Abstract
With the development of technologies such as microarrays and RNA deep sequencing, long noncoding RNAs (lncRNAs) have become the focus of cancer investigations. LncRNAs, nonprotein-coding RNA molecules longer than 200 nucleotides, are dysregulated in many human diseases, especially in cancers. Recent studies have demonstrated that lncRNAs play a key regulatory role in gene expression and cancer biology through diverse mechanisms, including chromosome remodeling and transcriptional and post-transcriptional modifications. The expression levels of specific lncRNAs are attributed to prognosis, metastasis, and recurrence of cancer. LncRNAs are often involved in various biological processes, such as regulation of alternative splicing of mRNA, protein activity, and epigenetic modulation or silencing of the microRNAs, via discrete mechanisms. Deregulated levels of lncRNAs are shown in diverse tumors, including breast cancer. Based on latest research data, the tissue-specific expression signature of lncRNAs may represent the potential to discriminate normal and tumor tissue or even the different stages of breast cancer, which makes them clinically beneficial as possible biomarkers in the diagnosis and prognosis or therapeutic targets. In this brief review, we summarize some recent researches in the context of lncRNAs’ roles in breast cancer pathogenesis and their potential to serve as diagnostic, predictive, and prognostic biomarkers and novel targets for breast cancer treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to American Cancer Society definition, breast cancer is a disease in which the malignant tumor originates from breast tissue. The cells with uncontrolled growth can invade surrounding tissue, but with early diagnosis and treatment, most people are able to resume their usual life. International Agency for Research on Cancer (2012) fact sheets show that breast cancer is the most commonly diagnosed cancer in women and ranks second as a cause of cancer death in women [1]. In accordance with the World Health Organization, comprehensive cancer control includes prevention, early detection, diagnosis, and treatment as well as rehabilitation and palliative care.
Breast cancer has a heterogeneous entity which can be divided by various methods into many molecular subgroups. Due to molecular differences between histologically similar tumors, it is hard to predict clinical outcomes [2]. This complexity exhibits a roadblock in the pathway to better molecular classification and personalized therapy that should be overcome. Hence, currently it is of pivotal importance to find noninvasive biomarkers with high sensitivity and specificity, which can be used for breast cancer detection at an early stage and monitoring of response to therapy [3].
Recent advances in technologies, such as microarray and high-throughput sequencing, represented a deeper understanding of molecular biology, especially noncoding RNA (ncRNA). It was found that there are only <2 % of the total genome sequence as protein-coding genes while at least 98 % of the genome are transcribed into ncRNA. In the past, nonprotein-coding RNAs were known as “transcription noise,” but now it is obvious that ncRNAs play a crucial regulatory role in cell differentiation and organism growth and metabolism [4]. The ncRNAs are composed of some well-known RNAs such as transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), ribosomal RNAs (rRNAs), and small nucleolar RNAs (snoRNAs) [5]. A significant and the greatest group of ncRNAs are long noncoding RNAs (lncRNAs). LncRNAs, which are recently detected in various cancer types, including breast cancer, are involved in multiple biological processes and can be used as a candidate for cancer diagnosis, prognosis, and treatment, which may result in defeating breast cancer in the near future [3, 6].
LncRNAs are endogenous cellular molecules with a mature length range from 200 nt to 100 kb that are not protein-coding [7]. LncRNAs and their biological roles are least described among the other ncRNAs. Hence, there is no adequate certain information about their functions. Mounting data indicate that most of the identified lncRNAs are transcribed by RNA polymerase II (RNA pol II). They may contain 5′ methyl cap and are often polyadenylated and spliced [5, 8]. Although there is not a satisfactory classification for lncRNAs due to wide size range, various locations in the genome, and wide range of functions, novel identified groups of lncRNAs are categorized as long intergenic noncoding RNAs, long intronic noncoding RNAs, lncRNAs with dual functions, telomere-associated lncRNAs, pseudogene RNAs, and transcribed-ultraconserved regions [9].
Despite the fact that many biological functions of lncRNAs are unknown yet, recent evidences depict a close link between lncRNAs and breast cancer occurrence and development. Some of lncRNAs were shown to have aberrant expression in breast cancer, including HOX transcript antisense intergenic RNA (HOTAIR), metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1), zinc finger antisense 1 (ZFAS1), growth arrest-specific 5 (GAS5), long stress-induced noncoding transcript 5 (LSINCT5), steroid receptor RNA activator 1 (SRA1), X-inactive specific transcript (XIST), H19, and BC200 [5, 10, 11]. In addition, certain lncRNAs have cell-type-specific expression signatures that may have potential use as biomarkers or therapeutic targets [12]. In this study, we aim to provide a brief review of the main roles of some certain lncRNAs which are involved in breast cancer pathogenesis and diagnosis. We also demonstrate the clinical applications of lncRNAs and discuss potential novel strategies for more effective treatments of human breast cancer at lncRNA molecular level.
