Abstract
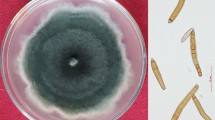
Cucumber (Cucumis sativus L.) is a nutritious domesticated vegetable which belongs to Cucurbitaceae family. India ranks 30th in global cucumber production and its total output is 1,61,000 metric tons annually. In the investigation a different kind of leaf spot disease was observed on 2 months old cucumber leaves samples from cucumber fields in Baruipur, West Bengal, India (Latitude: N 22° 22.262′, Longitude: E 88° 24.5086′). The spots appeared were large, greyish brown in colour surrounded by a yellow to brownish halo, measuring 2 × 1 cm in average. These lesions expanded gradually from circular to angular spots ultimately resulting in necrosis and defoliation. The isolated fungus showed greyish black to dark olive mycelial growth. Microscopic characterization revealed straight to slightly curved conidia and distoseptate with a distinctly protruding basal hilum. Through microscopic observation and ITS sequencing the organism was identified to be Exserohilum rostratum. E. rostratum was previously known for causing blight disease in maize as well as human immune system diseases. E. rostratum, E. longirostratum and E. mcginnisii are identified as human pathogens. Koch’s postulates were carried out and distinct symptoms were observed in cucumber. As because there is no such record, this is the first time the host pathogen combination is separated in India.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
India suffers a huge loss of agricultural produce due to bacterial and fungal attacks. Identification of these pathogens can help in prevention of these attacks. With the passage of time, new host-pathogen relationships are discovered which is mainly due to the ability of the pathogen to shift from one host to another. This ability to infect more than one host depends on the evolutionary history of both pathogen and its host. By investigating the host range variation within a family and using phylogenetic studies one can find out the frequency of host jump events in the course of evolution in that family (Navaud et al. 2018).
Cross kingdom pathogens are those capable of infecting hosts belonging to different kingdoms. For example, Burkholderia cepacia and Aspergillus flavus are potential phytopathogens who have evolved to infect humans. B. cepacia causes fibrosis and A. flavus causes fungal keratitis in humans (Jones et al. 2001; Krishnan et al. 2009). To be a cross kingdom pathogen it should evolve to a level where it can survive in different hosts. Cross kingdom gene transfers are believed to be the source of emergence of these cross kingdom pathogens. When searched for prokaryotic derived HGTs in 60 fully sequenced fungal genomes, 713 transferred genes were detected (Marcet-Houben and Gabaldón 2009). This reveals that cross kingdom HGTs must have played a role in evolution of fungi. Pathogen host jumps have become a major source of new infections in plants as well as humans.
Exserohilum is a dermatiaceous fungi which belongs to the family Pleosporaceae and is known to cause blight in plants. Exserohilum is an anamorphic fungi having the telemorphic or sexual reproductive stage i.e. Setosphaeria. Exserohilum reproduces by the formation of conidia with typically 4–12 septa (Bunkoed et al. 2014). The presence of a protruding hilum at the base of the conidia is an identifying feature of Exserohilum. The conidia are multi-distoseptate, fusiform, cylindrical or obclavate, straight or curved (Hernández-Restrepo et al. 2018).
Exserohilum has emerged to be a potential cross kingdom pathogen. Its relation with human diseases is well documented (Adler et al. 2006). A major outbreak took place in the United States in 2012 which resulted in 64 deaths and 753 cases in total. This outbreak included meningitis, paraspinal/spinal infection and peripheral joint infection. Exserohilum rostratum was identified to be the predominant fungus in patients(60.4%) followed by E. longirostratum(6.3%) and E. mcginnisii. Remaining 31.% were unidentified species. The source of these infections were found to be contaminated preservative-free methylprednisolone acetate (MPA) steroid injections (Katragkou et al. 2014). Cases of fungal keratitis caused by E. rostratum was reported in India and Thailand in 2012 and 2019 respectively (Joseph et al. 2012; Chaidaroon et al. 2019).
