Abstract
Renal cell carcinoma (RCC) is one of the leading causes of cancer mortality in adults, but there is still no acknowledged biomarker for its prognostic evaluation. Our previous proteomic data had demonstrated the dysregulation of some lipid metabolism enzymes in clear cell RCC (ccRCC). In the present study, we elucidated the expression of two lipid metabolism enzymes, hydroxyl-coenzyme A dehydrogenase, alpha subunit (HADHA) and acetyl-coenzyme A acetyltransferase 2 (ACAT2), using Western blotting analysis, then assessed the prognostic potential of HADHA and ACAT2 using immunohistochemistry (IHC) on a tissue microarray of 145 ccRCC tissues. HADHA and ACAT2 were downregulated in ccRCC (P < 0.05); further IHC analysis revealed that HADHA expression was significantly associated with tumor grade, stage, size, metastasis, and cancer-specific survival (P = 0.004, P < 0.001, P < 0.001, P = 0.049, P < 0.001, respectively) and ACAT2 expression was significantly associated with tumor stage, size, and cancer-specific survival (P < 0.001, P = 0.001, P < 0.001, respectively). In addition, a strong correlation was found between HADHA and ACAT2 expression (R = 0.655, P < 0.001). Further univariate survival analysis demonstrated that high stage, big tumor size, metastasis, and HADHA and ACAT2 down-expression were associated with poorer prognosis on cancer-specific survival (P = 0.007, P = 0.005, P = 0.006, P < 0.001, P = 0.001, respectively), and multivariate analysis revealed that HADHA, stage, and metastasis were identified as independent prognostic factors for cancer-specific survival in patients with ccRCC (P = 0.018, P = 0.046, P = 0.001, respectively). Collectively, these findings indicated that HADHA could serve as a promising prognostic marker in ccRCC, which indicated lipid metabolism abnormality might be involved in ccRCC tumorigenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Renal cell carcinoma (RCC) is one of the leading causes of cancer-related death in the world, which is characterized by heterogeneity of histological subtype, drug resistance, and absence of acknowledged molecular markers [1]. Clear cell RCC (ccRCC) is the most common subtype, and it accounts for 70∼80 % of all kidney neoplasm. Nephrectomy remains the best option for localized RCC, and systemic therapies are required for advanced RCC (including metastatic or recurrent tumors) as they are resistant to conventional chemotherapy and radiotherapy [2]. Although biological and targeted therapies have shown promising results for advanced tumors, these patients still have a dismal prognosis [3]. It is urgent to find more molecular therapeutic targets to improve the overall prognosis for advanced RCC patients, and further studies are also required to discover potential prognostic markers to monitor recurrence or progression for localized RCC patients [4–6].
Cancer is fundamentally a disorder of cell proliferation, and cancer cells harbor perturbed metabolism that allows them to accumulate metabolic intermediates and synthesize macromolecules, including nucleic acids, fatty acids, and proteins [7]. Glucose is the major energy source in normal cells, whereas it undergoes the metabolic switch from energy production to de novo synthesis of lipids, which are required for cell growth and proliferation in tumor cells [7]. In addition to the well-established Warburg effect (the abnormality in glucose metabolism), alterations in lipid metabolism, including fatty acid (FA) synthesis and oxidation, are increasingly being recognized in cancer cells [8–10]. Recently, there is a growing acceptance for a link between obesity and cancers, including breast, kidney, and liver cancers [11, 12], though the nature of this relationship remains to be fully elucidated. Lipid droplet formation in the cytoplasm is the typical characteristic for ccRCC, which has been recognized as abundant storage of lipid content [3]. Since the metabolism of cancer cells differs from that of normal ones, a better understanding of the metabolic changes facilitates to discover novel prognostic markers, intervene tumor progression, and prompt new approaches toward cancer therapy, thus improving the overall prognosis (including cancer-specific survival, tumor response rate, and health-related quality of life) of RCC patients [9, 10, 13].
