Abstract
The Runx1 transcription factor, known for its essential role in normal hematopoiesis, was reported in limited studies to be mutated or associated with human breast tumor tissues. Runx1 increases concomitantly with disease progression in the MMTV-PyMT transgenic mouse model of breast cancer. Compelling questions relate to mechanisms that regulate Runx1 expression in breast cancer. Here, we tested the hypothesis that dysregulation of Runx1-targeting microRNAs (miRNAs) allows for pathologic increase of Runx1 during breast cancer progression. Microarray profiling of the MMTV-PyMT model revealed significant downregulation of numerous miRNAs predicted to target Runx1. One of these, miR-378, was inversely correlated with Runx1 expression during breast cancer progression in mice and in human breast cancer cell lines MCF7 and triple-negative MDA-MB-231 that represent early- and late-stage diseases, respectively. MiR-378 is nearly absent in MDA-MB-231 cells. Luciferase reporter assays revealed that miR-378 binds the Runx1 3′ untranslated region (3′UTR) and inhibits Runx1 expression. Functionally, we demonstrated that ectopic expression of miR-378 in MDA-MB-231 cells inhibited Runx1 and suppressed migration and invasion, while inhibition of miR-378 in MCF7 cells increased Runx1 levels and cell migration. Depletion of Runx1 in late-stage breast cancer cells resulted in increased expression of both the miR-378 host gene PPARGC1B and pre-miR-378, suggesting a feedback loop. Taken together, our study identifies a novel and clinically relevant mechanism for regulation of Runx1 in breast cancer that is mediated by a PPARGC1B-miR-378-Runx1 regulatory pathway. Our results highlight the translational potential of miRNA replacement therapy for inhibiting Runx1 in breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most commonly diagnosed cancer worldwide, accounting for 25 % of all cancer cases and 15 % of all cancer-related deaths [1]. It has been recognized for over a decade that breast cancer progression is intimately coupled with specific complex molecular transcriptional programs and signaling pathways [2, 3]. Understanding the regulation of aberrant transcriptional programs in breast cancer is required to develop new strategies for prevention, better detection, and treatment of the disease.
Runx1, a runt-related transcription factor also known as AML1, CBFα2, or PEBP2αB, is essential for normal hematopoiesis and disrupted in leukemia [4]. Runx1 has normal development functions in the epithelial cells of several tissues [5], but is also associated with cancers arising from the epithelium of a number of recently reported tissues (skin, ovarian, and odontogenic tumors) [6–10]. The clinical relevance of Runx1 in breast cancer has come to the forefront in recent years, particularly with the publication of informative, genome-wide sequencing studies in human breast cancer [11–14]. These studies revealed Runx1 to be one of the most mutated and/or deleted genes in breast cancer, thus underscoring a potential role for Runx1 in this disease [11–13]. Notably, several studies have associated Runx1 with hormonal status [15–17]. van Bragt et al. identified Runx1 to be a key regulator of estrogen receptor-positive (ER+) luminal breast epithelial cells and proposed that disruption of Runx1 (in combination with the loss of specific tumor suppressors) may contribute to the development of ER+ luminal breast cancer [16]. Ferrari et al. examined Runx1 expression in samples from almost 500 patients with primary operable invasive ductal breast cancer and reported that high Runx1 protein was significantly associated with poorer cancer-specific survival in patients with invasive triple-negative breast cancer [15]. However, studies addressing the mechanisms of Runx1 dysregulation in breast cancer are limited [15, 16, 18, 19]. Here, we tested the hypothesis and present evidence that the expression of Runx1 in breast cancer is regulated by microRNAs.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that have emerged as key regulators of almost all normal biological and disease-related processes, including cancer progression and metastasis [20–23]. This regulatory control is through sequence-specific complementary binding to the 3′ untranslated region (3′UTR) of target messenger RNAs, resulting in translational repression or degradation of the target [24, 25]. MiRNAs are present in the genome as either independent miRNA genes or miRNA clusters, and depending on their location, miRNAs can be regulated by a dedicated promoter or by the regulatory machinery of the host gene [26]. Intriguingly, more than 60 % of protein-coding genes contain at least one conserved miRNA binding site, implying that most protein-coding genes may be under the control of miRNAs [24, 27, 28]. Of clinical relevance is the observation that miRNA dysregulation drives numerous pathological pathways and is often associated with breast cancer progression [20, 29–32]. Further, a myriad of studies has demonstrated the tumor suppressive, oncogenic, and even sometimes pleiotropic roles of individual or clusters of miRNAs in breast cancer. MiRNAs that are either abnormally expressed or lacking in the tumor cell while present in the normal epithelial cell have been characterized as either inhibiting or promoting tumor growth and progression. Thus, causal links between tumor progression and miRNA dysregulation have been established.
Regulatory interaction between transcription factors and miRNAs is well documented [33]. Indeed, miRNAs that target the Runx transcription factor family are known to be involved in cancer. Runx1 is required of normal hematopoiesis, while numerous Runx1 translocations cause multiple hematopoietic malignancies, serving as the nexus of a complex regulatory miRNA circuitry [34]. Runx2, an essential bone transcription factor, promotes metastasis to bone because it is abnormally expressed in cancer cells due to missing miRNAs that target Runx2 [21, 35], and Runx3, essential for nerve and gut development, is suppressed by several miRNAs that are elevated in cancer cells which results in promoting gastric cancer [36–38]. However, mechanisms contributing to deregulated Runx1 expression by miRNAs and the potential tumorigenic function of Runx1 are relatively understudied and thus require more in-depth characterization.
In the present study, we performed global miRNA profiling in the MMTV-PyMT transgenic mouse model of breast cancer, in which we previously demonstrated increased Runx1 during tumor progression [18]. Here, we find several miRNAs that target Runx1 to be depleted during tumor progression, with miR-378 as the most downregulated miRNA from onset to the end stage of tumor progression. Only a handful of studies have associated miR-378 with human cancer. Loss of miR-378 expression correlated with an aggressive disease phenotype in prostate and colorectal cancers and is associated with poor prognosis and poorer patient survival [39–44]. Our studies now demonstrate a functional inverse relationship between miR-378 and Runx1 in early-stage MCF7 human breast cancer cells that is amplified in aggressive late-stage MDA-MB-231 cells, observing a further reduction of miR-378 with concomitant higher Runx1 levels, together with increased invasion and migration of the triple-negative breast cancer cells. Moreover, inhibition of Runx1 levels modulated expression of the miR-378 host gene PPARGC1B as well as of pre-miR-378. Collectively, our study elucidates a clinically relevant and novel mechanism for miR-mediated dysregulation of Runx1 and highlights the therapeutic potential of targeting Runx1 in breast cancer using a miRNA-based approach.
