Abstract
Acquisition of gemcitabine resistance in breast cancer has not been fully clarified. Prior studies suggest that miRNAs are important to chemoresistance in solid tumors and we confirmed that miR-21 is involved in the development of gemcitabine resistance. Epithelial-to-mesenchymal transition (EMT) and AKT pathway activation were noted to be important to this resistance as well. PTEN, a direct target gene of miR-21, was significantly downregulated in gemcitabine-resistant breast cancer cells and restoration of PTEN expression blocked miR-21-induced EMT and gemcitabine resistance. Our data offer novel insight into gemcitabine resistance in breast cancer and suggest that miR-21 may be used to predict optimal breast cancer therapy and may be a potential therapeutic target for reversing gemcitabine resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Advanced breast cancer is treatable but often incurable and gemcitabine-based therapy is frequently used to treat such late-stage breast cancers [1]. However, gemcitabine resistance is increasing, limiting its utility. Our phase III trial (CBCSG006) revealed that ~30 % of patients did not respond to gemcitabine treatment due to drug resistance [2]. Thus, to improve therapeutic responses, we must understand the underlying mechanism of gemcitabine resistance.
Gemcitabine (2′, 2′-difluorodeoxycytidine, dFdC) is a nucleoside analogue that requires cellular uptake and intracellular phosphorylation for cytotoxicity [3]. After cytoplasmic influx by membrane transporters, gemcitabine undergoes complex intracellular phosphorylation to yield nucleotides: gemcitabine diphosphate (dFdCDP) and triphosphate (dFdCTP), which incorporate into DNA and RNA [3–5]. Previous studies indicate that alterations in gemcitabine transport pathways and abnormal kinase activity give rise to drug resistance.
Several molecular mechanisms may be responsible, such as reduced expression of nuclear transport protein hENT1 [5, 6], increased expression of cell membrane multidrug resistance protein 5 (MRP5 or ABCC5) [7], enhanced kinase activity within ribonucleotide reductase subunits M1 and M2 (RRM1, RRM2) [8, 9], cytidine deaminase (CDA) [10], liver kinase B1 (LKB1) [11], and reduced deoxycytidine kinase (dCK) activity [12]. Abnormal expression of membrane transporters or metabolic pathway dysfunction can diminish intracellular accumulation of gemcitabine and cause resistance, necessitating higher drug doses. Thus, understanding gemcitabine resistance is required to improve therapeutic outcomes.
MicroRNAs (miRNAs) are small, 20~22 nt noncoding RNA molecules that inhibit post-transcriptional activity and have pleiotropic roles in many cancer processes [13, 14], especially chemoresistance and EMT regulation [15–17]. MiRNA-21 (miR-21) is documented to be up-regulated in many human cancers including breast cancer [18]. miRNAs array data indicated that elevated expression of miR-21 occurred in gemcitabine-resistant MDA-MB-231 breast cancer cells [10] so miR-21 may be critical to gemcitabine resistance. To address this, we studied the relationship between miR-21 and gemcitabine resistance and investigated the underlying mechanism.
Materials and methods
Cell lines and culture conditions
MDA-MB-231 and MCF-7 human breast cancer cell lines were purchased from American Type Culture Collection (ATCC). MDA-MB-231 and MCF-7 cells were cultured with L-15 medium and DMEM medium (respectively) supplemented with 10 % FBS 100 U/mL penicillin and 100 μg/mL streptomycin in a humidified incubator at 37 °C under 5 % CO2. MDA-MB-231 gemcitabine-resistant cells (231/GEM) were a gift from Key Laboratory of Breast Cancer in Fudan University Shanghai Cancer Center and they were cultured for more than one year with gemcitabine (12–720 nM) [10].
Establishment of gemcitabine-resistant MCF-7/GEM sublines
MCF-7 cells were continuously exposed to gemcitabine (Eli Lilly) from 10 nM to 10 μM for more than 6 months according published methods [19]. Briefly, surviving cells were passaged and exposed to an ascending concentrations (0.01, 0.1, 0.25, 2, 4, 8, 10 μM) of gemcitabine when cells were 75 % confluent. A Cell Counting Kit-8 (CCK-8; Donjin Laboratories) was used to quantify gemcitabine-resistant MCF-7 (MCF-7/GEM) cell drug sensitivity.
Cell viability assay
Cell viability was assayed with a CCK-8 kit. In brief, 5 × 103 cells were seeded at equal densities into 96-well culture plates and incubated overnight. The next day, medium was replaced with medium containing different concentrations of gemcitabine and cells were incubated for 48 h. Then, 10 μL of CCK-8 kit reagent was added to each well, and 2 h later, plates were read under a microplate reader (Synergy H4, Bio-Tek) at 450 nm. Each experiment was performed in triplicate and cell viability was based on absorbance.