Increasing literatures have reported various prognostic factors, such as tumor stage, nuclear grade, P53, Ki-67, proliferating cell nuclear antigen (PCNA), Eg5, and TGF-β in RCC [14, 15], and cancer-specific survival (CSS) as one of the main end-points [16]. Recently, proteomic approach has been widely used to discover potential prognostic biomarkers and therapeutic targets in cancer biology [4–6, 15]. By mass spectrometry (MS)-based proteomic approach, we and others have identified some dysregulated metabolic pathways, such as glucose metabolic pathway, FA β-oxidation, and tryptophan degradation in RCC [5, 13]. ccRCC, which was characterized by typical “lipid drops” diffused in the cytoplasm, has been recognized as a chronic metabolic disease recently [7, 11, 17]. Our previous proteomic analysis displayed that some lipid metabolism enzymes, such as hydroxyl-coenzyme A dehydrogenase, alpha subunit (HADHA) and acetyl-coenzyme A acetyltransferase 1 (ACAT1), were downregulated in ccRCC, and ACAT1 dysregulation had been confirmed [5]. Bioinformatics analysis indicated HADHA and acetyl-coenzyme A acetyltransferase 2 (ACAT2) interacted tightly and played vital roles in lipid metabolism. According to above knowledge, we elucidated the expression of the two lipid metabolism enzymes, HADHA and ACAT2, in ccRCC specimens and analyzed the correlation between their expression and clinicopathological parameters, which aimed at evaluating the two proteins as potential prognostic factors or therapeutic targets in patients with RCC.
Materials and methods
Tissues
As approved by the ethical committees of Shandong Provincial Hospital Affiliated to Shandong University, a total of 145 ccRCC patients who underwent radical or palliative nephrectomy from March 2010 to January 2015 were chosen for immunohistochemistry (IHC) analysis. Clinical data were recorded, including their gender, age, tumor sizes, Fuhrman nuclear grade, pathological TNM stage, metastasis, and follow-up. They were all primary ccRCC, and their pathological characteristics were verified by two pathologists, separately. The patients consisted of 109 men and 36 women, with a mean age of 60.3 years old (range from 45 to 76). Twenty-four patients had distant metastases (to the lung in 3 patients, lymph nodes in 11 patients, and adrenal gland in 10 patients) at the time of surgery. Ninety-three (64.1 %) tumors were discovered incidentally, 32 (22.1 %) were locally symptomatic, and 20 patients (13.8 %) had systemic disease symptoms. The duration of follow-up was calculated from the date of surgery to death or last follow-up, and the median period of follow-up was 37.0 months, ranging from 8 to 65 months.
For Western blotting (WB) analysis, three cases of ccRCC and their adjacent tissues were collected from December 2011 to January 2013 and stored at −80 °C as described previously [5]. They were graded according to the Fuhrman nuclear grade (G1: 1 case, G2: 1 case, G3: 1 case) and staged according to the TNM classification (T1: 1 case, T2: 2 cases). Radiotherapy, chemotherapy, and immunotherapy were not performed before surgery, and these samples were verified by two pathologists after surgery. Kidney tissue samples were obtained with informed patient consent and approval of the hospital research ethics committee.
Reactome and STRING analyses
Cancer cells harbor abnormal lipid metabolism, especial for FA synthesis and β-oxidation [8, 18]. Our previous proteomic data had confirmed the dysregulation of ACAT1 in ccRCC; no one reported the expression of ACAT2, one enzyme for cholesterol synthesis and the cytosolic counterpart of ACAT1, in ccRCC. And the proteomic analysis also pinpointed HADHA, the alpha (α) subunit of trifunctional protein (TFP), which is involved in lipid metabolism and FA β-oxidation [5]. By Reactome “PathwayBrowser” analysis (http://www.reactome.org/PathwayBrowser), “fatty acid, triacylglycerol, and ketone body metabolism” pathway showed the vital proteins or protein complexes, small molecules, and their reactions, which were involved in the lipid metabolism.
Protein–protein interaction for HADHA and ACAT2 was visualized using STRING analysis (http://string-db.org/), which was based on “confidence mode.” HADHA was input into STRING database and one main interactive cluster was formed, which indicated the lipid metabolism proteins (or enzymes) interacted and formed a densely interconnected network.
