3).
Localization of NcPGRP12 in an adult female. Body parts of ten 3-day-old adult females were used for analysis. a Stained with Coomassie Brilliant Blue. b Western blotting using anti-NcPGRP12 antiserum. Anterior abdomen corresponds to the 1st to 4th segments of the abdomen and posterior abdomen corresponds to the 5th segments onwards to the tail. Anterior abdomen contains the bacteriome
Immune response in N. cincticeps
Bacterial inoculation into insects by microinjection usually upregulates PGRP gene expression together with an increase in antibacterial peptide gene expression (Irving et al. 2001; Lee et al. 2007). Therefore, E. coli was injected into female adults and the expression profiles of immune system-related genes were analyzed using the Nephotettix microarray (8 × 15 K, Agilent). The microarray included 297 PGRP gene probes from 150 PGRP genes; 2 probes were designed for most of the PGRP gene sequences. Total RNAs were isolated from the abdomen of 5 female adults.
Antibacterial protein genes were clearly upregulated by the bacterial challenge after 12 h (Online Resource 4); defencin gene (probe number NCNY2028) showed a 2.2-fold change at 3 h (p > 0.05, t test based on 3 biological replicates) and 8.3 at 12 h (p < 0.01, t test based on 4 biological replicates), and diptericin gene (probe numbers NCMG2043 and NCMG4985) 2.1 on average at 3 h (1.97–2.26, p > 0.05) and 5.0 on average at 12 h (3.46–6.62, p < 0.01) after bacterial injection. Although immune response pathways have not been elucidated in N. cincticeps, E. coli was recognized by the leafhopper. However, the E. coli challenge did not upregulate any NcPGRP genes after 3 h and 12 h with one exception among 297 probes on the microarray at 12 h (NC_Bac4333-1, p < 0.05, t test based on 4 biological replicates) (Online Resource 4).
Nephotettix cincticep used in the experiments harbors symbiotic Rickettsia in the whole body (Noda et al. 2012; Watanabe et al. 2014), which is considered to possess PGN in its outer membrane. A Rickettsia-uninfected colony was created by administrating rifampicin (Noda et al. 2001; Watanabe et al. 2014). Microarray analysis was performed for Rickettsia-infected and -uninfected young adults and the infection status of all examined individuals was confirmed by diagnostic PCR. Antimicrobial peptide genes (NCNY2028, NCMG2043, and NCMG4985) were slightly upregulated but the extent of upregulation was not significant (Online Resource 5). The expression level of many PGRP genes (246 out of 297 probes) were not affected by Rickettsia infection (p > 0.05, t test based on 4 biological replicates), although 45 probes were upregulated and 6 probes were downregulated (p < 0.05) (data not shown).
The effect of antibiotic treatments on symbiont populations and PGRP gene expression
Microarray analysis did not detect the activation of NcPGRP genes in the bacterial challenge. Together with the fact that most of NcPGRPs are considered to be located in the bacteriome, they may not participate in an immune response against invading microorganisms. Therefore, the bacteriome symbionts were exposed to antibiotics to disrupt the normal balance between the host and the symbionts in the bacteriome, and PGRP expression level was examined. Tetracycline, rifampicin, or ampicillin was administered from the 1st instar nymph stage and 16S rRNA gene copy numbers of Nasuia and Sulcia were calculated in 0-day-old 5th instar nymphs by qPCR. Sulcia population significantly decreased after tetracycline treatment (Online Resource 6A), and Nasuia population in the bacteriome was significantly decreased after tetracycline and rifampicin treatments (Online Resource 6B). The expression levels of 18 NcPGRP genes were examined by RRT-PCR in the antibiotic-treated 5th instar leafhoppers in comparison with untreated 5th instar leafhoppers. The expression levels of NcPGRP genes were reduced after antibiotic treatments, with the extent of reduction being in the following order: tetracycline > rifampicin > ampicillin (Online Resource 6C–E). Significant reduction was observed in 12, 11, and 7 PGRP genes after tetracycline, rifampicin, or ampicillin treatment, respectively. In tetracycline-treated samples, some PGRP genes showed less than 10% expression level compared to those of the untreated ones.