RNA extraction and quantitative RT-PCR for miRNA and mRNA assay
Total RNA was extracted from cultured cells using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. RNA quality was assessed by A260 absorption, and 500 ng of total RNA was used for first-strand DNA synthesis. Real-time PCR was performed in triplicate with SYBR Premix Ex Taq (TaKaRa, Dalian, China). miR-21 primers were purchased from Ribobio (Cat: ssD809230931, Guangzhou, China) and U6 was an endogenous control. For mRNA quantification, real-time PCR was performed in triplicate with SYBR Premix Ex Taq (Takara) and GAPDH was an internal control. Primers used for PCR amplification were synthesized by Sangon Biotech Co., Ltd., (Shanghai, China) as follows: 5′-AGCCCCGCCTTATGATTCTCTG-3′(forward) and 5′-TGCCCCATTCGTTCAAGTAGTCAT-3′(reverse) for E-cadherin; 5′-AGTCCACTGAGTACCGGAGAC-3′(forward) and 5′-CATTTCACGCATCTGGCGTTC-3′(reverse) for vimentin; and 5′-GCCAAAAGGGTCATCATCTC-3′(forward) and 5′-TGAGTCCTTCCACGATACCA-3′(reverse) for GAPDH. A comparative threshold cycle (CT) and a 2−ΔΔCt method were used to measure target genes.
Modulating miR-21 and PTEN in breast cancer cells
Human miR-21 gene was PCR-amplified from normal genomic DNA and cloned into a pGIPZ-shRNAmir-GFP plasmid for ectopic expression of miR-21. Primers used for amplification were 5′-CAACAGAAGGCTCGAGGATCTTAACAGGCCAGAAATG-3′ (sense) (Xho I site underlined) and 5′-ATTCTGATCAGGATCCCTAAGTGCCACCAGACAGAAG-3′ (antisense) (BamH I site underlined). The following primers were used for PCR to confirm insertion: 5′-ATGAGGCTTCAGTACTTTACAG-3′ (MIR30-F) and 5′-CATAGCGTAAAAGGAGCAACA-3′ (WPRE-R). A scrambled shRNA clone (empty vector) was a negative control. Plasmid construction and the lentiviral package were completed by Sunbio Company (Shanghai, China). MDA-MB-231 and MCF-7 cells were infected with negative control or miR-21-overexpressing constructs.
Two pooled shRNA sequences that offered the greatest reduction of miR-21 were as follows: 5′-aattcaaaaa TAGCTTATCAGACTGATGTTGA-3′ and 5′-ccgg TCAACATCAGTCTGATAAGCTAtttttg-3′ (stem is capitalized), and a scrambled sequence (5′-TTCTCCGAACGTGTCACGT-3′) as a negative control were cloned into a GV280-shRNAmir-GFP plasmid. The PCR primer to confirm insertion was 5′-CCATGATTCCTTCATATTTGC-3′ (pGCSIL-F). Lentiviral particles were prepared by Genechem Company (Shanghai, China) and used to infect 231/GEM and MCF-7/GEM cells. Targeted cells were selected with puromycin and pCDHCMV-MCS-EF1-puro plasmid (SBI, USA), a gift from Dr. Qin Y (Pancreatic Cancer Institute of Fudan University, Shanghai), was transfected into 231/miR21-ox cells to overexpress PTEN with LipofectamineTM 2,000 (Invitrogen) according to the manufacturer’s instructions.
Migration and invasion assay
Cell migration and invasion was assayed using a Transwell Permeable Support system with 8-μm pores (Corning). Cells were seeded on Transwell inserts coated with Matrigel (1:6; BD Biosciences) for the invasion assay and Matrigel-free wells were used for the migration assay. In brief, 2 × 104 MDA-MB-231 cells and sublines were seeded in serum-free medium and translocated to 10 % serum media for 24 h. For MCF-7 and its sublines, 5 × 104 cells were incubated for 48 h. After removal of non-migrated/non-invading cells, remaining cells were fixed in 4 % paraformaldehyde and then stained with Giemsa solution. Stained cells were counted in five different fields in each well under an inverted microscope.