Exserohilum is also a phytopathogen which causes leaf spot on grass (Brunings et al. 2009). It is also known to cause infections in both monocotyledonous and dicotyledonous plants which includes leaf spot disease in pineapple plants (Luo et al. 2012), rice brown spot (Toher et al. 2016), northern corn leaf blight (Bashir et al. 2018), banana leaf spot (Shan-Hai et al. 2011) and leaf spot on bottle gourd (Choudhary et al. 2018). Different parameters like relative humidity, rainfall and temperature were shown to favour maize infection by Exserohilum (Nwanosike and Mabagala 2017). Apart from its pathogenicity, its potential as a biocontrol agent is also verified (Tsukamoto et al. 1997; Kadir et al. 2007).
The main objective of this paper is to report a new host pathogen relationship in the South Bengal area of India. This new host pathogen interaction constitutes of Exserohilum as pathogen and Cucumis sativus L. (cucumber) as host. This indicates the potential of Exserohilum as a competent plant pathogen and raises concern regarding cross kingdom infections.
Materials and methods
Pathogen isolation from cucumber plants
The pathogenic fungus was isolated from diseased leaf sample collected from Baruipur in 2018. These were collected in airtight zip lock bags and stored at 4 °C.
The diseased tissue was randomly selected, cut into 1 × 1 cm pieces by flame sterilized forceps and washed in sterile water, dipped in 75% ethanol for 2–3 s, followed by HgCl2 solution (0.1% HgCl2 (w/v) in sterile distilled water) wash for 1 min. These leaf pieces were washed in sterile water and blotted dry with sterilised blotting paper to remove excess water. 3–4 pieces of surface sterilized leaf pieces were placed onto Potato dextrose agar (PDA) plates supplemented with streptomycin (HIMEDIA Streptomycin sulphate (TC035-5G) of 100 ppm concentration) and were incubated at 28 °C for 48 h for appearance of colonies. These were subcultured by inoculating fresh PDA slants with added streptomycin of same concentration and incubated at 28 °C until sporulation occurred followed by 4 °C storage.
Morphological characterization
The fungus was transferred to fresh Potato Dextrose Agar (PDA) plates and incubated for 10 days at 30 °C in a B.O.D. incubator. The colony growth, colour, conidial shape and size were examined in a Dewinter compound light microscope. Spore size were measured on the average of 100 spores and scale bars were added with the photographs taken during the morphological characterization.
Pathogenicity tests
The in vitro pathogenicity test was conducted using single cucumber leaf inoculated in Petri plate by spraying with conidial suspension of the isolate. The conidial suspension was prepared by gently scraping Exserohilum spores from the surface of media and then adding it to 10 ml sterile distilled water. The conidial concentration used for the test was 4.2 × 106 spores/ml. Negative control comprised of a leaf with no added inoculum and 100 μl of spore suspension was added to each of the experimental leaves in the form of droplets. These leaves were incubated in a covered sterilized Petri plate with adequate moisture. Lesions appeared in the experimental leaves after 3 days of inoculation.
The in vivo pathogenicity test was conducted using two sets of plants, first set was sprayed with conidial suspension (same as used for in vitro test) and the second set was sprayed with sterilized water. Both the upper and lower surface of each leaf of the healthy plants were sprayed 3–4 times and then these plants were carefully covered with UV-sterilized transparent plastic bags to ensure high relative humidity without causing any mechanical damage to the plant. Plants were incubated at 30 °C with a natural photoperiod. Number of plant replicates taken in each set were ten.
Leaf sectioning was observed via light microscopy (Dewinter Excel Microscope) to detect pathogen protusion through the cross sectional area. Re-isolation of fungus from the inoculated leaves was conducted to prove Koch’s postulates. Re-isolated fungus and the fungus isolated from the field plant was compared via microscopic observation.
18S rRNA and ITS (Internal transcribed spacer) Sequencing
18S rRNA and ITS Sequencing was performed for the identification of the fungus in this study by using universal primer (Wang et al. 2014).