An abundance of PGRP genes in N. cincticeps
The genes specifically expressed in the bacteriome were again examined after the analysis of the last EST library. A total of 38,309 ESTs were finally generated from 14 plasmid libraries made from 10 different organs or tissues (the mycetome, midgut, ovary, testis, salivary glands, compound eye, egg, nymphal whole body, cell line, and bacteria-challenged female adult) (https://ncest.dna.affrc.go.jp). The number of EST sequences obtained from the libraries of the bacteriome NcMYA and NcMYB was 3095. The 3095 ESTs from the bacteriome were clustered into 1293 groups containing 832 singletons (under the parameters of 95% identity and 50 bp coverage). The top 20 largest clusters are shown in Online Resource 7. The top 3 cluster genes were undefined and the 4th one was the PGRP gene (NcPGRP1). PGRP genes also ranked 18th and 20th on the list (NcPGRP2 and NcPGRP3, respectively). PGRP genes were only found in the libraries of bacteriome and bacteriome-containing tissues, except two PGRP genes, NCTE-1686 and NCTE-0899, which were found from the testis library. Other highly expressed genes in the bacteriome were those for bis5-nucleosyl-tetraphosphatase, arginine kinase, ferritin, lipoyltransferase, bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase, mitochondrial SarDH, and ribosomal protein L19, and 7 uncharacterized genes.
The abundance of PGRP genes in N. cincticeps was also confirmed by the RNA-Seq data of the whole leafhopper body. De novo assembly of the RNA-Seq sequences by Trinity (https://trinityrnaseq.sourceforge.net/) yielded 102,723 contigs with an average length of 1,115 bp and a median length of 456 bp. Tblastn analysis was performed against the contig nucleotide sequences using the putative amino acid sequences of the Drosophila 12 PGRP as queries (DmPGRP-LA [Q95T64], DmPGRP-LB [Q8INK6], DmPGRP-LC [Q9GNK5], DmPGRP-LD [Q9GN97], DmPGRP-LE [Q9VXN9], DmPGRP-LF [Q8SXQ7], DmPGRP-SA [Q9VYX7], DmPGRP-SB1 [NP_648917], DmPGRP-SB2 [NP_648916], DmPGRP-SC1 [NP_610407], DmPGRP-SC2 [NP_610410], and DmPGRP-SD [NP_648145]). The blast analysis found 572 contig sequences with PGRP similarity (E value < 10). Preliminary homology searches (blastx) of the 572 sequences against the non-redundant NCBI DNA database were performed to eliminate sequences that were not related to the PGRP gene, indicating that 554 sequences showed the best homology to PGRP. Among the 554 sequences, isoform sequences with the same Trinity gene ID but different isoform ID were combined. The longest or moderately sized contigs that included PGRP domain were selected, resulting in 317 independent sequences. Blastx homology searches were performed again against the most recent database of non-redundant GenBank CDS translations (e value < 1e−03). Finally, 307 contigs were selected as PGRP or PGRP-related genes of N. cincticeps (Online Resource 8). The corresponding genes in the EST libraries were blast-searched and 155 ESTs were found (Online Resource 8).
Eighteen NcPGRP did not possess signal peptides as previously described. The 307 sequences were then tested for signal peptides using SignalP 5.0. Two genes, comp41637_c0_seq1 and comp85737_c0_seq1 possessed signal peptides. These two genes were not found in ESTs (Online Resource 8).
Some NcPGRP sequences among the 307 contigs showed irregular features compared to the PGRP genes. For example, a plausible consensus amino acid sequence “YNFXXXXXXXXYEGRGW” or similar amino acid sequences were found in the putative amino acid sequences of most of the contigs (Online Resource 9). However, such sequences were not found in some contig sequences. Moreover, some contigs were too short as a PGRP gene or included stop codons in the PGRP domain. Such irregular features were found in at least 35 contigs, which suggests that these genes cannot be translated into functional PGRP proteins.