Western blot
Lysates were obtained from cultured cells with a mixture of RIPA buffer (Beyotime, Shanghai, China) and protease inhibitor cocktail (Sigma) and PhosSTOP (Roche). Cells at the logarithmic growth phase were harvested, washed with cold 1× PBS twice, and then lysed with cell lysis buffer on ice for 30 min. Cells were centrifuged at 12,000 rpm for 15 min at 4 °C. Protein concentration was measured with a BCA protein assay kit (Beyotime). Equal amounts (20 μg/well) of protein were separated by SDS-PAGE and transferred to PVDF membranes which were washed, blocked, and incubated with primary antibodies against E-cadherin (1:1000; all antibodies were from Cell Signaling Technology unless otherwise indicated), vimentin (1:1000), ZEB1 (1:1000), Twist1 (1:1000; Proteintech Group), Snail (1:1000; Proteintech Group), Slug (1:1000), PI3K(p85) (1:1000; Proteintech Group), PI3K(p110) (1:1000; Proteintech Group), PTEN (1:1000), AKT (1:1000), p-AKT (Ser473) (1:1000), β-catenin (1:2000; GeneTex), p-β-catenin (Ser33/37/Thr41) (1:1000), mTOR (1:1000), p-mTOR (Ser2448) (1:1000), and β-actin (1:2000) at 4 °C overnight. Afterwards, membranes were washed and incubated with goat anti-rabbit or anti-mouse IgG (1:2000 each; Biotech Well, Shanghai, China) for 1 h at room temperature. Signals were measured with a luminescent image analyzer (ImageQuant LAS4000 mini) and β-actin was a loading control.
Animal xenograft experiments
Four-week-old female BALB/c nude mice (Shanghai SLAC Laboratory Animal Center of Chinese Academy of Sciences, Shanghai, China) were purchased and randomly divided into two groups (n = 5/group) and subcutaneously injected (right axilla, sc) with 1 × 107 parental MDA-MB-231 and miR-21 stably overexpressing cells (231/miR21-ox) with matrigel (1:1)/100 μL per mouse. Tumor volume was measured twice weekly as follows: volume (mm3) = [width2 (mm2) × length (mm)]/2. Once tumor diameters reached 0.2–0.3 cm, mice received gemcitabine (10 mg/kg, ip, on days 1, 5, and 8). At the end of the study, mice were sacrificed and tumors were carefully removed. All procedures for animal care were approved by the Animal Management Committee of Fudan University.
Statistical analysis
Data were analyzed using GraphPad Prism 5.0 software [20] (San Diego, CA). Quantitative variables were expressed as means ± SEM and analyzed with the Student’s t test (P < 0.05 was considered statistically significant).
Results
Gemcitabine-resistant cell establishment and miR-21 expression
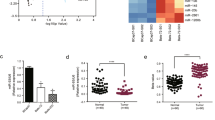
We initially established MCF-7 gemcitabine-resistant cells (MCF-7/GEM) and measured drug sensitivity in 231/GEM and MCF-7/GEM cells. Figure 1b shows that both drug-resistant cells were less sensitive to gemcitabine than corresponding parental cells. miR-21 in 231/GEM and MCF-7/GEM cells were 2.7 times greater than in 231/GEM cells and ~15 times greater in MCF-7/GEM cells than in MDA-MB-231 and MCF-7 cells (Fig. 1c). Thus, miR-21 is associated with gemcitabine resistance.
Establishment of gemcitabine-resistant cells and miR-21 expression. a MCF-7/GEM cells were created. b Gemcitabine sensitivity evaluated via CCK-8 assay in MDA-MB-231, MCF-7, 231/GEM, and MCF-7/GEM cells (n = 5 per triplicate experiments) *P < 0.05, **P < 0.01. c miR-21 expression in MCF-7/GEM cells (n = 3 per triplicate experiments). All data are means ± SEM
miR-21 promotes gemcitabine resistance in breast cancer
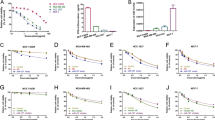
To understand the effect of miR-21 on gemcitabine resistance, loss and gain of function experiments in vitro and in vivo were conducted. First, we stably overexpressed miR-21 in MDA-MB-231 and MCF-7 cells (231/miR21-ox, MCF-7/miR21-ox) and stably downregulated miR-21 in gemcitabine-resistant 231/GEM and MCF-7/GEM cells (231GEM/miR21-kd, MCF-7GEM/miR21-kd) (Supplementary Fig S1). Data indicate that overexpression of miR-21 both in MDA-MB-231 and MCF-7 cells significantly decreased inhibitory rates in comparison with negative controls (NC and parental cell group; Fig. 2a). Interestingly, knockdown of miR-21 restored 231/GEM and MCF-7/GEM cell sensitivity to gemcitabine treatment (Fig. 2b). Thus, miR-21 can induce gemcitabine resistance in breast cancer cells in vitro.