Materials and methods
Mice
Animal studies were conducted in accordance with approved University of Vermont Animal Care and Use Committee (IACUC) protocols. Female FVB/NJ mice (Jackson Laboratory, Bar Harbor, ME) were crossed with male FVB mice that were transgenic (+/−) for PyMT antigen under the control of the MMTV promoter (a kind gift from LM Shaw, University of Massachusetts Medical School). Genotyping was performed by PCR as described previously for the PyMT transgene [45]. Female mice from this cross that were PyMT+/− were saved for further analysis. Beginning at 3 weeks of age, mice were palpated every 7 days to detect the onset of mammary tumor development. Three mice were sacrificed at 4, 8, and 13 weeks of age and whole mammary glands excised. The characteristics of a 4-week-old PyMT+/− are very similar to that of a PyMT−/− mouse and as such served as the appropriate control group in this study (Supplemental Figs. S1A and S1B). To avoid non-biological variation, mice were sacrificed (and processed) at random ages from different litters at different times. Tissues were either snap frozen for RNA extraction or fixed in 10 % zinc-formalin solution and paraffin embedded for histological analysis. Formalin-fixed, paraffin-embedded mammary glands from MMTV-PyMT mice were sectioned at 4 μm on a Leica 2030 paraffin microtome (Leica Microsystems, Buffalo, IL) and subsequently processed for routine hematoxylin and eosin staining [46].
Cell culture
The human breast cancer cell lines MCF7 and MDA-MB-231 (hereafter MDA-231) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Both cell lines were maintained in DMEM/F12 medium supplemented with 10 % fetal bovine serum and 1 % penicillin/streptomycin. HeLa cells (used for the luciferase assay described below) were also obtained from ATCC and were maintained in DMEM medium supplemented with 1 % glutamine, 10 % fetal bovine serum (FBS), and 1 % penicillin/streptomycin. Media and supplements were obtained from Life Technologies (Carlsbad, CA), and all cells were cultured at 37 °C in a humidified 5 % CO2 atmosphere. Cell lines were validated at the Vermont Cancer Center DNA Analysis Facility by STR DNA fingerprinting using the Promega GenePrint® 10 System according to manufacturer’s instructions. The STR profiles were compared to known ATCC fingerprints (http://ATCC.org/) and to the Cell Line Integrated Molecular Authentication (CLIMA) database version 2.0.201406 (http://bioinformatics.istge.it/clima/) [47]. The STR profiles of all cell lines matched (>85 %) known DNA fingerprints.
RNA extraction
Total RNA was isolated (1) from mouse mammary gland tissue specimens using Trizol (Life Technologies, Carlsbad, CA) and purified with the Zymo RNA purification kit (Zymo Research, Irvine, CA) and (2) from cells using the RNeasy plus mini Kit (Qiagen, Valencia, CA), both according to the manufacturer’s instructions. RNA quality and quantity were verified using the RNA 6000 Nano Kit with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA); RNA quantity was further assessed using both a Nanodrop2000 (Thermo Scientific, Lafayette, CO) and a Qubit® fluorometer (Life Technologies, Carlsbad, CA).
MicroRNA arrays and data analysis
RNA was isolated from mammary glands as described above from each of three mice at 4, 8, and 13 weeks of age. Subsequently, total RNA (500 ng per sample) was hybridized onto the Affymetrix GeneChip miRNA 3.0 Array (Affymetrix, Santa Clara, CA) and processed according to Affymetrix technical protocols by the Vermont Genetics Network Microarray Core. Scanned images of individual microarray chips were analyzed by Affymetrix GCOS software. Probe-level intensities were calculated using the Robust Multichip Average (RMA) algorithm, including background correction, normalization (quantile), and summarization (median polish), for each probe set and sample, as implemented in Partek Genomic Suites®, version 6.6 (Partek Inc., St. Louis, MO). Sample quality was assessed based on the relative log expression (RLE) and normalized unscaled standard error. Principal component analysis (PCA) was also used to look for outlier samples that would potentially introduce latent variation into the analysis of differential expression across sample groups. For identification of differential expression, linear modeling of sample groups was performed using ANOVA as implemented in Partek Genomics Suite. The magnitude of the response (fold change calculated using the least square mean) and the p value associated with each probe set and binary comparison are calculated, as well as the step-up and adjusted p value for the purpose of controlling the false discovery rate [48]. Data presented in heatmaps were filtered based on the following parameters: (1) Mus probe sets (251119 to 1966 probes) and (2) presence/absence calls (1966 to 775 probes) before being further filtered based on statistical parameters, as indicated. Heatmaps were visualized using the heatmap; 2 function in the R language package (http://www.r-project.org/). The results from the microRNA array analysis have been deposited to the National Center for Biotechnology Information (NCBI)’s GEO database accession number: GSE68630.
Gene and precursor miRNA expression analysis
Total RNA was isolated as described above. The complementary DNA (cDNA) was synthesized using the Superscript III First-Strand Synthesis System according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA). Quantitative RT-PCR (qPCR) was performed using SYBR Green Master Mix (Bio-Rad, Hercules, CA) and gene-specific primers (Supplemental Table S1) in an Applied Biosystems Viia 7 system (Life Technologies, Carlsbad, CA). After normalization to the reference gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH), relative expression levels of each target gene were calculated using the comparative C T (ΔΔCT) method.
Mature miRNA expression analysis
Total RNA was isolated as described above. The miScript II RT kit was used for cDNA synthesis according to the manufacturer’s instructions (Qiagen, Valencia, CA). Quantitative PCR was performed using miScript SYBR green PCR kit (Qiagen, Valencia, CA) with a universal reverse primer and a specific forward primer designed for each mature miRNA of interest (Supplemental Table S1) in an Applied Biosystems Viia 7 system (Life Technologies, Carlsbad, CA). U6 was used as the reference and relative expression was calculated as described above.
Transfections
MDA-231 cells were transfected at 70–80 % confluence with 100 nM control non-silencing siRNA (+siNS) or On-Target Plus Human Runx1 siRNA smartpool (+siRunx1) (Thermo Scientific, Lafayette, CO), or 50 nM mirVana negative control #1 (+miR-C) or miR-378a-3p (miR-378) mimic (+miR-378) using Oligofectamine, as indicated, according to the manufacturer’s guidelines (Life Technologies, Carlsbad, CA). A mock transfection (transfection reagent and media only) was included as a control in each experiment. For inhibition studies, MCF7 cells were transfected at 90 % confluence with 50 nM of miRIDIAN control or hsa-miR-378a-3p inhibitors (Thermo Scientific, Lafayette, CO), using Lipofectamine 2000, according to the manufacturer’s directions (Life Technologies, Carlsbad, CA). Subsequently, cells were harvested for RNA and protein analyses as described 48–72 h after transfection. Cells used in functional analyses were transfected for 24 h before proceeding with protocols (migration, invasion, proliferation) as described.