Phylogenetic analysis
ITS sequences of 38 different species of Exserohilum including the isolated sequence of E.rostratum MN337265 was used for phylogenetic analysis (Table 1). Multiple sequence alignments were conducted by MAFFT (Katoh and Standley 2013), followed by Gblocks for aligned sequence optimization (Talavera and Castresana 2007). PhyML+SMS software was used for design of phylogenetic tree in which aBayes was the selected method for bootstrapping (Lefort et al. 2017; Guindon et al. 2010; Lemoine et al. 2018). The produced phylogenetic tree bearing the evolutionary distances among Exserohilum sp. isolates was viewed in Newick display with the help of iTOL (Junier and Zdobnov 2010; Letunic and Bork 2019).
Sporulation study in relation to salt stress
The fungal pathogen was cultured in salt stress induced media. These were grown in PDA using 1%, 2%, 3%, 5%, 7% and 10% NaCl concentrations and incubated for 7 days at 35 °C. This was observed by lactophenol cotton blue stained light microscopy (Dewinter microscope digital camera, DIGI510, 5.1 MP 1/ 2.5 ´´CMOS sensor).
Influence of culture media on growth characteristics
Six millimeter discs of test fungi obtained from 7 days old pure cultures were transferred at the centre of sterile Petri plates containing four different media: Potato dextrose agar (PDA)(HIMEDIA M096, suspend 39 g in 1000 ml distilled water), Czapek Dox agar (CZA)(HIMEDIA M075, suspend 49.01 g in 1000 ml distilled water), Oat meal agar (OMA)(HIMEDIA M397, suspend 72.5 g in 1000 ml distilled water) and Malt extract agar (MEA)(HIMEDIA M1913, suspend 61 g in 1000 ml distilled water). The petri plates were then incubated at 25 °C and observed regularly. Sporulation was observed via staining under microscope.
Results
Microscopic examination
Cottony white colonies appeared after 48 h of incubation of isolated fungus. Colony eventually turns grayish brown to black after 7 days of incubation. Mycelia showed brown colouration under microscope. Conidia were straight or slightly curved ellipsoidal to fusoid, with typically 4–12 distosepta as revealed by lactophenol cotton blue staining by light microscopy (Fig. 1a, b). The conidia were characterized by the presence of a strongly protruding beak shaped basal hilum. The septa above the hilum is much thicker than that of the intercalary ones and retains dark stain. Apart from conidial structures there was no other kind of spores. Spore size was measured to be 40–160 μm in length and 12 μm in breadth. In few cases the fungus was found to undergo sexual reproduction by means of gametangial contact.
Pathogenicity tests
The in vitro pathogenicity test showed irregular shaped brown outlined olive greenish to grayish brown spots scattered throughout the leaf surface and brown spots along the periphery (Fig. 1c). The control sample showed no spots after the incubation.
Under in vivo treatment, inoculated leaves of cucumber plants showed similar spots with distinct yellow hallow 7 days after spray application. Leaves sprayed with sterilized water showed no change in appearance. The pathogen isolated from the infected leaves showed ellipsoidal conidia with truncated hilum as observed by light microscopic study of field plant isolated fungus (Fig. 2).
Sequencing
ITS sequencing data via BLAST revealed 98.94% sequence similarity with Exserohilum rostratum strain AUMC 14373 ITS sequence. To specify the pathogen isolated from Cucumis sativus, lactophenol cotton blue staining followed by light microscopy (Dewinter microscope digital camera, DIGI510, 5.1 MP 1/ 2.5 ´´CMOS sensor) was conducted. Lactophenol cotton blue stained fungal conidia via light microscopy revealed darkly pigmented basal and distal septa which characterise E. rostratum.
Phylogenetic analysis
Exserohilum rostratum (MN337265) is evolutionally more diverged from Bipolaris maydis (NR_138224) as compared to Setosphaeria rostrata (KT 265248). The most diversified among all the 40 selected fungal species is Setosphaeria rostrata (MH201152) having Triticum aestivum as the host, found in Uttar Pradesh, India. The two Setosphaeria monoceras (KR263036 & KR 263035), originated from Iran has more similarities as compared to the Indian/ American species listed (Fig. 3).