miR-21 promotes gemcitabine resistance in breast cancer. a, b Gemcitabine sensitivity was decreased in MDA-MB-231 and MCF-7 cells overexpressing miR-21 (a) and increased (b) in 231/GEM and MCF-7/GEM cells that under-expressed miR-21 (n = 5 per triplicate experiments) *P < 0.05, **P < 0.01. c Animal tumors were inhibited in MDA-MB-231 animals after gemcitabine treatment for 3 days and this effect was modest with miR-21 overexpression (231-miR-21-ox) from day 3 to 5 after gemcitabine exposure and progressed until day 11 after treatment (left panel). Tumor volumes in miR-21 overexpressing animals were greater than in MDA-MB-231 animals at the experiment end (right panel). (n = 5/group). Data are means ± SEM
To understand whether tumors that ectopically express miR-21 reduces their sensitivity to gemcitabine, nude mice were treated with MDA-MB-231 and 231/miR21-ox cells (right axillary, sc administration) and exposed to gemcitabine (10 mg/kg). Data indicate that MDA-MB-231 and 231/miR21-ox tumors were inhibited after 3 days of gemcitabine treatment but tumor inhibition in the MDA-MB-231 group was greater. After 11 days of gemcitabine treatment, 231/miR21-ox group tumors progressed and MDA-MB-231 group tumors were inhibited (Fig. 2c, left panel). MDA-MB-231 tumors were significantly smaller than in the 231/miR21-ox group at the study end (Fig. 2c, right panel). Thus, in vitro and in vivo studies confirmed that miR-21 promotes gemcitabine resistance in breast cancer.
EMT properties acquired in gemcitabine-resistant breast cancer cells
Gemcitabine-resistant breast cancer cells had more EMT-like properties compared to parental cells. 231/GEM cells had altered morphology, changing from short rod-like shapes to irregular and elongated shapes. MCF-7/GEM cells changed from round pebble-shaped cells into long shuttle-strip cells (Fig. 3a). Morphological changes indicated that gemcitabine resistant cells may acquire a more aggressive mesenchymal phenotype. Transwell assays confirmed that gemcitabine-resistant cells were more motile and invasive than parental cells (Fig. 3b, c). The epithelial molecular marker E-cadherin in protein and mRNA was reduced and the mesenchymal molecular marker vimentin (VIM) in protein and mRNA was increased in drug-resistant cells (Fig. 3d, e). As shown in Supplementary Fig S2a, Twist1 and Snail were increased in 231/GEM cells and MCF-7/GEM cells, respectively. ZEB1 was downregulated in 231/GEM cells, while lost in MCF-7 and MCF-7/GEM cells. No significant difference was observed in Slug. Therefore, Twist1 and Snail were further assessed under the condition of miR-21 loss and gain expression. Taken together, phenotypic changes, alterations in mobility, and changes to EMT-associated molecular markers confirmed that gemcitabine-resistant breast cancer cells acquired EMT traits.
Gemcitabine-resistant breast cancer cells acquired EMT properties. a Typical EMT-like morphological changes occurred in 231/GEM and MCF-7/GEM cells. b Representative images from migration and invasion assays, bars 100 μM. c Motility and invasiveness were promoted in 231/GEM and MCF-7/GEM cells. (n = 3 per triplicate experiments). d, e E-cadherin mRNA and protein in 231/GEM and MCF-7/GEM cells was significantly downregulated and vimentin was upregulated compared to 231 and MCF-7 cells (n = 3 per triplicate experiments). Data are means ± SEM
miR-21 regulates EMT in gemcitabine-resistant breast cancer cells
miR-21 overexpression promoted acquisition of gemcitabine resistance with EMT traits in breast cancer cells. We noted typical EMT-like morphological changes such as irregular elongated shapes in MDA-MB-231 and MCF-7 cells after ectopic expression of miR-21 compared to negative controls (Fig. 4a). When miR-21 expression was suppressed in gemcitabine-resistant 231/GEM and MCF-7/GEM cells, the elongated morphology returned to short rod-like shapes in MDA-MB-231 cells and cobblestone patterns in MCF-7 cells (Fig. 4b). After these morphological changes, MDA-MB-231 and MCF-7 cells that overexpressed miR-21 had increased migratory and invasive capacities (Fig. 4c, d), whereas these features were reduced in 231/GEM and MCF-7/GEM cells after miR-21 was downregulated (Fig. 4e, f). Finally, upregulating miR-21 (Fig. 4g) reduced E-cadherin and increased vimentin in MDA-MB-231 and MCF-7 cells. With knockdown of miR-21 in gemcitabine-resistant cells, the opposite occurred (Fig. 4h). As for transcriptional factors, Twist1 expression increased after upregulating miR-21 in MDA-MB-231 cells and this occurred with Snail in MCF-7 cells. Downregulation of Twist1 occurred after knock down miR-21 in 231/GEM cells and the same event occurred with Snail in MCF-7/GEM cells (Supplementary Fig S2b). Thus, overexpression of miR-21 induced EMT associated with gemcitabine-resistance in breast cancer cells.