Discussion
The present study on N. cincticeps revealed 2 unusual features of the PGRP genes. One, NcPGRP genes were mostly expressed in the bacteriome. Most NcPGRP genes were found in the libraries of bacteriome and bacteriome-containing tissues and a few in those of other tissues in EST analyses. Their expression in other tissues was highly insignificant as far as at least 18 genes identified are concerned (Fig. 2). High level of PGRP gene expression in Sulcia and Nasuia bacteriocytes and low level of expression in the insect body could be actually found in an RNA-Seq study reporting the expression levels of 11 PGRP genes from the aster leafhopper Macrosteles quadrilineatus Forbes (Mao et al. 2018, Table S2 in supplementary information). The other unusual feature is that an extremely large variety of PGRP genes were expressed in N. cincticeps (Online Resource 8, 9). This large number of PGRP genes in leafhoppers has not been reported so far. These inexplicable facts of PGRP genes posed some questions that have been discussed below.
It is well established that the most important role of PGRPs in insects is to recognize the invading bacteria. However, it is questionable whether NcPGRPs work as PRR molecules against the bacteria. Many NcPGRP genes were expressed in the bacteriome. NcPGRP molecules, which did not have a signal peptide, seem to work in the cytoplasm of bacteriome cells. Actually, Western blot analysis showed the presence of NcPGRP12 in the bacteriome (Fig. 3). NcPGRPs, therefore, do not easily reach the invading bacteria in the hemolymph. Two genes in the contigs of RNA-Seq analysis, comp41637_c0_seq1 and comp85737_c0_seq1, which possessed a signal peptide, require further examination. PGRP genes are usually known to be upregulated in response to invading bacteria, for example in Drosophila (Irving et al. 2001), Bombyx (Lee et al. 2007), the maize weevil Sitophilus zeamais Motschulsky (Anselme et al. 2006), and the brown planthopper Nilaparvata lugens (Stål) (Bao et al. 2013). No significant upregulation was observed in 150 NcPGRP genes used for microarray analysis upon the bacterial challenge (Online Resource 4). No noticeable upregulation in many PGRP genes including the 18 highly expressed genes was also observed in Rickettsia-infected leafhopper (Online Resource 5). The Rickettsia symbiont infects the cytoplasm and nuclei of most leafhopper tissues including the bacteriome (Mitsuhashi and Kono 1975; Noda et al. 2012; Watanabe et al. 2014), apparently having PGN as well as other rickettsiae.
The PGRP genes expressed in the symbiotic organs are so far reported in 2 groups of insects: weevils (Anselme et al. 2006, 2008; Heddi et al. 2005; Maire et al. 2019; Vigneron et al. 2012) and the tsetsefly (Wang and Aksoy 2012; Wang et al. 2009). The maize weevil S. zeamais expressed a PGRP gene in the bacteriome, which is located at the junction of foregut and midgut (Nardon and Grenier 1989). The weevil PGRP gene, an ortholog of Drosophila PGRP-LB, shows a high steady-state level expression in the bacteriome and its expression is upregulated by Gram-negative bacteria (Anselme et al. 2006). Symbiont infection in the bacteriome also causes upregulation of the PGRP gene. Moreover, the symbionts outside the bacteriome are recognized as microbial intruders by the host, inducing antibacterial peptide production (Anselme et al. 2008). A notable point is that overexpression of the PGRP gene in the bacteriome would prevent the activation of the immune pathway by its putative enzyme activity as a scavenger, similar to the Drosophila PGRP-LB gene (Anselme et al. 2006). A similar case has been reported in the tsetse fly Glossina morsitans morsitans Westwood, which possesses the obligate endosymbiont Wigglesworthia in the midgut bacteriome. The antibacterial peptides of this fly killed the symbionts. The tsetse PGRP-LB gene is considered to degrade the PGN released by the bacterial endosymbionts (Bing et al. 2017; Wang and Aksoy 2012; Wang et al. 2009) through N-acetylmuramyl-l-alanine amidase activity of the PGRP molecule (Zaidman-Remy et al. 2006), resulting in preventing the activation of the host’s immune responses, which damages the symbionts in the bacteriome. The involvement of PGRP in microbial association in animals has also been observed in some invertebrates such as the bobtail squid (Troll et al. 2009) wherein a PGRP neutralizes a PGN-derived toxin of the luminescence-producing bacterial symbiont, Vibrio fischeri (Troll et al. 2010). All these examples indicate that PGRPs exhibit enzymatic activity and degrade the PGN of symbionts that may trigger the host’s immune response.