Influence of different salt concentrations on sporulation
The pathogen showed no sporulation and very scanty growth at 1% and 10% salt concentration respectively. Sporulation was observed at 2%, 3%, 5%, 7% salt concentrations via lactophenol cotton blue staining followed by light microscopy (Fig. 4). Under different NaCl concentration the mycelia possesses few special structures. Chlamydospores were observed distinctly in 5% NaCl concentration. With the increasing salt concentration conidial structure was significantly altered. Arthroconidia was found when grown under 2%, 3%, 5% and 7% salt concentration. Number of spores increases but they were greatly reduced in size, ellipsoidal with slightly appressed middle portion. All the spores contained granular structures when grown under salt stress.
0% salt concentration: conidia (a), presence of chlamydospore (b), gametangial contact (c). 2% salt concentration: conidia in 40x (d) and 100x (e). 3% salt concentration: conidia in 40x (f) and 100x (g). 5% salt concentration: conidia in 40x (h), presence of oil/lipid bodies and intercalary chlamydospores (i), conidia in 100x (j). 7% salt concentration: conidia and thick walled mycelia in 40x (k) and 100x magnification (l)
Influence of culture media on growth, colony character and sporulation
All three culture media (PDA, CZA, OMA and MEA) supported the growth of test fungi to various degrees. Out of all these media, the test fungi showed maximum mycelial growth on CZA (75 mm) after 6 days of incubation period (Table 2). In both OMA and MEA, the test fungi showed good growth (72 mm and 71 mm respectively) whereas the least growth was observed in PDA (48 mm). The mycelial colour varied greatly in different media which was further observed and tabulated. Despite of the poor mycelial growth in comparison with other culture media PDA shows early and heavy sporulation (Fig. 5).
Discussion
Disease caused by pathogenic fungus is a major limiting factor in order to produce healthy cucumber plant and increasing the yield. During our current study it has been observed that the whole cucumber field was severely affected by leaf spot disease. Plants were mostly drooping down and under the threat of rapid defoliation. Exserohilum rostratum (Drechs. ex.type of E. macginnisii) Leonard and Suggs was identified as the causal organism for the destructive leaf spot disease based on morphological characteristics (Ahmadpour et al. 2013; Shan-Hai et al. 2011). The ITS sequence has been already submitted in GenBank with accession number MN337265 and that sequence has highest similarity with other E. rostratum ITS sequences. Koch’s postulates proved the occurrence of the disease in cucumber plant. Microscopic observation showed conidial growth in the mesophyll layer of the infected leaf and partial necrosis of plant tissue. E. rostratum was previously known to cause leaf spot disease in bottle gourd of Cucurbitaceae family (Choudhary et al. 2018). But this is for the first time this host pathogen interaction is found in India where the host plant is Cucumis sativus L. and the pathogen is E. rostratum. In order to characterize the isolated pathogenic strain of E. rostratum and to check its conidia production rate the fungus was further grown in different fungal specific media like OMA, MEA, CZA and PDA (Zhang and Watson 1997). All the substrates shown better mycellial growth than that of in PDA. In case of OMA and Czapekdox media luxuriant mycelial growth found may be due to the presence of adequate nitrogenous compound. MEA facilitate typical mycelial growth and pigmentation due to the presence of mycological peptone (Jorgensen et al. 2015). Heavy sporulation was found in OMA as this particular media is rich in Nitrogen, Carbon, Proteins and nutrient (Murray et al. 2003). PDA and MEA also shows moderate to heavy sporulation as these fungal media also stimulate sporulation (MacFaddin 1985).