miR-21 modulated EMT in gemcitabine-resistant breast cancer cells. a, b Representative images of morphological changes after manipulating miR-21 status in gemcitabine-sensitive and resistant breast cancer cells. c, e Representative images of migration and invasion assay, bars 100 μM. d, f Motility and invasiveness were promoted by miR-21 in 231 and MCF-7 cells and this was inhibited after miR-21 knock down in 231/GEM and MCF-7/GEM cells (n = 3 per triplicate experiments). g, h Western blot quantification of E-cadherin and vimentin protein after manipulating miR-21 in 231 and MCF-7 cells and gemcitabine-resistant cells
AKT pathway activation during gemcitabine resistance in breast cancer cells
To understand molecular mechanisms behind EMT and gemcitabine resistance in breast cancer, several important regulators of EMT and chemoresistance were assayed. PI3K(p85) was increased while PI3K(p110) was not altered in two gemcitabine resistant cells. Little change of mTOR was observed and p-mTOR was overexpressed (Supplementary Fig S3a). AKT and p-AKT (Ser473) in gemcitabine-resistant cells were increased compared to parent cells (Fig. 5a, b), and PTEN was decreased (Fig. 5a, b). Interestingly, a downstream factor of the AKT pathway, β-catenin was elevated with a concomitant decrease in p-β-catenin followed by activation of AKT (Fig. 5a, b). Therefore, AKT pathway activation may be pivotal to EMT and gemcitabine resistance in breast cancer.
miR-21 induced EMT and gemcitabine resistance through the PTEN/AKT pathway. Western blot quantification of PTEN, AKT, p-AKT, β-catenin, and p-β-catenin. a, b p-AKT and β-catenin were upregulated; PTEN and p-β-catenin were downregulated in gemcitabine-resistant breast cancer cells. Ectopic expression of miR-21 in 231 and MCF-7 cells upregulated p-AKT and β-catenin and downregulated PTEN and p-β-catenin (c). Knock down of miR-21 in gemcitabine-resistant breast cancer cells rescued PTEN, AKT, p-AKT, β-catenin, and p-β-catenin (d). e Morphological changes after treatment. f Representative images of migration and invasion, bars 100 μM. g Cells quantified in migration and invasion assays. Data are means ± SEM (n = 3 per triplicate experiments). h Gemcitabine sensitivity was restored after rescued expression of PTEN in miR-21 overexpressing cells. Data are means ± SEM (n = 5 per triplicate experiments). i E-cadherin, vimentin, PTEN, AKT, p-AKT, β-catenin, and p-β-catenin protein was restored to wild-type status after rescued expression of PTEN in miR-21 overexpressing cells. j Diagram of miR-21 function within the PTEN/AKT pathway
miR-21 induced EMT and gemcitabine resistance in breast cancer by targeting PTEN
To verify whether miR-21 exerts its effect on EMT regulation and gemcitabine resistance by targeting PTEN, we manipulated miR-21 and measured PTEN and AKT pathway changes. When miR-21 was upregulated in MDA-MB-231 and MCF-7 cells, PTEN downregulation was observed and then p-AKT and β-catenin increased whereas p-β-catenin decrease (Fig. 5c). Suppressing miR-21 in gemcitabine-resistant cells enhanced PTEN and elevated p-AKT and β-catenin subsequently decreased and p-β-catenin increased (Fig. 5d). However, little change in PI3K (p85) was observed in parent and drug-resistant cells. The up- and downregulation of p-mTOR were found just in MCF-7/miR21-ox and MCF-7GEM/miR21-kd cells not in MDA-MB-231 and 231/GEM cells. (Supplementary Fig S3b). Therefore, miR-21 may not exert its function through the PI3K/mTOR pathway.