PGRPs that function as PGN scavengers are known to have 5 important amino acid residues for their enzymatic activity, namely, His-17, Tyr-46, His-122, Lys-128, and Cys-130 compared with the T7 lysozyme amino acid structure (Cheng et al. 1994; Mellroth et al. 2003; Persson et al. 2007). Drosophila PGRPs that exhibit amidase activity, e.g., DmPGRP-LB, -SB1, -SB2, -SC1a, -SC1b, and SC2, all possess 4 amino acids, namely, His-17, Tyr-46, His-122, and Cys-130. The abovementioned PGRPs of the weevils, tsetsefly, and squid possess most of these amino acids (Anselme et al. 2006; Wang et al. 2009; Troll et al. 2010). In contrast, NcPGRPs do not have these amino acid residues; Tyr-46 was found in some NcPGRPs (NcPGRP1, 3, 7, 8, 9, 10, 11, 12, 14, 15, 16, and 17) and His-122 in NcPGRP16, which suggests that NcPGRPs do not work as PGN scavengers for protecting symbionts from the host’s immune attack. NcPGRPs appear to have a function different from that of PGRPs of the weevils, tsetsefly, and squid. An important feature of the bacteriome symbionts of Nephotettix when compared with the symbionts of the weevils, tsetsefly, and squid is the absence of PGN; Nasuia and Sulcia do not possess PGN. The peptidoglycan biosynthesis pathway is not found in the five species of Sulcia (Bennett and Moran 2013; McCutcheon and Moran 2007, 2010; McCutcheon et al. 2009; Woyke et al. 2010) and the Nasuia species of Macrosteles quadrilineatus (Bennett and Moran 2013), thus lacking most of the related enzymes. Because of no possession of PGN, Nasuia and Sulcia are amorphous in shape (Mitsuhashi and Kono 1975; Nasu 1965; Noda et al. 2012) (Online Resource 1C and D). The presence of Rickettsia, a probable facultative symbiont in N. cincticeps, also appeared to be unrelated to the high level of expression of PGRP genes (Online Resource 5). These results indicate that N. cincticeps does not need PGN clearance mediated by PGRPs in the bacteriome.
In Nephotettix, bacterial challenge caused the induction of antimicrobial peptide genes, indicating that this species has innate immune pathway(s) as other insect species, although the details of the pathway(s) remain unclear. The expression of many PGRP genes in this leafhopper remained unaffected in the face of a bacterial invasion (Online Resource 4), although this observation does not necessarily deny the presence of PGRP gene(s) that are involved in the immune response pathway. Among hemipteran insects, the brown planthopper N. lugens is reported to possess 2 PGRP genes (Bao et al. 2013) while the pea aphid A. pisum does not have any PGRP genes (Gerardo et al. 2010). A. pisum also lacks some genes of the Imd pathway. In N. cincticeps, 3 genes involved in the Imd pathway, imd, Dredd, and Relish, were blast-surveyed into Nephotettix RNA-Seq sequences using Drosophila gene sequences as queries. Although the gene imd was not found (least e value = 0.023), Dredd (e value 2e−13) and Relish (2e−53) genes were found in the RNA-Seq data (data not shown). N. cincticeps may possess the Imd or Imd-related pathway, and if so, the PRRs working for the pathway should be explored.