Western blotting analysis
WB analysis was performed as previously described [5]. Three pairs of fresh frozen ccRCC and adjacent tissues were homogenized in RIPA lysis buffer (Millipore, USA) containing protease inhibitor cocktail (Roche, Switzerland). After protein concentration was determined using BCA protein assay (Bio-Rad, USA), the tissue lysates (50 μg each) were separated on 10 % polyacrylamide gel, and the proteins in the gel were transferred to polyvinylidene difluoride (PVDF; Bio-Rad, USA) membranes. The membranes were detected with desired antibodies: anti-HADHA (dilution 1:1000, rabbit; ABcom, USA), anti-ACAT2 (dilution 1:1000, goat; Santa Cruz, USA), and anti-tubulin (dilution 1:10,000, mouse; Sigma-Aldrich, USA), then fluorescence-conjugated secondary antibodies were used to visualize the detected proteins. Finally, the blots were developed by enhanced chemiluminescence (LAS4000; GE, USA).
Immunohistochemistry analysis
Kidney tissue hematoxylin–eosin (H&E)-stained sections were reviewed by a pathologist to identify representative areas of the tumors. After tissue microarrays (TMA) were constructed [19], IHC analysis was performed as described previously [14]. The slides were stained with anti-HADHA (dilution 1:200) and anti-ACAT2 (dilution 1:100) antibodies using an Envision+-based detection system and developed with diaminobenzidine (DAB; Zhongshan, China). Negative controls were performed by omitting the primary antibody.
Evaluation of immunohistochemical staining
Immunostained slides were scanned and analyzed with the fluorescent microscope (Olympus, Japan). To ensure accuracy, two pathologists, blinded to the clinicopathological parameters, independently evaluated immunostaining of the TMA slides. Immunostaining was evaluated by recording the percentage of cells staining for four random areas and at least 50 tumor cells were counted in each area, then the mean percentage of the positive staining cells were calculated. According to the median percentage of positive cells for each antibody, the protein expression results were classified into two groups: low group (no staining or less than the median value) and high group (equal to or greater than the median value). The median values were 72 % for HADHA and 60 % for ACAT2, respectively.
Statistical analysis
SPSS 17.0 software (SPSS Inc., USA) was used for statistical analysis. For WB analysis, the protein expression of HADHA and ACAT2 was analyzed using paired t test. For IHC analysis, the associations between the expression of the two proteins and clinicopathological parameters were explored using Pearson chi-square test, and correlations between HADHA and ACAT2 were carried out using Spearman correlation analysis. The survival curves for CSS was analyzed by Kaplan–Meier method, and Cox proportional hazards regression model was performed to define the risk factors (including HADHA, ACAT2, and clinicopathological parameters) for tumor patient death. P values less than 0.05 were considered statistically significant.
Results
Pathway analyses pinpointed HADHA and ACAT2
Fatty acid, triacylglycerol, and ketone body metabolism pathway analysis showed the lipid metabolism-related proteins or protein complexes, small molecules, and their reactions, among which HADHA was located in the core of the network (Fig. 1a). Mitochondrial TFP is a hetero-octamer composed of four α- (HADHA) and four β-subunits (HADHB), and HADHA is more abundant than HADHB in kidney tissue. STRING analysis demonstrated HADHA interconnected with ACAT1, ACAT2, HADHB, et al., and HADHA interacted tightly with ACAT2 in the cluster (confidence mode, Fig. 1b). Other lipid-related proteins, such as ACADS, ACADL, and ACADM, were scattered in both pathway networks.
Bioinformatic analysis pinpointed two lipid metabolism enzymes, HADHA and ACAT2. a Reactome analysis showed fatty acid, triacylglycerol, and ketone body metabolism pathway (http://www.reactome.org/PathwayBrowser), which trifunctional proteins (TFA, including four HADHA and four HADHB), ACAT1, ACADS, ACADL, and ACADM were involved in the pathway. b STRING analysis displayed protein–protein interactions of the lipid metabolism enzymes or proteins in the literatures (confidence mode, http://string-db.org/). HADHA was input into STRING and a main cluster was formed. Blue lines represent interactions between proteins and the thickness denotes the confidence level associated with each interactions. Within this cluster, HADHA and ACAT2, which were located in the key nodes and interacted with each other, were chosen to be validated
Validation of HADHA and ACAT2 differential expression between ccRCC and adjacent tissues
Our previous proteomic data had demonstrated the dysregulation of some lipid metabolism enzymes (including HADHA) in ccRCC [5]. This time, we detected the expression of two lipid metabolism enzymes, HADHA and ACAT2, then evaluated their prognostic significance in ccRCC.