As the field area from where the pathogen was isolated is not so far from the coastal region and the biggest mangroves of the world, the salt tolerance of the fungus was checked. The fungus showed mycelial growth even in 10% salt (NaCl) concentration. Different spore morphology found in different salt concentrations which reveals that the pathogen is well adapted to combat salt stress. In lower salt concentration some reproductive structures was found. This fungus undergoes sexual reproduction by means of gametangial contact. In 5% salt concentration chlamydospore was found. Depending upon the environmental condition fungus produces different resting spores. The formation of chlamydospores indicates the ability of the pathogen to survive in adverse environmental condition (Levy 1995). So depending upon the morphological characterization it can be concluded as, E. rostratum is a potent phytopathogen which is really tough to control by altering physical factors. Further different biological control methods can be adapted in order to eradicate this destructive plant pathogen.
References
Adler A, Yaniv I, Samra Z, Yacobovich J, Fisher S, Avrahami G, Levy I (2006) Exserohilum: an emerging human pathogen. Eur J Clin Microbiol Infect Dis 25(4):247–253. https://doi.org/10.1007/s10096-006-0093-3
Ahmadpour A, Karami S, Heidarian Z, Javan-Nikkhah M (2013) Exserohilum rostratum causing sugarcane leaf spot in Iran. Aust Plant Dis Notes 8(1):97–99. https://doi.org/10.1007/s13314-013-0105-y
Bashir KA, Kamaruzaman S, Khairulmazmi A (2018) First report of northern corn leaf blight disease caused by Exserohilum turcicum on Zea mays in Malaysia. J Mol Genet Med 12(4):387. https://doi.org/10.4172/1747-0862.1000387
Brunings AM, Datnoff LE, Palmateer AJ, Locke JC, Krause CR (2009) Exserohilum leaf spot on tiger grass. Plant Health Progress. https://doi.org/10.1094/PHP-2009-1215-01-RS
Bunkoed W, Kasam S, Chaijuckam P, Yhamsoongnern J, Prathuangwong S (2014) Sexual reproduction of Setosphaeria turcica in natural corn fields in Thailand. Kasetsart J (Nat Sci) 48:175–182
Chaidaroon W, Phaocharoen N, Srisomboon T, Vanittanakom N (2019) Exserohilum rostratum keratitis in a patient with human immunodeficiency virus. Case Rep Ophthalmol 10:127–133. https://doi.org/10.1159/000499688
Choudhary M, Sardana HR, Bhat MN, Gurjar MS (2018) First report of leaf spot disease caused by Exserohilum rostratum on bottle gourd in India. Plant Dis 102(10):2042. https://doi.org/10.1094/PDIS-02-18-0315-PDN
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Hernández-Restrepo M, Madrid H, Tan YP, Da Cunha KC, Gené J, Guarro J, Crous PW (2018) Multi-locus phylogeny and taxonomy of Exserohilum. Persoonia - Molecular Phylogeny and Evolution of Fungi 41(38):71–108. https://doi.org/10.3767/persoonia.2018.41.05
Jones AM, Dodd ME, Webb AK (2001) Burkholderia cepacia: current clinical issues, environmental controversies and ethical dilemma. Eur Respir J 17(2):295–301
Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS et al. (Eds.) (2015) Manual of clinical microbiology (11th ed.), ASM Press, Washington DC, pp. 1253–1273
Joseph NM, Kumar MA, Stephen S, Kumar S (2012) Keratomycosis caused by Exserohilum rostratum. Indian J Pathol Microbiol 55:248–249
Junier T, Zdobnov EM (2010) The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26:1669–1670. https://doi.org/10.1093/bioinformatics/btq243
Kadir JB, Ahmad A, Sariah M, Juraimi AS (2007) Fungal pathogen of Rottboellia cochinchinensis and its potential as bioherbicide. Asian J Plant Sci 6(1):21–28. https://doi.org/10.3923/ajps.2007.21.28