The remaining questions are why N. cincticeps possesses more than 300 PGRP genes and what is the function of the NcPGRPs. The phylogenetic tree of insect PGRPs (Fig. 1) suggests that multiple gene duplication has occurred in an evolutionary process in the leafhopper. New genes are often created by gene duplication, and some duplicated genes are maintained in the genome for a long time (Nei and Rooney 2005). Montaño et al. (2011) indicates that the mode of PGRP gene evolution was characterized by birth and death processes. The high level of expression of NcPGRP genes in the bacteriome strongly suggests that PGRPs have an important function in the bacteriome. However, some NcPGRP genes show irregular features. Nei and Rooney (2005) indicated that some duplicate genes become nonfunctional through deleterious mutations and are deleted from the genome. Even if we suppose that a large number of PGRP genes is the result of birth, why has the death of PGRP genes, especially in truncated or nonfunctional genes, not occurred in the past evolutionary process?
Antibiotic treatments that killed some of the symbionts suppressed PGRP gene expression (Online Resource 6), indicating an as-yet-unknown relationship among symbionts and PGRPs in the bacteriome. Many PGRP genes are retained in the leafhopper and mostly expressed in the bacteriome. These unusual features of NcPGRP genes are apparently related to the function of this specialized organ for endosymbiosis of the leafhopper. However, the physiological function of the bacteriome is less well understood. Studies of other genes highly expressed in the bacteriome undoubtedly contribute to elucidating the function of the PGRPs in N. cincticeps, because some highly- and bacteriome specifically expressed genes are expected to have novel function important for bacteriome maintenance.
References
Anselme C, Vallier A, Balmand S, Fauvarque MO, Heddi A (2006) Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl Environ Microb 72:6766–6772
Anselme C, Perez-Brocal V, Vallier A, Vincent-Monegat C, Charif D, Latorre A, Moya A, Heddi A (2008) Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol 6:43
Bao YY, Qu LY, Zhao D, Chen LB, Jin HY, Xu LM, Cheng JA, Zhang CX (2013) The genome- and transcriptome-wide analysis of innate immunity in the brown planthopper Nilaparvata lugens. BMC Genom 14:160
Bennett GM, Moran NA (2013) Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol 5:1675–1688
Bing X, Attardo GM, Vigneron A, Aksoy E, Scolari F, Malacrida A, Weiss BL, Aksoy S (2017) Unravelling the relationship between the tsetse fly and its obligate symbiont Wigglesworthia: transcriptomic and metabolomic landscapes reveal highly integrated physiological networks. Proc Biol Sci 284:20170360
Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J (2004) Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol 5:1175–1180
Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J (2006) Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog 2:e14 139–e14 147
Boneca IG (2009) Mammalian PGRPs in the spotlight. Cell Host Microbe 5:109–111
Cheng XD, Zhang X, Pflugrath JW, Studier FW (1994) The Structure of bacteriophage-T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA-polymerase. Proc Natl Acad Sci USA 91:4034–4038
Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV (2002) Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in Drosophila. Science 296:359–362
Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, Von Mering C, Xu JN, Zheng LB, Bork P, Kafatos FC (2002) Immunity-related genes and gene families in Anopheles gambiae. Science 298:159–165
Dziarski R (2004) Peptidoglycan recognition proteins (PGRPs). Mol Immunol 40:877–886
Dziarski R, Gupta D (2006) The peptidoglycan recognition proteins (PGRPs). Genome Biol 7:232
Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, Kanost M, Thompson GJ, Zou Z, Hultmark D (2006) Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol Biol 15:645–656
Ferrandon D, Imler JL, Hetru C, Hoffmann JA (2007) The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol 7:862–874
Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, De Vos M, Duncan EJ, Evans JD, Gabaldon T, Ghanim M, Heddi A, Kaloshian I, Latorre A, Moya A, Nakabachi A, Parker BJ, Perez-Brocal V, Pignatelli M, Rahbe Y, Ramsey JS, Spragg CJ, Tamames J, Tamarit D, Tamborindeguy C, Vincent-Monegat C, Vilcinskas A (2010) Immunity and other defenses in pea aphids Acyrthosiphon pisum. Genome Biol 11:R21
Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J (2002) The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416:640–644
Guan RJ, Mariuzza RA (2007) Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol 15:127–134
Heddi A, Vallier A, Anselme C, Xin H, Rahbe Y, Wackers F (2005) Molecular and cellular profiles of insect bacteriocytes: mutualism and harm at the initial evolutionary step of symbiogenesis. Cell Microbiol 7:293–305
Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffmann JA, Hetru C (2001) A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci USA 98:15119–15124
Kimura I, Omura T (1988) Leafhopper cell cultures as a means for phytoreovirus research. Adv Dis Vector Res 5:111–135
Kim MS, Byun MJ, Oh BH (2003) Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol 4:787–793
Kurata S (2014) Peptidoglycan recognition proteins in Drosophila immunity. Dev Comp Immunol 42:36–41
Lee JH, Lee IH, Noda H, Mita K, Taniai K (2007) Verification of elicitor efficacy of lipopolysaccharides and peptidoglycans on antibacterial peptide gene expression in Bombyx mori. Insect Biochem Mol Biol 37:1338–1347
Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B (2003) The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol 4:478–484
Maire J, Vincent-Monegat C, Balmand S, Vallier A, Herve M, Masson F, Parisot N, Vigneron A, Anselme C, Perrin J, Orlans J, Rahioui I, Da Silva P, Fauvarque MO, Mengin-Lecreulx D, Zaidman-Remy A, Heddi A (2019) Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc Natl Acad Sci USA 116:5623–5632
Mao M, Yang X, Bennett GM (2018) Evolution of host support for two ancient bacterial symbionts with differentially degraded genomes in a leafhopper host. Proc Natl Acad Sci USA 115:E11691–E11700
Mccutcheon JP (2010) The bacterial essence of tiny symbiont genomes. Curr Opin Microbiol 13:73–78
Mccutcheon JP, Moran NA (2007) Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA 104:19392–19397
Mccutcheon JP, Moran NA (2010) Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol 2:708–718
Mccutcheon JP, Moran NA (2012) Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26
Mccutcheon JP, Mcdonald BR, Moran NA (2009) Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA 106:15394–15399
Mellroth P, Karlsson J, Steiner H (2003) A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem 278:7059–7064
Michel T, Reichhart JM, Hoffmann JA, Royet J (2001) Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414:756–759
Mitsuhashi J, Kono Y (1975) Intercellular microoganisms in green rice leafhopper, Nephotettix cincticeps Uhler (Hemiptera; Deltocephalidae). Appl Entomol Zool 10:1–9
Montaño AM, Tsujino F, Takahata N, Satta Y (2011) Evolutionary origin of peptidoglycan recognition proteins in vertebrate innate immune system. BMC Evol Biol 11:79
Nardon P, Grenier AM (1989) Endocytobiosis in coleoptera: Biological, biochemical, and genetic aspects. In: Schwemmler W, Gassner G (eds) Insect endocytobiosis: morphology, physiology, genetics, evolution. CRC Press Inc, Boca Raton, pp 175–216
Nasu S (1965) Electron microscopic studies on transovarial passage of rice dwarf virus. Jpn J Appl Entomol Zool 9:225–237
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Noda H, Koizumi Y, Zhang Q, Deng KJ (2001) Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem Mol Biol 31:727–737
Noda H, Kawai S, Koizumi Y, Matsui K, Zhang Q, Furukawa S, Shimomura M, Mita K (2008) Annotated ESTs from various tissues of the brown planthopper Nilaparvata lugens: a genomic resource for studying agricultural pests. BMC Genom 9:117
Noda H, Watanabe K, Kawai S, Yukuhiro F, Miyoshi T, Tomizawa M, Koizumi Y, Nikoh N, Fukatsu T (2012) Bacteriome-associated endosymbionts of the green rice leafhopper Nephotettix cincticeps (Hemiptera: Cicadellidae). Appl Entomol Zool 47:217–225