As shown in Fig. 2a, significant downregulation of HADHA expression in ccRCC was detected in all the three samples, compared with adjacent tissues (0.724-fold, P = 0.017). Meanwhile, there was also a prominent downregulation of ACAT2 expression in ccRCC (0.524-fold, P = 0.049).
Validation of protein dysregulation in ccRCC by Western blotting (WB). a WB for HADHA and ACAT2 expression of three individual ccRCC and adjacent normal tissues. Tubulin as a loading control (left). b The statistic results of gray value ratios of the WB bands in ccRCC (right). *P < 0.05; **P < 0.01. N normal kidney tissue, T ccRCC
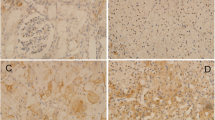
IHC also confirmed the low expression of HADHA and ACAT2 in ccRCC (Fig. 3). To be specific, strong cytoplasmic staining for HADHA was confined in kidney tubule epithelium cells instead of normal glomerular epithelial cells, and different degrees of positive staining in the majority of the ccRCC cells, which verified both the MS and immunoblotting results. ACAT2-positive cytoplasmic staining was seen in normal kidney tubule epithelium cells instead of normal glomerular cells and weak to middle positive staining in the ccRCC cells, which was also consistent with the immunoblotting results.
Representative IHC photomicrographs showed HADHA (upper) and ACAT2 (lower) expression in ccRCC and adjacent normal tissues (original magnification, ×400). a Positive HADHA staining in normal kidney tissues, b null or low HADHA staining in ccRCC, c high HADHA staining in ccRCC, d positive ACAT2 staining in normal kidney tissues, e null or low ACAT2 staining in ccRCC, f high ACAT2 staining in ccRCC. N normal kidney tissue, T ccRCC
HADHA and ACAT2 expression and their correlation with clinicopathological parameters
The protein expression and clinicopathological parameters of the patients are summarized in Table 1 and Fig. 4. In ccRCC tissues, positive staining signals of HADHA were detected in 76 (52.414 %) with low immunoreactivity and 69 (47.586 %) with high immunoreactivity, respectively. The relationship between HADHA expression and tumor grade, stage, size, and metastasis was statistically significant (P = 0.004, P < 0.001, P < 0.001, P = 0.049, respectively). There was no significant correlation between HADHA expression and patients’ gender and age (P = 0.960, P = 0.982).
At the same time, positive cytoplasm staining signals of ACAT2 were detected in 81 (55.862 %) with low immunoreactivity and 64 (44.138 %) with high immunoreactivity, respectively (Table 1, Fig. 4). The relationship between ACAT2 expression and tumor stage and size was statistically significant (P < 0.001, P = 0.001, respectively). There was no significant correlation between ACAT2 expression and patients’ gender, age, grade, and metastasis (P = 0.265, P = 0.367, P = 0.960, P = 0.475, respectively).
During a median follow-up of 37.0 months, we also found that patients with low HADHA expression (<72 % positive cells) or ACAT2 expression (<60 % positive cells) had poorer survival rates, or a worse outcome, than patients with high HADHA expression (≥72 % positive cells) or ACAT2 expression (≥60 % positive cells), and the differences were statistically significant (P < 0.001, P < 0.001, respectively, Table 1).
Associations between HADHA and ACAT2 expression
A significant positive correlation was found between HADHA and ACAT2 expression (R = 0.655, P < 0.001). A further test was performed between patients with different metastatic status. In non-metastatic ccRCC, the relationship between HADHA and ACAT2 expression was statistically significant (R = 0.625, P < 0.001), and in metastatic ccRCC, the relationship between HADHA and ACAT2 expression was remarkably significant (R = 0.828, P < 0.001).
Survival analysis
The postoperative follow-up time for the 145 ccRCC patients ranged from 8 to 56 months (median 37.0 months). In the univariate analysis, the Kaplan–Meier survival curves demonstrated that CSS with low HADHA or ACAT2 expression was significantly shorter than that with high expression (P < 0.001, P = 0.001, respectively, Fig. 5, Table 2). And univariate Cox regression analysis indicated that high pathological stage (hazard ratio (HR) 3.319, confidence interval (CI) = 1.318–8.356, P = 0.007), big tumor size (HR 1.986, CI = 1.218–3.239, P = 0.005), and metastasis (HR 3.667, CI = 1.372–9.805, P = 0.006) were all associated with shorter CSS (Table 2).