Western blot
Whole cell protein lysates were generated using an SDS lysis buffer as described previously [49]. Western blots were performed and quantitated in biological triplicates and blots representative of each experiment are shown. Briefly, lysates were separated in a 10 % polyacrylamide gel and immobilized on PVDF membranes (Millipore, Billerica, MA). Blots were blocked using 5 % bovine serum albumin (Sigma-Aldrich, St. Louis, MO) before being incubated overnight at 4 °C with the following primary antibodies: anti-AML1 (rabbit polyclonal, 1:800); anti-GAPDH (rabbit monoclonal, 1:5000) (Cell Signaling, Danvers, MA); or anti-PPARGC1B (rabbit polyclonal, 1:1000) (Novus Biologics, Littleton, CO). GAPDH was used as a loading control. Secondary antibodies conjugated to HRP (Santa Cruz Biotechnology, Dallas, TX) were used to detect proteins in conjunction with an enhanced chemiluminescence kit and Chemidoc XRS+ imaging system (both Bio-Rad, Hercules, CA).
Chromatin immunoprecipitation, high-throughput sequencing, and data analysis
Preparation of samples
Cells were grown (as described) to 80 % confluence. Cells were washed twice with phosphate buffered saline (PBS) and crosslinked with 0.8 % formaldehyde (Thermo Scientific, Lafayette, CO) in PBS for 10 min at room temperature with continuous shaking. Formaldehyde was neutralized by the addition of 2.5 M glycine (final concentration 0.125 M) for 5 min. Cells were then washed twice with ice-cold PBS plus complete protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) and collected by scraping and subsequent centrifugation, flash frozen, and stored at −80 °C. Nuclei were isolated using a modification of the Dignam method [50], and pellets were sonicated using a Covaris S-220 Ultrasonic Processor to obtain sheared chromatin ranging from 200 to 800 bp with an average peak size of 500 bp. For immunoprecipitation, 30 μg of sheared chromatin was incubated with 10 μg of anti H3K4me3 (Abcam, Cambridge, MA) overnight at 4 °C and then incubated with 50 μl Protein-G Dynabeads (Life Technologies, Carlsbad, CA) for 4 h at 4 °C. Complexes were washed with 1 ml cold RIPA buffer (150 mM NaCl) for a total of three washes, followed by two washes with 1 ml cold RIPA buffer (500 mM NaCl), and a final wash with TE + NaCl (10 mM Tris, 1 mM EDTA, and 50 mM NaCl). Recovered chromatin was then treated with 1 % SDS overnight at 65 °C to reverse crosslinks, treated with RNase A (0.2 mg/ml final concentration) (Life Technologies, Carlsbad, CA) for 2 h at 37 °C followed by proteinase K treatment (Life Technologies, Carlsbad, CA) for 2 h at 55 °C. Samples were recovered by phenol extraction/ethanol precipitation.
Library and sequencing preparation
Briefly, end-repair, A-tailing, and paired-end adapter ligation were performed using 8 μg of ChIP-DNA at 150 pg/μl using the TruSeq ChIP sample preparation kit (Illumina, San Diego, CA), following the manufacturer’s instructions. Excess adapters were removed by sequential Ampure XP (Beckman Coulter, Pasadena, CA) purifications and recovered; ligated fragments were then amplified by PCR. Libraries were then run on a 2 % agarose/TAE gel to select for fragments in the 350–400-bp range. Final libraries were quantified using the Qubit fluorimeter (Life Technologies, Carlsbad, CA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and by qPCR-based library quantification (Kapa Biosystems, Wilmington, MA). Barcoded libraries were loaded onto an Illumina HiSeq 1500, and single-end 100-base (SE100) sequencing was performed at the Advanced Genome Technologies Core Massively Parallel Sequencing Facility at UVM.
Analysis of sequence data
Base calls were generated on the HiSeq 1500 instrument, and fastq conversion and demultiplexing were done by bcl2fastq (Illumina, v1.8.4). Fastq files were evaluated (Fastqc) and processed to remove low-quality reads and trimmed (FastX toolkit). Reads were mapped to the human genome (hg38) using STAR aligner (version 2.4) with splicing disabled [51]. Wig tracks and enriched regions (peak calls) for each replicate were generated by MACS2 [52], and replicates were then evaluated by wigCorrelate and IDR before being combined and wig tracks regenerated for combined signal. Specific signals (log2FE) for individual gene regions were compared on the UCSC Genome Browser. The ChIP-seq data for H3K4Me3 studies is located at NCBI’s GEO in accession number GSE69377.
Luciferase assay
For this assay, residues 3348–6708 of the human Runx1 3′UTR (NM_001001890.2) were cloned into the pMir-Report miRNA expression reporter vector (Life Technologies, Carlsbad, CA) between the SacI and MluI sites in the multiple cloning site region to obtain the luciferase-Runx1 3′UTR reporter plasmid according to the manufacturer’s instructions. Primers used for cloning are listed in Supplemental Table S1. The final construct was sequenced for validation purposes. Subsequently, HeLa cells were plated in 6-well plates and transfected at 70–80 % confluence with the Runx1 3′UTR construct (500 ng) and either the mirVana miR-378a-3p mimic or negative control #1 (Life Technologies, Carlsbad, CA) at a final concentration of 50 nM as indicated using the X-tremegene HP transfection reagent (Roche Diagnostics, Indianapolis, IN). A Renilla luciferase-containing plasmid (100 ng) was co-transfected as an internal control in each assay (Promega, Madison, WI). Concomitantly, transfections carried out with the Runx1 3′UTR construct only, or the Renilla construct only, or mock transfection (transfection reagent and media only) were included as negative experimental controls. The Dual-Glo™ Luciferase Assay System (Promega, Madison, WI) was carried out according to the manufacturer’s instructions using a Victor X4 Multilabel Plate Reader (Perkin Elmer, Waltham, MA) 48 h after transfection. Ratios of Renilla luciferase readings to firefly luciferase readings were taken for each experiment and triplicates were averaged. Bars represent the average of three independent assays.
Migration assays
For the scratch assays, after transfection (described above for MDA-MB-231 and MCF7 cells), cells were seeded in duplicate and, when they reached 95–100 % confluence, were serum starved with 0.1 % FBS-containing media for 12 h. Subsequently, a scratch was made across the cell layer using a pipette tip, and cell migration was monitored by recording images at indicated time points post-scratch. The area of the scratch was quantified using the MiToBo plug-in for ImageJ software and plotted as a percentage of total area. For the transwell migration assay, 24 h after transfection (as described), cells were trypsinized and re-seeded in triplicate in migration chambers (BD Biosciences, Bedford, MA) in serum-free medium. Twenty-four hours after cell seeding, the experiment was performed and results quantified as previously described [18].
Invasion
For the invasion assay, 24 h after transfection (as described), cells were trypsinized and re-seeded in triplicate in growth factor-reduced Matrigel invasion chambers (BD Biosciences, Bedford, MA) in serum-free medium. Twenty-four hours after cell seeding, the experiment was performed and results quantified as previously described [18].