A rescue experiment to confirm miR-21 activation of the AKT/β-catenin pathway through PTEN was performed and PTEN was restored in miR-21 overexpressing 231/miR21-ox breast cancer cells as evidenced by morphological changes in what from spindle-like to short rod-like structures (Fig. 5e), changes in migration and invasion, and changes in drug sensitivity to gemcitabine recovery to that of MDA-MB-231 wild-type cells (Fig. 5f–h). Also, E-cadherin, vimentin, AKT, p-AKT, β-catenin, and p-β-catenin protein all returned to the MDA-MB-231 wild-type level (Fig. 5i). Thus, miR-21 regulates the AKT pathway and consequent EMT and gemcitabine resistance in breast cancer cells by suppressing PTEN.
Discussion
Drug resistance slows in breast cancer treatment advances with gemcitabine, so to overcome this drug resistance and improve gemcitabine application, the underlying mechanism underlying resistance should be clarified. Previously, miRNAs arrays showed that miR-21 was overexpressed in gemcitabine-resistant breast cancer cells [10], suggesting an association with resistance. miR-21 is involved in multiple biological events and regulation of signaling pathways [21, 22]. Later, recent studies confirmed that miR-21 overexpression was associated with cisplatin resistance in ovarian cancer cells [23] and gastric cancer cells [24], EGFR-TKI resistance in non-small cell lung cancer [25], and mediated resistance to trastuzumab therapy for breast cancer [26]. Wang’s group [27] reported that serum miR-21 may predict gemcitabine sensitivity for advanced pancreatic cancer, but the role for miR-21 in gemcitabine resistance in breast cancer is not clear.
In the present study, we confirmed that miR-21 was significantly overexpressed in gemcitabine-resistant breast cancer cells and this overexpression appears to increase breast cancer cell survival after drug exposure, whereas reduced expression of miR-21 rescued breast cancer cell sensitivity to gemcitabine. Therefore, miR-21 may regulate multidrug resistance in breast cancer. Drug resistance appears to be more complex than initially thought [28], possibly involving alterations in transport pathways and metabolic cascades that decrease drug accumulation. Likely, miR-21 is involved and this may be tied to EMT and cell reprograming, in which epithelial cells lose polarity, cell-cell adhesion, and tight junctions and acquire mesenchymal-like cell traits with increased mobility, less drug sensitivity, and facilitated metastatic potential [29, 30].
We found that gemcitabine resistance in breast cancer cells was accompanied by EMT changes in morphology, biomarkers, motility, and invasiveness. Moreover, miR-21 expression manipulation changed gemcitabine sensitivity in wild type and gemcitabine-resistant breast cancer cells and EMT transformation occurred as well but the contribution of each of these events to gemcitabine resistance is unclear. Cellular heterogeneity is a histological hallmark of breast cancer [31], as tumors consist of morphologically distinct subpopulations with varied molecular features and these may contribute to gemcitabine resistance. Therefore, we speculate that differential expression of miR-21 can induce gemcitabine resistance by regulating EMT reprogramming in some breast cancer patients.
Accumulating evidence indicates that miRNAs are crucial regulators of EMT [32–35] and we report that adaptive expression of miR-21 regulated EMT and consequent gemcitabine resistance in breast cancer but how many signaling agents participate in this process is not certain. PTEN and AKT pathways were associated with miR-21-induced EMT and other work confirms that PTEN is an miR-21 direct-targeted gene [22, 36, 37] and thought to be an inhibitor of the PI3K/AKT pathway which is central to EMT regulation [29]. Thus, we studied whether miR-21 regulates EMT by the PTEN/AKT pathway.
We measured PTEN during miR-21-associated EMT and noted that PTEN and p-AKT were correlated with miR-21 and that forced expression of PTEN in gemcitabine-resistant cancer cells caused mesenchymal-like traits to return to epithelial phenotypes, alterations to E-cadherin and vimentin, and a restoration of gemcitabine sensitivity. Thus, the PTEN/AKT pathway is a signaling agent, at least partially, within the miR-21-induced EMT program.
The canonical EMT program is characterized by complicated gene expression changes with complex signaling networks, among which β-catenin is important [32]. During EMT, reduced E-cadherin expression triggered by inducers of EMT caused cytoplasmic accumulation of β-catenin and formation of a transcriptional complex that promotes robust gene expression [38, 39]. Within the signaling networks, AKT activation may promote cytoplasmic accumulation of β-catenin by repressing β-catenin degradation via phosphorylation inhibition [40]. We noted that miR-21 overexpression caused AKT activation followed by reduced p-β-catenin and increased β-catenin, and these observations suggest that miR-21 can initiate EMT by AKT pathway regulation. Detailed mechanisms underlying this are not clear but the PTEN/AKT pathway may be pivotal for regulating miR-21-induced EMT.