Parkinson J, Guiliano DB, Blaxter M (2002) Making sense of EST sequences by CLOBBing them. BMC Bioinform 3:31
Persson C, Oldenvi S, Steiner H (2007) Peptidoglycan recognition protein LF: a negative regulator of Drosophila immunity. Insect Biochem Mol Biol 37:1309–1316
Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, RaB E (2002) Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E-coli. Nature 416:644–648
Royet J, Dziarski R (2007) Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol 5:264–277
Royet J, Reichhart JM, Hoffmann JA (2005) Sensing and signaling during infection in Drosophila. Curr Opin Immunol 17:11–17
Royet J, Gupta D, Dziarski R (2011) Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol 11:837–851
Steiner H (2004) Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev 198:83–96
Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S (2002) Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA 99:13705–13710
Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M (2008) A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem Mol Biol 38:1087–1110
Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, Mcfall-Ngai MJ (2009) Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol 11:1114–1127
Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, Mcfall-Ngai MJ (2010) Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol 12:2190–2203
Vigneron A, Charif D, Vincent-Monegat C, Vallier A, Gavory F, Wincker P, Heddi A (2012) Host gene response to endosymbiont and pathogen in the cereal weevil Sitophilus oryzae. BMC Microbiol 12:S14
Wang JW, Aksoy S (2012) PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse’s offspring. Proc Natl Acad Sci USA 109:10552–10557
Wang JW, Wu YN, Yang GX, Aksoy S (2009) Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci USA 106:12133–12138
Watanabe K, Yukuhiro F, Matsuura Y, Fukatsu T, Noda H (2014) Intra-sperm vertical symbiont transmission. Proc Natl Acad Sci USA 111:7433–7437
Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, Lapidus A, Wu DY, Mccutcheon JP, Mcdonald BR, Moran NA, Bristow J, Cheng JF (2010) One bacterial cell, one complete genome. PLoS ONE 5:e10314
Yano T, Kurata S (2011) Intracellular recognition of pathogens and autophagy as an innate immune host defence. J Biochem 150:143–149
Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S (2008) Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol 9:908–916
Yoshida H, Kinoshita K, Ashida M (1996) Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem 271:13854–13860
Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B (2006) The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24:463–473
Zaidman-Remy A, Poidevin M, Herve M, Welchman DP, Paredes JC, Fahlander C, Steiner H, Mengin-Lecreulx D, Lemaitre B (2011) Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS ONE 6:e17231
Zou Z, Evans JD, Lu ZQ, Zhao PC, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang HB (2007) Comparative genomic analysis of the Tribolium immune system. Genome Biol 8:R177
Acknowledgements
We thank Y. Koizumi, J. Shiba, M. Ebihara, and Y. Matsumoto for EST analysis, M. Hattori for salivary gland sample preparation, K. Yamamoto and M. Shimomura for EST database construction, A. Jouraku for RNA-Seq, F. Yukuhiro for electron microscopy, T. Omura for provision of the leafhopper cell line, and K. Watanabe and H. Tanaka for their valuable suggestions.
Funding
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industry (BRAIN) and by JSPS KAKENHI Grant Number 25292201.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest directly relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yoshitaka Suetsugu: Deceased.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tomizawa, M., Nakamura, Y., Suetsugu, Y. et al. Numerous peptidoglycan recognition protein genes expressed in the bacteriome of the green rice leafhopper Nephotettix cincticeps (Hemiptera, Cicadellidae). Appl Entomol Zool 55, 259–269 (2020). https://doi.org/10.1007/s13355-020-00680-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13355-020-00680-z