Proliferation
After 24 h of transfection (as described), cells were trypsinized and re-seeded in 96-well plates at a density of 3000 cells per well in technical quintuplicate before performing the proliferation assay at time points indicated using the CellTiter 96 Aqueous One Solution kit according to the manufacturer’s instructions (Promega, Madison, WI).
Statistical analysis
All experiments were carried out in biological triplicate, and all data are expressed as mean ± standard deviation of the mean, unless otherwise stated. Statistically significant differences were determined using unpaired Student’s t tests with Welch’s correction with GraphPad Prism 6 software (GraphPad Software, La Jolla, CA) unless otherwise stated. In all cases, p ≤ 0.05 was considered significant.
Results
Reciprocal expression of Runx1 and miRNAs predicted to target Runx1 in the MMTV-PyMT transgenic mouse model of breast cancer
We tested the hypothesis that dysregulation of Runx1-targeting miRNAs contributes to altered levels of Runx1 expression in breast cancer. Thus, we performed miRNA microarrays to investigate the relationship between Runx1 and miRNAs during progression of breast cancer in the MMTV-PyMT mouse model. Samples in triplicate were analyzed from time points representing early (4 weeks), mid (8 weeks), and late (13 weeks) stages of disease progression, recognized to be highly analogous with human breast cancer (Fig. 1; Supplemental Fig. S1) [53]. Statistical analyses of the results combined with hierarchical clustering of miRNA profiles revealed that 775 miRNAs were differentially expressed throughout disease progression based on presence/absence call (Supplemental Fig. S1C); 31 miRNAs met strict statistical criteria (p value <0.0025, fold change >2), and 81 % (25/31) of these were downregulated during disease progression (Fig. 1a). In contrast to the striking most down- and upregulated miRNAs, different patterns of expression clustered into six different groups (Supplemental Fig. S1C).
Correlation of the expression of Runx1-targeting miRNAs with Runx1 expression during breast cancer progression in MMTV-PyMT transgenic mice. a Heatmap depicting statistically significant mature miRNA expression changes from miRNA microarrays performed on three biological replicates of mammary gland at three time points at early, mid, and late breast cancer progression in MMTV-PyMT mice. Red represents high expression and white represents low expression with degree of change indicated by intensity of color. All 31 miRNAs showed a p value ≤0.0025 for differential expression over time and all miRNAs passed a false discovery rate <0.05. Bolded miRNA names represent miRNAs predicted to target Runx1. b Table showing specific microarray signal values (log2), fold change over time, and p values for indicated Runx1-targeting miRNAs; red highlighting denotes miR-378 as the most significantly downregulated miRNA during breast cancer progression. c Trend in miR-378 expression similar over time as determined by either qPCR (left y-axis) or microarray (right y-axis). d Runx1 gene expression was evaluated by qPCR in the same samples that microarray was performed on. Values were normalized to GAPDH and data is presented as fold change in Runx1 expression from early to late disease. e Model summarizing the demonstrated reciprocal relationship between Runx1 and Runx1-targeting miRNA expression during disease progression in the MMTV-PyMT transgenic mouse model of breast cancer. Data (c, d) are presented as mean ± SEM of each group
To initially identify and focus on miRNAs with the potential to regulate Runx1, we queried three independent databases, including TargetScan (http://www.targetscan.org/), DIANA LAB (http://diana.imis.athena-innovation.gr/DianaTools/index.php), and miRDB (http://mirdb.org/miRDB/). This analysis revealed that 20 % (5/25) of the significantly downregulated miRNAs were predicted to target Runx1 (miR-378, miR-378b, miR-335-5p, miR-370, and miR-329) (Fig. 1a, b). Furthermore, these miRNAs are conserved between mouse and human. Initially, expression of miR-378, as well as six other differentially expressed miRNAs, was validated by qPCR and found to be decreased in late-stage breast cancer, consistent with the result of the microarray analysis (Fig. 1c and Supplemental Fig. S2). Quantitative PCR evaluation of Runx1 expression showed a seven fold increase in Runx1 gene expression between early and late time points (Fig. 1d). Taken together, these data demonstrate an inverse correlation between expression of Runx1 and specific Runx1-targeting miRNAs over time in the MMTV-PyMT model (summarized in Fig. 1e) and suggest that loss of specific Runx1-targeting miRNAs during breast cancer progression may be an important regulatory mechanism contributing to the dysregulation of Runx1 in breast cancer.
Inverse correlation between Runx1 and miR-378 expression in human breast cancer cells
We utilized knowledge gained from interrogation of the murine model of breast cancer progression to inform more clinically relevant studies in human breast cancer. Expression of Runx1 and miR-378 was examined in the well-characterized non-metastatic MCF7 and highly metastatic MDA-231 breast cancer cell lines, representative of early and late/advanced breast cancer, respectively. Of note, these cell lines also possess some molecular characteristics (e.g., ER/PR status) that mirror early- and late-stage diseases in the MMTV-PyMT mouse model (Supplemental Fig. S1B [54, 55]).
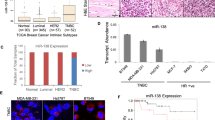
Runx1 protein expression is markedly elevated in the late-stage MDA-231 cells when compared with MCF7 cells (Fig. 2a, inset). An inverse correlation between miR-378 and Runx1 expression was observed, with significantly less miR-378 detected in MDA-231 cells than MCF7 cells (Fig. 2b). Additionally, chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-seq) for histone H3 lysine 4 trimethylation (H3K4me3), which is associated with transcriptionally active or poised genes [56, 57], showed that the H3K4me3 signal in MCF7 and MDA-231 cells corresponded to Runx1 and miR-378 expression patterns (Fig. 2c, d). Greater enrichment of the H3K4me3 signal at the promoter region of the three Runx1 transcripts (as evidenced by larger peaks) was observed in MDA-231 cells as compared with MCF7 cells (Fig. 2c). MicroRNA-378 has been reported to be co-expressed with peroxisome proliferator-activated receptor gamma co-activator 1 beta (PPARGC1B) in human breast cancer cells [58]. At the promoter region of the miR-378 host gene PPARGC1B, enrichment of H3K4me3 was similar in both cell lines; however, in the genomic region where miR-378 is located, K4 methylation was present in MCF7 but absent in MDA-231 cells (Fig. 2d). This finding suggests that absence of the transcriptional activation mark in MDA-MB-231 cells contributes to decreased miR-378 levels (Fig. 2d). Collectively, genetic and epigenetic evidence demonstrates an inverse correlation between Runx1 and miR-378 expression in human breast cancer cell lines and is consistent with findings from the murine model of breast cancer.