In the multivariate analysis, the Cox proportional hazard model also revealed that HADHA (HR 0.359, CI = 0.153–0.842, P = 0.018), stage (HR 3.031, CI = 1.021–8.999, P = 0.046), and metastasis (HR 6.691, CI = 2.089–21.431, P = 0.001) were identified as independent prognostic factors in patients with ccRCC (Table 2), while ACAT2 and tumor size were excluded as independent predictors with no statistical significance (P = 0.909, P = 0.411).
Discussion
Renal cell carcinomas (RCCs) are a heterogeneous group of tumors, which are associated with a high potential of metastases or recurrence and therapeutic resistance. Clinically, localized RCC patients have a favorable prognosis after radical nephrectomy (5-year survival rate ∼85 %), while advanced RCC patients still have a dismal prognosis (5-year survival rate ∼10 %) [2]. Although considerable progress has been made in the treatment of RCC, some aspects of the prognosis still remain unclear, especially concerning the prognostic markers of patient, so it is urgent to identify the potential prognosis markers to predict or improve patient outcomes. Recently, proteomic approaches have been used in RCC research to discover diagnosis or prognosis markers and novel therapeutic targets [4–6, 15]. Our previous proteomic data indicated HADHA, a lipid metabolism enzyme, was involved in the dysregulated proteins network that had been implicated in ccRCC [5]. Bioinformatics analysis demonstrated that HADHA interacted directly with ACAT2 among the lipid metabolism enzymes. Based on these observations, we speculated the two lipid metabolism enzymes, HADHA and ACAT2, played a vital role in ccRCC pathogenesis and might be the prognostic markers or therapeutic targets. To date, very few data elucidated the relationship between lipid metabolism enzymes and RCC tumorigenesis.
HADHA is the alpha subunit of the mitochondrial trifunctional protein (TFP), which catalyzes the last three steps of mitochondrial β-oxidation of long chain fatty acids. The mitochondrial TFP is composed of four α (HADHA) and four β (HADHB) subunits: HADHA is responsible for catalyzing the long-chain 3-hydroxyacyl-coenzyme A dehydrogenase (LCHAD) and enoyl-coenzyme A hydratase (ECH) activities while HADHB catalyzes the 3-ketoacyl-coenzyme A thiolase (KACT) activity [20]. Mutations in HADHA gene or deficiency in HADHA enzymatic activity (<50 % of normal activity) is associated with life-threatening manifestation of the LCHAD deficiency, which include hypoglycemia, cardiomyopathy, and sudden death [21]. Besides FA β-oxidation, HADHA also participates in other lipid metabolism processes, such as lipid biosynthesis, ketogenesis, and ketolysis.
To date, no one had confirmed HADHA dysregulation in ccRCC yet. In the present study, we first verified HADHA differential expression between ccRCC and adjacent kidney tissues, then provided evidence that HADHA was an independent prognostic factor in patients with ccRCC. The immunoblotting indicated a significant decrease of HADHA expression in ccRCC tissues (Fig. 2), and IHC also demonstrated that HADHA decreased in high grade or stage, metastatic, and low survival rate ccRCC tissues (Table 1). We therefore hypothesized that there might be a compromised metabolism of long-chain FAs in ccRCC due to a relative deficiency of HADHA, and lipid droplets were accumulated in the cytoplasm, which was a typical histological characteristic of ccRCC. During the follow-up period, multivariate survival analysis indicated HADHA, together with tumor stage and metastasis, was an independent prognostic factor for cancer-specific survival in patients with ccRCC. And in an epidemiological context, our study showed that dysregulation of HADHA in the kidney tissue might herald the onset of carcinogenesis, and HADHA could also be a potential target for RCC prevention and treatment [22]. Together, these observations strongly supported the finding that HADHA could be an independent prognostic marker of ccRCC, and it also might be a potential molecular target for ccRCC prevention and treatment.
Several previous studies had reported that HADHA was involved in the pathogenesis, prognosis, and therapy resistance of human cancers. By meta-analysis, Manju reported HADHA down-expression in breast cancer, especially in metastatic and recurring breast cancers [22]. Kim also observed a decrease of HADHA in hepatocellular cancer compared to non-neoplastic controls [23], and Kageyama reported HADHA might be a potential predictor of response to platinum-based chemotherapy for lung cancer [24]. Recent reports also indicated HADHA was involved in the processing of miRNAs [25], autophagy, and apoptosis [26, 27].