In the study, we first demonstrate that miR-21 participated in gemcitabine resistance promotion in breast cancer through EMT process regulation, which is a novel mechanism distinguished from the previous studies reported. Together with the previous evidences, our findings further support that miR-21 may be a promising predictor of gemcitabine efficacy or as a target for reversing drug resistance.
References
Zhang J, Wang Z, Hu X, Wang B, Wang L, Yang W, et al. Cisplatin and gemcitabine as the first line therapy in metastatic triple negative breast cancer. Int J Cancer. 2015;136(1):204–11. doi:10.1002/ijc.28966.
Hu XC, Zhang J, Xu BH, Cai L, Ragaz J, Wang ZH, et al. Cisplatin plus gemcitabine versus paclitaxel plus gemcitabine as first-line therapy for metastatic triple-negative breast cancer (CBCSG006): a randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2015;16(4):436–46. doi:10.1016/S1470-2045(15)70064-1.
Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17 Suppl 5:v7–v12. doi:10.1093/annonc/mdj941.
Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2′,2′-difluorodeoxycytidine (gemcitabine). Drug Resist Updat. 2002;5(1):19–33.
Andersson R, Aho U, Nilsson BI, Peters GJ, Pastor-Anglada M, Rasch W, et al. Gemcitabine chemoresistance in pancreatic cancer: molecular mechanisms and potential solutions. Scand J Gastroenterol. 2009;44(7):782–6. doi:10.1080/00365520902745039.
Kim MP, Gallick GE. Gemcitabine resistance in pancreatic cancer: picking the key players. Clin Cancer Res. 2008;14(5):1284–5. doi:10.1158/1078-0432.CCR-07-2247.
Hagmann W, Jesnowski R, Lohr JM. Interdependence of gemcitabine treatment, transporter expression, and resistance in human pancreatic carcinoma cells. Neoplasia. 2010;12(9):740–7.
Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64(11):3761–6. doi:10.1158/0008-5472.CAN-03-3363.
Rha SY, Jeung HC, Choi YH, Yang WI, Yoo JH, Kim BS, et al. An association between RRM1 haplotype and gemcitabine-induced neutropenia in breast cancer patients. Oncologist. 2007;12(6):622–30. doi:10.1634/theoncologist.12-6-622.
Ye FG, Song CG, Cao ZG, Xia C, Chen DN, Chen L, et al. Cytidine deaminase axis modulated by miR-484 differentially regulates cell proliferation and chemoresistance in breast cancer. Cancer Res. 2015;75(7):1504–15. doi:10.1158/0008-5472.CAN-14-2341.
Xia C, Ye F, Hu X, Li Z, Jiang B, Fu Y, et al. Liver kinase B1 enhances chemoresistance to gemcitabine in breast cancer MDA-MB-231 cells. Oncol Lett. 2014;8(5):2086–92. doi:10.3892/ol.2014.2446.
Sebastiani V, Ricci F, Rubio-Viqueira B, Kulesza P, Yeo CJ, Hidalgo M, et al. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: relationship to molecular mechanisms of gemcitabine resistance and survival. Clin Cancer Res. 2006;12(8):2492–7. doi:10.1158/1078-0432.CCR-05-2655.
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi:10.1038/nature02871.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Garg M. Targeting microRNAs in epithelial-to-mesenchymal transition-induced cancer stem cells: therapeutic approaches in cancer. Expert Opin Ther Targets. 2015;19(2):285–97. doi:10.1517/14728222.2014.975794.
Wang Z, Li Y, Ahmad A, Azmi AS, Kong D, Banerjee S, et al. Targeting miRNAs involved in cancer stem cell and EMT regulation: an emerging concept in overcoming drug resistance. Drug Resist Updat. 2010;13(4-5):109–18. doi:10.1016/j.drup.2010.07.001.
Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210(4):789–803. doi:10.1084/jem.20120153.
Shi Z, Zhang J, Qian X, Han L, Zhang K, Chen L, et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73(17):5519–31. doi:10.1158/0008-5472.CAN-13-0280.
Wu Q, Wang R, Yang Q, Hou X, Chen S, Hou Y, et al. Chemoresistance to gemcitabine in hepatoma cells induces epithelial-mesenchymal transition and involves activation of PDGF-D pathway. Oncotarget. 2013;4(11):1999–2009.
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi:10.1038/nature06487.
Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–803. doi:10.1038/sj.onc.1210083.
Bao L, Yan Y, Xu C, Ji W, Shen S, Xu G, et al. MicroRNA-21 suppresses PTEN and hSulf-1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways. Cancer Lett. 2013;337(2):226–36. doi:10.1016/j.canlet.2013.05.007.