Biology of long noncoding RNAs
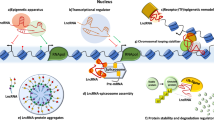
LncRNAs show developmental and tissue-specific expression patterns and have been associated with numerous biological processes, for example, alternative splicing, regulation of protein activity, and alternation of protein localization, as well as epigenetic regulations (DNA methylation, chromatin remodeling, histone modification, and gene silencing). Some of the lncRNAs can be precursors of small RNAs and tools for silencing the miRNAs. In a recent study, it has been shown that lncRNAs can function as competitive inhibitors of microRNAs called “miRNA sponges” to decrease miRNA levels [10].
New researches demonstrated that these molecules can act as signals, guides, decoys, or as scaffolds [13]. LncRNAs can function as signal molecules; when cells endure particular stimuli, lncRNAs will represent a corresponding tissue specificity. LncRNAs can serve as decoy for miRNA target sites. It can also instigate and bind to specific proteins and then impede the reaction of its downstream sequence. LncRNAs which play a guiding role can recruit chromatin-modifying enzymes or other proteins to target genes. LncRNA can form a structure for protein complex to assemble two superficial modification enzymes [7].
The effect of long noncoding RNAs in pathogenesis of breast cancer
LncRNAs were found to be deregulated in many cancers, such as lung, glioma, liver, prostate, and breast cancers. LncRNAs are also involved in cancer development as new regulators [14]. Analogically to protein-coding genes and miRNAs, lncRNAs can participate in cancer progression as oncogenes or tumor suppressors. In addition, certain lncRNAs demonstrate distinct expression patterns between primary tumors and metastases. So far, a few lncRNAs were shown to have aberrant expression in breast cancer, including HOTAIR, MALAT-1, ZFAS1, GAS5, LSINCT5, SRA1, XIST, H19, and BC200. Here, we discuss some of significant lncRNAs in the context of breast cancer in detail [5, 10, 11, 15].
H19
H19 is the first known lncRNA with a vital role in genomic imprinting during growth and development. It is a 2.3-kb transcript and is located on chromosome 11p15.5. The regulation of H19 is closely related to its reciprocally imprinted neighbor, insulin-like growth factor 2 (IGF2), but these alleles, the maternal H19 and paternal IGF-2, are selectively expressed. It has been proposed that H19 is involved in many various processes, varying from transcriptional and post-transcriptional regulation of expression to tumor suppression and oncogenesis in different cancer types especially in breast cancer. In a study on breast cancer cells, lncRNA H19 was overexpressed except in MDA-MB-231 cells [16]. H19 can be regulated by known oncogenes c-myc and Gli1 in a positive manner. C-myc can directly induce transcription of H19 by allele-specific binding, and H19, a target gene of E2F, can regulate cell cycle progression, especially the transition of G1 to S. The tumor suppressor gene p53 has been shown to decrease H19 levels. Furthermore, it is approved that lncRNA H19 is the precursor of miR-675 whose direct target is tumor suppressor retinoblastoma [17]. H19 functions as a riboregulator so it can participate in the regulation of translation. H19 under hypoxic circumstances has been shown to repress caveolin-1, a breast cancer metastasis suppressor. Further study focusing on the isogenic model MCF-7/MCF-7Ras showed that overexpression of H19 intensified the aggressive phenotype of tumor cells and is a marker for cell proliferation, which clarified the function of H19 as an oncogene in breast cancer [5, 18].
HOTAIR
HOTAIR, a 2.2-kb carcinogenic lncRNA, is transcribed from HOXC locus on chromosome 12q13.13. It binds to polycomb and directs it to HOXD genes on chromosome 2, finally resulting in subsequent transcriptional silencing. Upregulation of HOTAIR leads to altered gene expression, including the silencing of tumor suppressor genes JAM2 and PCDH. It has been demonstrated that HOTAIR is highly expressed in primary breast tumors and metastases, and its expression level in primary tumors is a potential predictor of eventual metastasis and death [19]. HOTAIR also plays a role in breast cancer through competitive binding with tumor suppressor genes such as BRCA1 which is an important breast cancer tumor suppressor gene. Decreased expression levels of BRCA1 lets HOTAIR to bind to polycomb repressive complex 2 (PRC2), which results in reprogramming promotion of breast epithelial cells to aggressive cancer cells [20].