ACAT2, also known as acetyl-coenzyme A transferase-like protein or cytosolic acetoacetyl-coenzyme A thiolase, like its mitochondrial counterpart, ACAT1, belongs to the thiolase family. Both acetoacetyl-coenzyme A-specific thiolases, ACAT1 and ACAT2, catalyze the formation of acetoacetyl-coenzyme A from two acetyl-coenzyme A molecules. Little is known about ACAT2 expression in RCC. We provided evidence that ACAT2 downregulated in ccRCC, and ccRCC tissues with low stage, small tumor size, and high survival rate displayed higher ACAT2 expression than their counterparts, which indicated it was also involved in ccRCC tumorigesis. Although multivariate survival analysis did not indicate that ACAT2 was an independent prognostic factor for ccRCC patients, the Kaplan–Meier survival curves demonstrated that high ACAT2 expression was positively associated with cancer-specific survival in ccRCC patients (Fig. 5; Table 2). Based on these observations, ACAT2 might be a potential predictor of favorable prognosis for patients with ccRCC. The relationship between ACAT2 and pancreatic or hypopharynx cancers had been reported, and ACAT2 was a potential biomarker and therapeutic target in hypopharynx cancer [28] or pancreatic cancer [29]. Recently, Shan identified ACAT2 as an upstream acetyltransferase of 6-phosphogluconate dehydrogenase (6PGD), which activated 6PGD and promoted oxidative pentose phosphate pathway [30].
ACAT2 is involved in lipid metabolism, and STRING pathway analysis indicated its interaction with some vital lipid metabolism enzymes, such as HADHA, ACAT1, ACADM, ACADVL, et al. Our ccRCC data also showed a positive significant correlation between ACAT2 and HADHA expression (P < 0.001), both of which were inversely associated with ccRCC stage, tumor size, and cancer-specific survival (Table 1). During the follow-up duration, only HADHA as an independent prognostic marker was concluded, and the follow-up period should be prolonged and samples be increased to elucidate whether ACAT2 might be an independent prognostic marker of ccRCC.
Reprogramming of cellular energy metabolism is one of the main hallmarks of cancer, and cancer cells rely on FAs as building blocks for cell proliferation, so disturbing the crucial enzymes or regulators in cellular metabolism is a promising novel approach for cancer therapies [8, 31]. Etomoxir, the carnitine palmitoyltransferase 1 (CPT1) inhibitor in the mitochondria, was reported as a therapeutic target for prostate cancer [32]. The acyl-coenzyme A synthetase/stearoyl-coenzyme A desaturase (ACSL/SCD) network caused an epithelial–mesenchymal transition (EMT) program that promoted migration and invasion of colon cancer cells and might represent a new therapeutic opportunity for colon cancer [33]. Both HADHA and ACAT2 were reportedly involved in cancer chemotherapeutic or radiotherapeutic resistance, while no one demonstrated the role of the two enzymes in cancer-targeted therapy yet. Future researches about the roles of HADHA and ACAT2 in ccRCC-targeted therapy and therapy resistance were in progress.
Although extensive researches about cancer and its initiation are beginning to unravel the role of lipid metabolism in cancer, the lipid metabolism enzymes that dictate their characteristics for ccRCC remain largely ill-defined. ccRCC, which is characterized by lipid accumulation in the cytoplasm, has been recognized as a chronic metabolic disease recently. By IHC analysis of HADHA and ACAT2, we found the two lipid metabolism enzymes were involved in ccRCC tumorigenesis, which facilitated to understand tumor biology, develop novel targeted anticancer medicines, and increase overall survival in ccRCC patients.
Conclusion
Collectively, our study suggested that two lipid metabolism enzymes, HADHA or ACAT2, could serve as potential prognostic markers, and they might be promising therapeutic targets in ccRCC, which indicated lipid metabolism abnormality might be involved in ccRCC tumorigenesis. Further researches are to be undertaken to investigate the abnormal or dysregulated lipid metabolism process in RCC, which will shed light on potential prognostic biomarkers and therapeutic molecular targets for clinical intervention of RCC.