Chan JK, Blansit K, Kiet T, Sherman A, Wong G, Earle C, et al. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol Oncol. 2014;132(3):739–44. doi:10.1016/j.ygyno.2014.01.034.
Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162–8. doi:10.1016/j.tox.2013.02.014.
Li B, Ren S, Li X, Wang Y, Garfield D, Zhou S, et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer. 2014;83(2):146–53. doi:10.1016/j.lungcan.2013.11.003.
Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286(21):19127–37. doi:10.1074/jbc.M110.216887.
Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7(3):334–45. doi:10.1016/j.molonc.2012.10.011.
Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–26. doi:10.1038/nrc3599.
Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15(6):740–6.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. doi:10.1016/j.cell.2009.11.007.
Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;405–9. doi:10.1038/nature11154.
Tomaskovic-Crook E, Thompson EW, Thiery JP. Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res. 2009;11(6):213. doi:10.1186/bcr2416.
Zhao X, Lu Y, Nie Y, Fan D. MicroRNAs as critical regulators involved in regulating epithelial-mesenchymal transition. Curr Cancer Drug Targets. 2013;13(9):935–44.
De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi:10.1038/nrc3447.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi:10.1038/ncb1722.
Zhu W, Xu B. MicroRNA-21 identified as predictor of cancer outcome: a meta-analysis. PLoS One. 2014;9(8):e103373. doi:10.1371/journal.pone.0103373.
Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372(1-2):35–45. doi:10.1007/s11010-012-1443-3.
Zhang JX, Mai SJ, Huang XX, Wang FW, Liao YJ, Lin MC, et al. MiR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of beta-catenin signaling. Ann Oncol. 2014;25(11):2196–204. doi:10.1093/annonc/mdu439.
Li J, Zhou BP. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer. 2011;11:49. doi:10.1186/1471-2407-11-49.
Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J Biol Chem. 2007;282(15):11221–9. doi:10.1074/jbc.M611871200.
Acknowledgments
We thank Xiaoli Yang from the Key Laboratory of Breast Cancer at Fudan University Shanghai Cancer Center for generously providing the MDA-MB-231 gemcitabine-resistant cell subline and Yi Qin from the Pancreatic Cancer Institute of Fudan University for the gift of PTEN-overexpressing plasmid.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures for animal care were approved by the Animal Management Committee of Fudan University.
Conflicts of interest
None
Grant support
This work was funded by grants from the National Natural Science Foundation of China (Grant No: 81372846 and 81402188).
Additional information
Zhen-Hua Wu and Zhong-Hua Tao contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
Validation of miR-21 over- and under-expressed in breast cancer cells. (A-B) miR-21 expression was increased in MDA-MB-231 and MCF-7 cells infected with an miR-21 over-expressing lentiviral vector (A) and decreased in gemcitabine-resistant cells after treatment with miR-21 under-expressing lentiviral vector (B). (n = 3 per triplicate experiments). All data are means ± SEM. (TIFF 2834 kb)
Supplementary Figure S2
ZEB1, Twist1, Snail and Slug expression. (A) Twist1 and Snail were increased in 231/GEM and in MCF-7/GEM cells, respectively. ZEB1 was down-regulated in 231/GEM cells and negatively expressed in MCF-7 and MCF-7/GEM cells. No significant difference was observed in Slug. (B) Twist1 expression increased after up-regulating miR-21 in MDA-MB-231 cells and this occurred with Snail in MCF-7 cells. Down regulation of Twist1 occurred after knock down miR-21 in 231/GEM cells and the same event occurred with Snail in MCF-7/GEM cells. (TIFF 1764 kb)
Supplementary Figure S3
PI3K and mTOR expression in parent- and drug-resistant cells. PI3K and mTOR expression. (A) PI3K(p85) was increased in two drug-resistant cells, and PI3K(p110) was not changed. Little change of mTOR was observed and p-mTOR was overexpressed in drug-resistant cells. (B) PI3K(p85) was not changed after manipulating miR-21 in parent and drug-resistant cells. P-mTOR was up-regulated in MCF-7/miR21-ox cells and down-regulated in MCF-7GEM/miR21-kd cells. However, p-mTOR changed little after up- or down-regulation of miR-21 in MDA-MB-231 and 231/GEM cells. (TIFF 2380 kb)
Rights and permissions
About this article
Cite this article
Wu, ZH., Tao, ZH., Zhang, J. et al. MiRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumor Biol. 37, 7245–7254 (2016). https://doi.org/10.1007/s13277-015-4604-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4604-7