Inverse expression of Runx1 and miR-378 expression in human breast cancer cells. a Endogenous Runx1 mRNA expression in human breast cancer cell models representative of early-stage (MCF7) and late-stage metastatic (MDA-231) disease assessed by qPCR and (a, inset) Runx1 protein expression as demonstrated by representative western blot. b Endogenous expression of miR-378 in MCF7 and MDA-231 cells examined by qPCR using U6 as the internal control. c, d Adapted UCSC Genome Browser view of H3K4me3 enrichment tracks for MCF7 (green) and MDA-231 (orange) cells; blue boxes highlight H3K4me3 enrichment at genomic loci of the three Runx1 transcripts for Runx1 (c) and at the miR-378A locus (d). Gene names are consistent with NCBI Reference Sequence records. Gene annotation follows standard display conventions used by the UCSC Genome Browser (exons, solid boxes; introns, solid lines; direction of transcription, arrows). Data (a, b) are presented as mean ± SD of each group. *p < 0.05 (Student’s t test)
MiR-378 directly targets and inhibits Runx1 in human breast cancer cells
Considering the reciprocal relationship between miR-378 and Runx1 expression in both murine and human breast cancer models, coupled with the prediction of Runx1 as a target of miR-378, we investigated whether miR-378 directly targets and represses Runx1. Analysis of the Runx1 mRNA sequence revealed a potential binding site for miR-378 at position 1731–1740 in the Runx1 3′UTR (Fig. 3a). To confirm Runx1 as a direct target of miR-378, a luciferase reporter construct containing the wild-type Runx1 3′UTR was generated. Luciferase reporter assays revealed that co-expression of the luciferase-Runx1 3′UTR with a miR-378 mimic suppressed luciferase activity as compared with expression of a scrambled miRNA sequence (miR-C) (Fig. 3b). Thus, miR-378 can inhibit Runx1 via the 3′UTR.
Regulation of Runx1 by miR-378 in human breast cancer cells. a Schematic showing the potential binding site for the human miR-378 in the 3′UTR of Runx1. The miR-378 seed sequence is highlighted in bold. b To determine if miR-378 could directly target the Runx1 3′UTR, miR-C or miR-378 mimic and the Runx1 3′UTR luciferase construct were transfected into HeLa cells and luciferase activity was normalized to co-transfected Renilla luciferase, with data presented as relative light units produced. c Ectopic expression of miR-378 after transfection of MDA-231 cells with miR-378 mimic (or miR-C non-targeting control) confirmed by qPCR. d Runx1 mRNA expression in MDA-231 cells administered miR-C or miR-378 mimic. e Representative western blot (upper) showing endogenous Runx1 protein expression in MDA-231 cells after transfection with miR-C or miR-378 mimic and (lower) Runx1 protein quantitation (relative to GAPDH control). All data are presented as mean ± SD of each group. *p < 0.05, **p < 0.01 (Student’s t test)
We next evaluated functional activity of miR-378 on the expression of Runx1 in human breast cancer cells (MDA-231). Ectopic expression of miR-378 in the cells was confirmed by qPCR after transfection with a miR-378 mimic or miR-C (Fig. 3c). Subsequent evaluation of Runx1 RNA levels in the same cells demonstrated no change in gene expression (Fig. 3d), but importantly, a significant decrease in protein expression (Fig. 3e), indicating that miR-378 does not degrade the mRNA but selectively reduces Runx1 protein production. Overall, these data demonstrate a functional role for miR-378 in repressing Runx1 in human breast cancer cells and support our hypothesis that loss of miR-378 in breast cancer is a mechanism contributing to increased Runx1 levels.
Inhibition of Runx1 can modulate expression of the miR-378 host gene PPARGC1B as well as pre-mir-378
In addition to miRNA regulation of transcription factors including Runx1, many transcription factors also regulate miRNA expression in a tissue- or developmental-specific manner, often generating an autoregulatory feedback (reviewed in [59]). We hypothesized that a feedback loop exists between Runx1 and miR-378 in the context of breast cancer, whereby Runx1 regulates the miR-378 host gene PPARGC1B. Therefore, motif analyses were performed to determine the presence of the most commonly observed Runx1 core DNA binding motif TGTGGT in the promoter region (±2 kb from the transcription start site) of PPARGC1B (Fig. 4a) [60, 61]. These initial investigations revealed three such sites, indicating the potential for Runx1 to bind and regulate the miR-378 host gene PPARGC1B (Fig. 4a). Subsequently, the expression of the three isoforms of PPARGC1B in the early (MCF7) and advanced (MDA-231) breast cancer cell lines was determined. There was significantly less PPARGC1B transcript (isoforms 1 and 2) in MDA-231 cells than in MCF7 cells; isoform 3 was not detected (Fig. 4b). Similarly, expression of pre-miR-378 was analyzed and as expected, there was significantly lower expression in the MDA-231 cells than in the MCF7 cells (Fig. 4c). This result is consistent with the expression of miR-378 and inversely correlated with the expression of Runx1 in the same cells (Fig. 2a).
Effect of inhibition of Runx1 on expression of the miR-378 host gene PPARGC1B and pre-mir-378. a Adapted UCSC Genome Browser view of the three PPARGC1B transcripts, with promoter regions (+2 kb from the transcription start site) highlighted by red boxes. Gene names are in line with NCBI Reference Sequence records. Gene annotation follows standard display conventions used by the UCSC Genome Browser (exons, solid boxes; introns, solid lines; direction of transcription, arrows). Zoomed insets highlight positions (solid bar, black) of Runx1 core binding motif sites (TGTGGT, red) within the promoter region of PPARGC1B. b, c Endogenous expression of each of the three isoforms of PPARGC1B (b) and of precursor miR-378 (pre-miR-378) (c) determined by qPCR in both MCF7 and MDA-231 cells. d–f Gene expression of Runx1 (d), PPARGC1B (e), and pre-miR-378 (f), as determined by qPCR in siNS- or siRunx1-transfected MDA-231 cells. g Representative western blot showing changes in expression of Runx1 and PPARGC1B protein expression upon transfection of MDA-231 cells with siNS or siRunx1. GAPDH was used as the loading control. In all cases, data are shown relative to GAPDH and are presented as ±SD of each group. ND not determined. *p < 0.05, **p < 0.01 (Student’s t test)
To determine if modulation of Runx1 would impact upon expression of PPARGC1B, pre-miR-378, or miR-378, Runx1 was depleted in MDA-231 cells using siRNAs resulting in at least a 50 % decrease in both RNA and protein expression (Fig. 4d, g). Upon Runx1 depletion, PPARGC1B expression significantly increased over controls (cells transfected with non-silencing siRNA) at both the RNA and protein levels (Fig. 4e, g). Moreover, the expression of pre-miR-378 doubled upon Runx1 depletion (Fig. 4f). However, no significant change in miR-378 was detected upon Runx1 depletion (Supplemental Fig. S3). Further functional analysis on the direct interaction between Runx1 and PPARGC1B would be required to definitively conclude that a feedback mechanism is in place in this context. Although positive feedback regulation of miR-378 by Runx1 could not be fully demonstrated in this system, these data suggest a predominant feed-forward regulation of Runx1 by miR-378.