References
Raimondo F, Salemi C, Chinello C, Fumagalli D, Morosi L, Rocco F, et al. Proteomic analysis in clear cell renal cell carcinoma: identification of differentially expressed protein by 2-D DIGE. Mol BioSyst. 2012;8(4):1040–51. doi:10.1039/c2mb05390j.
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi:10.3322/caac.21208.
Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–32. doi:10.1016/S0140-6736(09)60229-4.
Atrih A, Mudaliar MA, Zakikhani P, Lamont DJ, Huang JT, Bray SE, et al. Quantitative proteomics in resected renal cancer tissue for biomarker discovery and profiling. Br J Cancer. 2014;110(6):1622–33. doi:10.1038/bjc.2014.24.
Zhao Z, Wu F, Ding S, Sun L, Liu Z, Ding K, et al. Label-free quantitative proteomic analysis reveals potential biomarkers and pathways in renal cell carcinoma. Tumour Biol J Int Soc Oncodev Biol Med. 2015;36(2):939–51. doi:10.1007/s13277-014-2694-2.
White NM, Masui O, Desouza LV, Krakovska O, Metias S, Romaschin AD, et al. Quantitative proteomic analysis reveals potential diagnostic markers and pathways involved in pathogenesis of renal cell carcinoma. Oncotarget. 2014;5(2):506–18.
Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Gen Dev. 2009;19(1):32–7. doi:10.1016/j.gde.2009.01.002.
Currie E, Schulze A, Zechner R, Walther TC, Farese Jr RV. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18(2):153–61. doi:10.1016/j.cmet.2013.05.017.
Tanaka M, Masaki Y, Tanaka K, Miyazaki M, Kato M, Sugimoto R, et al. Reduction of fatty acid oxidation and responses to hypoxia correlate with the progression of de-differentiation in HCC. Mol Med Rep. 2013;7(2):365–70. doi:10.3892/mmr.2012.1201.
Swierczynski J, Hebanowska A, Sledzinski T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World J Gastroenterol WJG. 2014;20(9):2279–303. doi:10.3748/wjg.v20.i9.2279.
Lee JV, Shah SA, Wellen KE. Obesity, cancer, and acetyl-CoA metabolism. Drug Discov Today Dis Mech. 2013;10(1–2):e55–61. doi:10.1016/j.ddmec.2013.03.005.
Zhu Y, Wang HK, Zhang HL, Yao XD, Zhang SL, Dai B, et al. Visceral obesity and risk of high grade disease in clinical t1a renal cell carcinoma. J Urol. 2013;189(2):447–53. doi:10.1016/j.juro.2012.09.030.
Wettersten HI, Hakimi AA, Morin D, Bianchi C, Johnstone ME, Donohoe DR, et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015;75(12):2541–52. doi:10.1158/0008-5472.CAN-14-1703.
Sun D, Lu J, Ding K, Bi D, Niu Z, Cao Q, et al. The expression of Eg5 predicts a poor outcome for patients with renal cell carcinoma. Med Oncol. 2013;30(1):476. doi:10.1007/s12032-013-0476-0.
Lebdai S, Verhoest G, Parikh H, Jacquet SF, Bensalah K, Chautard D, et al. Identification and validation of TGFBI as a promising prognosis marker of clear cell renal cell carcinoma. Urol Oncol. 2015;33(2):69 e11-8. doi:10.1016/j.urolonc.2014.06.005.
Kramar A, Negrier S, Sylvester R, Joniau S, Mulders P, Powles T, et al. Guidelines for the definition of time-to-event end points in renal cell cancer clinical trials: results of the DATECAN projectdagger. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2015. doi:10.1093/annonc/mdv380.
Pantano F, Santoni M, Procopio G, Rizzo M, Iacovelli R, Porta C, et al. The changes of lipid metabolism in advanced renal cell carcinoma patients treated with everolimus: a new pharmacodynamic marker? PLoS One. 2015;10(4):e0120427. doi:10.1371/journal.pone.0120427.
Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227–32. doi:10.1038/nrc3483.
Liu Z, Fu Q, Lv J, Wang F, Ding K. Prognostic implication of p27Kip1, Skp2 and Cks1 expression in renal cell carcinoma: a tissue microarray study. J Exp Clin Cancer Res CR. 2008;27:51. doi:10.1186/1756-9966-27-51.