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Katragkou A, Pana ZD, Perlin DS, Kontoyiannis DP, Walsh TJ, Roilides E (2014) Exserohilum infections: review of 48 cases before the 2012 United States outbreak. Med Mycol 52(4):376–386. https://doi.org/10.1093/mmy/myt030
Krishnan S, Manavathu EK, Chandrasekar PH (2009) Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses 52(3):206–222. https://doi.org/10.1111/j.1439-0507.2008.01642.x
Lefort V, Longueville JE, Gascuel O (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424. https://doi.org/10.1093/molbev/msx149
Lemoine F, Domelevo Entfellner JB, Wilkinson E, Correia D, Dávila Felipe M, De Oliveira T, Gascuel O (2018) Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556:452–456. https://doi.org/10.1038/s41586-018-0043-0
Letunic I, Bork P (2019) Interactive tree of life(iTOL) v4: recent updates and new developments. Nucleic Acids Res 47(W1):W256–W259. https://doi.org/10.1093/nar/gkz239
Levy Y (1995) Inoculum survival of Exserohilum turcicum on corn between and during growing periods. Can J Plant Pathol 17:144–146
Luo ZW, He F, Fan HY, Wang XH, Hua M, Hu FC, Li XH, Liu ZX, Yu NT (2012) First report of leaf spot disease caused by Exserohilum rostratum on pineapple in Hainan Province, China. Plant Dis 96(3):458. https://doi.org/10.1094/PDIS-11-11-0979
MacFaddin JF (1985) Media for the isolation-cultivation-identification-maintenance of medical bacteria. Williams and Wilkins, Baltimore
Marcet-Houben M, Gabaldón T (2009) Acquisition of prokaryotic genes by fungal genomes. Trends Genet 26(1):5–8. https://doi.org/10.1016/j.tig.2009.11.007
Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH (2003) Manual of clinical microbiology, Washington, D.C.: ASM Press, 8
Navaud O, Barbacci A, Taylor A, Clarkson JP, Raffaele S (2018) Shifts in diversification rates and host jump frequencies shaped the diversity of host range among Sclerotiniaceae fungal plant pathogens. Mol Ecol 27(5):1309–1323. https://doi.org/10.1111/mec.14523
Nwanosike MRO, Mabagala RB (2017) Influence of metrological parameters on the development of Exserohilum turcicum (Pass.) Leonard and Suggs on maize in Tanzania. International Journal of Agricultural and Food Research [IJAFR] 6(3):1–9
Shan-Hai L, Huang SL, Li Q, Hu CJ, Fu G, Qin L, Ma Y, Xie LZ, Cen Z, Yan W (2011) Characterization of Exserohilum rostratum, a new causal agent of banana leaf spot disease in China. Australas Plant Pathol 40(3):246–259. https://doi.org/10.1007/s13313-011-0037-y
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. https://doi.org/10.1080/10635150701472164
Toher ASM, Ahmad ZAM, Wong MY (2016) First report of Exserohilum rostratum as pathogen of rice brown spot in Malaysia. Plant Dis 100(1):226. https://doi.org/10.1094/PDIS-03-15-0276-PDN
Tsukamoto H, Gohbara M, Tsuda M, Fujimori T (1997) Evaluation of fungal pathogens as biological control agents for the paddy weed, Echinochloa species by drop inoculation. Jpn J Phytopathol 63(5):366–372. https://doi.org/10.3186/jjphytopath.63.366
Wang Y, Tian RM, Gao ZM, Bougouffa S, Qian P-Y (2014) Optimal eukaryotic 18S and universal 16S/18S ribosomal RNA primers and their application in a study of symbiosis. PLoS One 9(3):e90053. https://doi.org/10.1371/journal.pone.0090053
Zhang WM, Watson K (1997) Characterization of growth and conidia production of Exserohilum monoceras on different substrates. Biocontrol Sci Tech 7(1):75–86. https://doi.org/10.1080/09583159731063
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dhara, B., Maity, A., Mondal, P. et al. First report of Exserohilum leaf spot: a unique halophilic pathogen in Cucumis sativus in the South Bengal area of India. Australasian Plant Pathol. 49, 257–266 (2020). https://doi.org/10.1007/s13313-020-00705-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-020-00705-9