Modulation of the miR-378-Runx1 regulatory pathway impacts on human breast cancer cell migration and invasion
Based on miR-378 expression patterns in the MMTV-PyMT model of breast cancer as well as the consistent high expression in non-metastatic MCF7 and low expression in aggressive MDA-231 human breast cancer cells, we postulated that miR-378 influences the tumorigenic properties of the cells. To test this hypothesis, a miR-378 mimic and corresponding non-targeting control miRNA (miR-C) were ectopically expressed in MDA-231 cells before examining cancer-related phenotypic changes. Treating the MDA-231 cells with miR-378 markedly suppressed breast cancer cell migration, as assessed by a scratch/wound healing assay (Fig. 5a). Consistent with these findings, migration evaluated using a transwell assay was also significantly suppressed by exogenous expression of miR-378 (Fig. 5b). Similarly, the ability of the cells to invade was significantly diminished upon exogenous expression of miR-378 (Fig. 5c). To determine if ectopic expression of miR-378 impacted the growth rate of the cells, a proliferation assay was performed. This assay revealed that expression of miR-378 had no significant effect on the proliferation rate of the cells (Fig. 5d). Thus, the loss of miR-378 expression in triple-negative breast cancer cells contributes to tumorigenic properties of migration and invasion.
Changes in human breast cancer cell phenotype upon modulation of the miR-378-Runx1 regulatory pathway. MDA-231 cells treated with non-targeting control miRNA (+miR-C) or miR-378 mimic (+miR-378) were assayed as follows: a Representative phase contrast images (mag. ×10) of MDA-231 cells treated as above were subjected to a scratch assay for times indicated. The area of the scratch was plotted as a percentage of total area for three independent experiments carried out in duplicate. b Light microscopy images (mag. ×12) of stained cells from a representative transwell migration assay experiment with MDA-231 cells treated as above; inset images show an overview of all cells stained in the well at lower magnification (×2.5) (left). Quantitation of migrated cells assessed by measurement of the absorbance of solubilized crystal violet stain retained by migrated cells from two independent experiments carried out in triplicate (right). c Light microscopy images (mag. ×12) of stained cells from a representative Matrigel invasion assay experiment with MDA-231 cells treated as above; inset images show an overview of all cells stained in the well at lower magnification (×2.5) (left). Quantitation of invaded cells assessed by measurement of the absorbance of solubilized crystal violet stain retained by invaded cells from two independent experiments carried out in triplicate (right). d Cell proliferation assessed in MDA-231 cells treated as above for time points indicated. All quantitative data are depicted as mean ± SD per group. NS no significant difference. *p < 0.05, **p < 0.01 (Student’s t test)
To strengthen this conclusion, we examined whether inhibition of miR-378 in MCF7 cells could result in a more aggressive phenotype and induce the expression of Runx1. Indeed, MCF7 cells treated with a miR-378 inhibitor exhibited increased (27 %) migration as evidenced by a wound healing assay (Fig. 6a), and this was associated with a concomitant increase (32 %) in Runx1 expression (Fig. 6b). These complementary findings establish that modulation of the miR-378-Runx1 regulatory pathway significantly impacts the tumorigenic properties of breast cancer cells and supports the notion that the tumor suppressive effects of miR-378 on aggressive MDA-231 breast cancer cells are, at least in part, mediated through translational repression of Runx1 by miR-378 (summarized in Fig. 6c).
Inhibition of miR-378 increases Runx1 and migration of MCF7 cells. MCF7 cells treated with control inhibitor (+miR-C) or hsa-miR-378a-3p hairpin inhibitor (+miR-378 inhibitor) were assayed as follows: a Representative phase contrast images (mag. ×20) of MCF7 cells subjected to a scratch assay for time points indicated. The area of the scratch was quantified using the MiToBo plug-in for ImageJ software and plotted as a percentage of total area for three independent experiments performed in triplicate. b Runx1 mRNA expression in MCF7 cells treated with miR-control or miR-378 inhibitor. Data are shown relative to GAPDH and are presented as ±SD per group. c Schematic summarizing the miR-378-Runx1 regulatory pathway in human breast cancer cells. Schematic key: pointed arrowhead, promotes expression; blunt-end arrowhead, inhibits expression; dashed line, assumed—not supported by experimental evidence in this system; solid lines, proven—supported by experimental evidence in this system. *p < 0.05, **p < 0.01 (Student’s t test)
Discussion
The present studies have provided several lines of evidence that support the involvement of miR-mediated regulation of Runx1 in contributing to the tumorigenic properties of breast cancer cells. We showed that Runx1 is significantly increased in late-stage metastatic human MDA-MB 231 when compared to early-stage MCF7 cells. This finding is consistent with our recent report that demonstrated continuously increased Runx1 expression in diseased mammary glands during progression of tumor growth from early to late stages in the MMTV-PyMT mouse, a model recognized to be analogous to human breast cancer [18]. Using microarrays, interrogation of the MMTV-PyMT model revealed differential expression of miRNAs throughout disease progression, many of which were up- or downregulated, consistent with previously published studies in human breast cancer [62]. Subsequent statistical analysis demonstrated miR-378 to be the most significantly downregulated Runx1-targeting miRNA over time.
We have demonstrated that miR-378, as a novel Runx1-targeting miRNA in breast cancer, was inversely correlated with Runx1 expression and was evidenced to be capable of inhibiting Runx1 and suppressing breast cancer cell migration and invasion. Notably, four other miRNAs targeting Runx1 are also depleted from mouse tumors, further suggesting that the progression to advanced breast cancer may be closely linked to increased Runx1 levels. Expression analysis in human breast cancer cell line models revealed a downregulation of miR-378 in late (MDA-231) vs. early (MCF7) human breast cancer which inversely correlated with Runx1 expression. Luciferase assays demonstrated that miR-378 could directly target the Runx1 3′UTR. Finally, reintroduction of miR-378 in metastatic breast cancer cells decreases Runx1 expression and inhibits their aggressive migratory and invasive phenotype. Consistent with this finding, inhibition of miR-378 in early-stage breast cancer cells increased Runx1 migration. Thus, we have identified a mechanism for increased Runx1 expression in human breast cancer cells through loss of miR-378 and, importantly, that the aggressive tumorigenic cancer phenotype of MDA-231 cells can be inhibited upon ectopic expression of miR-378. Indeed, in our initial studies [18], these same cancer-related properties were demonstrated by inhibition of Runx1 itself in late-stage murine breast cancer cells. From these data, we conclude that the decline in miR-378 expression with disease progression unblocks Runx1 translation in breast cancer, permitting a pathologic increase in Runx1 protein levels.