Orii KE, Orii KO, Souri M, Orii T, Kondo N, Hashimoto T, et al. Genes for the human mitochondrial trifunctional protein alpha- and beta-subunits are divergently transcribed from a common promoter region. J Biol Chem. 1999;274(12):8077–84.
Spiekerkoetter U. Mitochondrial fatty acid oxidation disorders: clinical presentation of long-chain fatty acid oxidation defects before and after newborn screening. J Inherit Metab Dis. 2010;33(5):527–32. doi:10.1007/s10545-010-9090-x.
Mamtani M, Kulkarni H. Association of HADHA expression with the risk of breast cancer: targeted subset analysis and meta-analysis of microarray data. BMC Res Notes. 2012;5:25. doi:10.1186/1756-0500-5-25.
Kim SY, Lee PY, Shin HJ, do Kim H, Kang S, Moon HB, et al. Proteomic analysis of liver tissue from HBx-transgenic mice at early stages of hepatocarcinogenesis. Proteomics. 2009;9(22):5056–66. doi:10.1002/pmic.200800779.
Kageyama T, Nagashio R, Ryuge S, Matsumoto T, Iyoda A, Satoh Y, et al. HADHA is a potential predictor of response to platinum-based chemotherapy for lung cancer. Asian Pac J Cancer Prevent APJCP. 2011;12(12):3457–63.
Kakumani PK, Shanmugam RK, Kaur I, Malhotra P, Mukherjee SK, Bhatnagar RK. Association of HADHA with human RNA silencing machinery. Biochem Biophys Res Commun. 2015;466(3):481–5. doi:10.1016/j.bbrc.2015.09.055.
Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi:10.1038/nature09204.
Zhang L, Zhang W, Wang YF, Liu B, Zhang WF, Zhao YF, et al. Dual induction of apoptotic and autophagic cell death by targeting survivin in head neck squamous cell carcinoma. Cell Death Dis. 2015;6:e1771. doi:10.1038/cddis.2015.139.
Xu CZ, Shi RJ, Chen D, Sun YY, Wu QW, Wang T, et al. Potential biomarkers for paclitaxel sensitivity in hypopharynx cancer cell. Int J Clin Exp Pathol. 2013;6(12):2745–56.
Souchek JJ, Baine MJ, Lin C, Rachagani S, Gupta S, Kaur S, et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br J Cancer. 2014;111(6):1139–49. doi:10.1038/bjc.2014.385.
Shan C, Elf S, Ji Q, Kang HB, Zhou L, Hitosugi T, et al. Lysine acetylation activates 6-phosphogluconate dehydrogenase to promote tumor growth. Mol Cell. 2014;55(4):552–65. doi:10.1016/j.molcel.2014.06.020.
Ooi AT, Gomperts BN. Molecular pathways: targeting cellular energy metabolism in cancer via inhibition of SLC2A1 and LDHA. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21(11):2440–4. doi:10.1158/1078-0432.CCR-14-1209.
Sadeghi RN, Karami-Tehrani F, Salami S. Targeting prostate cancer cell metabolism: impact of hexokinase and CPT-1 enzymes. Tumour Biol J Int Soc Oncodev Biol Med. 2015;36(4):2893–905. doi:10.1007/s13277-014-2919-4.
Sanchez-Martinez R, Cruz-Gil S, Gomez de Cedron M, Alvarez-Fernandez M, Vargas T, Molina S et al. A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget. 2015.
Acknowledgments
This study was supported in part by the grants from the Shandong Key Research and Development Project (No. 2015GSF118055 and 2012YD18049), the Natural Science Foundation of Shandong Province (No. 2014ZRB14513 and 2014ZRB14081), and the Medicine and Healthcare Technology Development Project of Shandong Province (No. 2014WS0341).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures were consistent with the National Institutes of Health Guide and approved by the institutional board with patients’ written consent. This study was evaluated and approved by the Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong University.
Conflicts of interest
None
Additional information
Zuohui Zhao and Jiaju Lu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhao, Z., Lu, J., Han, L. et al. Prognostic significance of two lipid metabolism enzymes, HADHA and ACAT2, in clear cell renal cell carcinoma. Tumor Biol. 37, 8121–8130 (2016). https://doi.org/10.1007/s13277-015-4720-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4720-4