The potential for positive feedback regulation of miR-378 by Runx1 was investigated because of studies supporting coordination of miRNAs and transcription factors, including Runx factors, to form autoregulatory feedback loops [33, 63, 64]. Specifically, Runx1 was shown to regulate the miR-23a∼27a∼24-2 cluster in normal hematopoiesis and that Runx1 and miR-27a are engaged in a feedback loop where miR-27a expression is positively regulated by Runx1 during megakaryopoiesis [65, 66]. MicroRNA-378-3p is embedded within the Peroxisome Proliferator-Activated Receptor Gamma, Coactivator 1 Beta (PPARGC1B) gene locus [58]. Our data initially suggested that a feedback loop did exist between Runx1 and miR-378 in breast cancer, where inhibition of Runx1 resulted in significantly increased expression of the miR-378 host gene PPARGC1B as well as the pre-miR-378. Two putative Runx consensus sites occur in the host gene and one in proximity to the pre-miRNA; however, no change in the expression of the mature miR-378 was observed, leaving the feedback loop incomplete. Given the complex, multi-step nature of miRNA biogenesis, it is possible that there may be a defect in processing of the pre-miR-378 to mature miR-378 in the experimental system used. For example, defects with Drosha-mediated pre-miRNA processing, or exportin 5-mediated nuclear transport of the pre-miRNA, or Dicer-mediated cleavage of the pre-miRNA upon export to the cytoplasm are all possible [26]. More experiments would be required to further elucidate if indeed a miR-378-Runx1-PPARGC1B positive feedback loop exists in the context of human breast cancer. Nonetheless, our finding that miR-378 targets Runx1, a master transcription factor in tumor cells that promotes a more aggressive cancer cell phenotype, has important therapeutic implications.
A relatively small number of studies have associated miR-378 with breast cancer [58, 67]. Knezevic and colleagues reported that inhibition of miR-378 resulted in apoptosis resistance and promoted expression of IGF1R and Akt, components of signaling pathways intimately associated with breast cancer progression [68–70]. Another study investigating expression of miR-378 in breast cancer patient samples found higher expression of miR-378 in cancer vs. normal tissues; however, the limited patient population was biased towards early-stage, ER+/PR+ cases which may have inadvertently impacted the findings [67]. There is a need for more unbiased studies with larger breast cancer patient cohorts to investigate the potential for miR-378 as a biomarker and therapeutic target in breast cancer.
During the preparation of this manuscript, another study has considered that miR-378 may be related to specific subtypes of breast cancer, including triple-negative breast cancer [71]. Distinct expression profiles and functional roles have been attributed to the -3p and -5p strands [72]. Notably, Eichner and colleagues demonstrated that miR-378* (commonly referred to as miR-378-5p, a mature miRNA derived from pre-miR-378 along with miR-378-3p [73]) demonstrated that mir-378* mediated a metabolic shift in breast cancer [58]. In our study, we focused on expression patterns of the non-miR* strand and identified an inverse correlation between Runx1 and a Runx1-targeting miRNA in (1) a luminal mouse model that loses ER expression with disease progression [53, 74] and (2) human cell models that represent early ER+ and late-stage ER− disease, with tumor suppressive phenotypic effects seen after miR-378-mediated inhibition of Runx1 [54, 55]. Given this knowledge, it seems there is duality with respect to the role of Runx1, i.e., a tumor suppressor in normal mammary epithelial cells and an oncogene in tumor cells. These Runx1 activities may be dependent on the ER status, stage, and/or subtype of the cancer cells. Thus, it is possible that miR-378 and miR-378* have equally important yet distinct roles in breast cancer.
The importance of miRNA regulation of Runx1 and associated normal and abnormal physiological processes is well documented in hematopoiesis and leukemia, respectively [34]. However, the significance of these regulatory interactions is unknown in breast cancer. Our study describes, for the first time, a clinically relevant mechanism for control of Runx1 in breast cancer through miRNA regulation. These studies showing that miR-378 expression inhibits breast cancer cell activities are relevant to other cancers where Runx1 is associated with tumorigenesis [6–8]; also, loss of miR-378 is associated with cancers [42–44], thereby becoming a viable therapeutic strategy. MicroRNAs are emerging as therapeutic molecules [23, 75], and a replacement therapy using a miR-34 mimic (called MRX34), a master tumor suppressor, was first administered to cancer patients in a multicenter, open-label phase I clinical trial, due to be completed later this year ([76, 77]: http://clinicaltrials.gov/ct2/show/NCT01829971).
Taken together, our major and significant finding that miR-378 targets the Runx1 transcription factor to inhibit its post-transcriptional expression in metastatic cells highlights a novel and promising opportunity for miR-378-mediated therapeutic inhibition of otherwise difficult to target nuclear proteins such as Runx1 in breast cancer.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Toss A, Cristofanilli M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015;17:60.
Ichikawa M, Yoshimi A, Nakagawa M, Nishimoto N, Watanabe-Okochi N, Kurokawa M. A role for runx1 in hematopoiesis and myeloid leukemia. Int J Hematol. 2013;97:726–34.
Scheitz CJ, Tumbar T. New insights into the role of runx1 in epithelial stem cell biology and pathology. J Cell Biochem. 2013;114:985–93.
Hoi CS, Lee SE, Lu SY, McDermitt DJ, Osorio KM, Piskun CM, et al. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol Cell Biol. 2010;30:2518–36.
Heikinheimo K, Kurppa KJ, Laiho A, Peltonen S, Berdal A, Bouattour A, et al. Early dental epithelial transcription factors distinguish ameloblastoma from keratocystic odontogenic tumor. J Dent Res. 2015;94:101–11.
Keita M, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, et al. The runx1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle. 2013;12:972–86.
Wang X, Zhao Y, Qian H, Huang J, Cui F, Mao Z. The mir-101/runx1 feedback regulatory loop modulates chemo-sensitivity and invasion in human lung cancer. Int J Clin Exp Med. 2015;8:15030–42.
Jacques C, Guillotin D, Fontaine JF, Franc B, Mirebeau-Prunier D, Fleury A, et al. DNA microarray and miRNA analyses reinforce the classification of follicular thyroid tumors. J Clin Endocrinol Metab. 2013;98:E981–9.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9.
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–60.
Chimge NO, Frenkel B. The runx family in breast cancer: relationships with estrogen signaling. Oncogene. 2013;32:2121–30.
Ferrari N, Mohammed ZM, Nixon C, Mason SM, Mallon E, McMillan DC, et al. Expression of runx1 correlates with poor patient prognosis in triple negative breast cancer. PLoS One. 2014;9, e100759.
van Bragt MP, Hu X, Xie Y, Li Z. Runx1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. Elife. 2014;3, e03881.
Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, et al. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–55.
Browne G, Taipaleenmaki H, Bishop NM, Madasu SC, Shaw LM, van Wijnen AJ, et al. Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J Cell Physiol. 2015;230:2522–32.
Tang W, Yu F, Yao H, Cui X, Jiao Y, Lin L, et al. Mir-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene. 2014;33:2629–38.
Acunzo M, Romano G, Wernicke D, Croce CM. Microrna and cancer—a brief overview. Adv Biol Regul. 2015;57:1–9.
Browne G, Taipaleenmaki H, Stein GS, Stein JL, Lian JB. MicroRNAs in the control of metastatic bone disease. Trends Endocrinol Metab. 2014;25:320–7.
van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015;17:21.
Nana-Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15:445.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33.
Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24.
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105.
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73.
Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55.
Le Quesne J, Caldas C. Micro-RNAs and breast cancer. Mol Oncol. 2010;4:230–41.
Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–36.
Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215–22.
Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. MiRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288:77–87.
Rossetti S, Sacchi N. Runx1: a microRNA hub in normal and malignant hematopoiesis. Int J Mol Sci. 2013;14:1566–88.
Taipaleenmaki H, Browne G, Akech J, Zustin J, van Wijnen AJ, Stein JL, et al. Targeting of runx2 by mir-135 and mir-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75:1433–44.
Liu Z, Chen L, Zhang X, Xu X, Xing H, Zhang Y, et al. Runx3 regulates vimentin expression via mir-30a during epithelial-mesenchymal transition in gastric cancer cells. J Cell Mol Med. 2014;18:610–23.
Lai KW, Koh KX, Loh M, Tada K, Subramaniam MM, Lim XY, et al. MicroRNA-130b regulates the tumour suppressor runx3 in gastric cancer. Eur J Cancer. 2010;46:1456–63.
Wang M, Li C, Yu B, Su L, Li J, Ju J, et al. Overexpressed mir-301a promotes cell proliferation and invasion by targeting runx3 in gastric cancer. J Gastroenterol. 2013;48:1023–33.
Lee DY, Deng Z, Wang CH, Yang BB. Microrna-378 promotes cell survival, tumor growth, and angiogenesis by targeting sufu and fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–5.
Ma J, Lin J, Qian J, Qian W, Yin J, Yang B, et al. Mir-378 promotes the migration of liver cancer cells by down-regulating fus expression. Cell Physiol Biochem. 2014;34:2266–74.
Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang L, et al. MicroRNA-378 functions as an onco-mir in nasopharyngeal carcinoma by repressing tob2 expression. Int J Oncol. 2014;44:1215–22.
Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. Mir-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109.
Avgeris M, Stravodimos K, Scorilas A. Loss of mir-378 in prostate cancer, a common regulator of klk2 and klk4, correlates with aggressive disease phenotype and predicts the short-term relapse of the patients. Biol Chem. 2014;395:1095–104.
Wang KY, Ma J, Zhang FX, Yu MJ, Xue JS, Zhao JS. MicroRNA-378 inhibits cell growth and enhances l-ohp-induced apoptosis in human colorectal cancer. IUBMB Life. 2014;66:645–54.
Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle t oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61.
Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008;2008:pdb prot4986.
Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B. Cell line data base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res. 2009;37:D925–32.
Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Methodol. 1995;57:289–300.
Gutierrez S, Javed A, Tennant DK, van Rees M, Montecino M, Stein GS, et al. Ccaat/enhancer-binding proteins (c/ebp) beta and delta activate osteocalcin gene transcription and synergize with runx2 at the c/ebp element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–23.
Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase ii in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying CHiP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–40.
Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26.
Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32:35–48.
Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215.
Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–45.
Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, et al. H3k4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–88.
Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, et al. Mir-378(*) mediates metabolic shift in breast cancer cells via the pgc-1beta/errgamma transcriptional pathway. Cell Metab. 2010;12:352–61.
Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610.
Bowers SR, Calero-Nieto FJ, Valeaux S, Fernandez-Fuentes N, Cockerill PN. Runx1 binds as a dimeric complex to overlapping runx1 sites within a palindromic element in the human GM-CSF enhancer. Nucleic Acids Res. 2010;38:6124–34.
Speck NA, Terryl S. A new transcription factor family associated with human leukemias. Crit Rev Eukaryot Gene Expr. 1995;5:337–64.
Serpico D, Molino L, Di Cosimo S. MicroRNAs in breast cancer development and treatment. Cancer Treat Rev. 2014;40:595–604.
Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, et al. Mir-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem. 2012;287:42084–92.
Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, et al. A network connecting runx2, satb2, and the mir-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107:19879–84.
Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between mir-27a and runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106:238–43.
Zaidi SK, Dowdy CR, van Wijnen AJ, Lian JB, Raza A, Stein JL, et al. Altered runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a mir-24/MKP-7/MAPK network. Cancer Res. 2009;69:8249–55.
Yin JY, Deng ZQ, Liu FQ, Qian J, Lin J, Tang Q, et al. Association between mir-24 and mir-378 in formalin-fixed paraffin-embedded tissues of breast cancer. Int J Clin Exp Pathol. 2014;7:4261–7.
Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS, et al. A novel cardiomyocyte-enriched microRNA, mir-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem. 2012;287:12913–26.
Christopoulos PF, Msaouel P, Koutsilieris M. The role of the insulin-like growth factor-1 system in breast cancer. Mol Cancer. 2015;14:43.
Pande S, Browne G, Padmanabhan S, Zaidi SK, Lian JB, van Wijnen AJ, et al. Oncogenic cooperation between pi3k/akt signaling and transcription factor runx2 promotes the invasive properties of metastatic breast cancer cells. J Cell Physiol. 2013;228:1784–92.
Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH, Chang YC, et al. Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol Cancer. 2015;14:36.
Jiang L, Huang Q, Zhang S, Zhang Q, Chang J, Qiu X, et al. Hsa-mir-125a-3p and hsa-mir-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318.
Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5, e11387.
Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, et al. Gata-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–52.
Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65.
Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577.
Bader AG. Mir-34—a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120.
Acknowledgments
This work was supported by the National Cancer Institute (Nos. P01 CA082834 and R03 CA167726), National Institute of Arthritis and Musculoskeletal and Skin Diseases (No. R01 AR039588), National Institute of Dental and Craniofacial Research (No. R37 DE012528), Pfizer (WS2049100), and grants from the University of Vermont Cancer Center and Lake Champlain Cancer Research Organization. The microarray research reported was supported by the National Institute of General Medical Sciences (No. P20 GM103449). The authors thank all members of our laboratories, especially Philip W. L. Tai, for valuable suggestions throughout the study. The authors are grateful to the UVM Cancer Center Advanced Genome Technologies Core, supported by UVM Cancer Center, Lake Champlain Cancer Research Organization, and the UVM College of Medicine for data pertaining to cell line authentication, as well as the Vermont Genetics Network Microarray Facility for miRNA target preparation, hybridization, and scanning processing. The authors are also grateful to the Molecular Bioinformatics Shared Resource of the University of Vermont College of Medicine for microarray data analysis. Finally, the authors thank the Advanced Genome Technologies Core Massively Parallel Sequencing Facility for sequencing data. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Browne, G., Dragon, J.A., Hong, D. et al. MicroRNA-378-mediated suppression of Runx1 alleviates the aggressive phenotype of triple-negative MDA-MB-231 human breast cancer cells. Tumor Biol. 37, 8825–8839 (2016). https://doi.org/10.1007/s13277-015-4710-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4710-6