GAS5
GAS5 is located in Chr1q25.1, and the length of GAS5 is approximately 0.6–1.8 kb. GAS5 can modulate apoptosis and differentiation in mammalian cells. GAS5 is downregulated in breast cancer and breast cancer cells and is significantly low compared with normal breast epithelial tissue from the same patients. This decrease was observed in both stages I and II in breast cancer, which indicates that decline in GAS5 expression is an early event in oncogenesis [21]. GAS5 acts as “riborepressor” of the glucocorticoid receptor (GR), which regulates cell survival and metabolic activities during starvation by interacting with the activity of the GR in transcription. GAS5 transcript interacts with the DNA-binding domain of GRs and inhibits the steroids’ interaction with their receptors [22]. Via this mechanism, GAS5 decreases the expression of various genes, including cellular inhibitor of apoptosis 2 (cIAP2), and makes the cells vulnerable to apoptosis [23]. Furthermore, the resistance to apoptosis was observed due to silencing of endogenous GAS5 levels in breast cancer cells [21].
MALAT-1
MALAT1 is a highly evolutionarily conserved, nuclear speckle-associated lncRNA which is expressed locally in the nucleus. It is a 7-kb lncRNA that was observed in nonsmall cell lung cancer for the first time [24]; since then, it has been detected in a high rate in different types of tumors, including breast cancer. A research by Guffanti et al. showed that MALAT1 is significantly upregulated in breast cancer samples and may demonstrate a different regulation in estrogen receptor-positive (ER+) tamoxifen-treated and untreated samples [5]. MALAT1 possesses mutations and deletions in luminal breast cancer. These mutations may cause alteration in the function of MALAT1, which may result in splicing deregulations [25]. Deregulated expression levels of MALAT1 were observed in breast cancer tissue in comparison with normal breast tissue. Besides, MALAT1 locus is often altered in breast cancer and other cancer types [26]. In a recent study, Sun et al. have identified that MALAT1 is reduced in proliferating mammary glands of c-myc transgenic mice and is significantly decreased in mammary tumors [4].
XIST
One of the most studied lncRNAs is XIST, transcribed from the X inactivation center of the X chromosome that is involved in its inactivation in female mammals. The X chromosome includes various oncogenes; thus, the activation of both X chromosomes may be favorable to the cancer cell. XIST expression is found to be dysregulated in various female cancers, including breast, cervical, and ovarian cancer [27]. XIST’s mechanism of action in breast cancer is not identified yet, and there are some discrepancies in results of studies. Some studies showed the interaction of the tumor suppressor gene BRCA1 with XIST, and that the loss of BRCA1 is related to the loss of XIST [28]. In another study on breast cancer tissue from patients with germline BRCA1 mutations, it was demonstrated that XIST is expressed in a significant number of samples [29]. More studies are required to elucidate whether XIST interactions with BRCA1 modulate cancer progression or if genetic instability related to BRCA1 expression loss is responsible for the decrease in XIST expression [30].
Other important lncRNAs which function in breast cancer are mentioned in Table 1 [4].
Long noncoding RNAs as diagnostic tools in breast cancer
LncRNA transcripts can be detected with various methods, but their biological functions remain, in most cases, unknown. Methods used in the detection of lncRNAs include microarrays [35, 36], RNA sequencing, quantitative reverse transcription PCR (qRT-PCR) [37–39], northern blot, Fluorescence in situ hybridization (FISH), RNA interference (RNAi), and methods designed for detection of RNA–protein interactions (RNA immunoprecipitation (RIP), RNA-binding protein immunoprecipitation-chip (RIP-CHIP)) [10, 40, 41].
Deregulated expression levels of lncRNAs have been linked with various cancers [42]. For instance, differential display code 3 (DD3 or PCA3), a prostate-specific lncRNA, is identified as a reliable marker for early detection of prostate cancer [43]. Furthermore, high levels of DD3 in urine sediments demonstrated more sensitivity than blood for discriminating among cancerous and noncancerous individuals [44].
HOTAIR expression levels were increased in primary and metastatic breast cancer tissue compared with normal breast tissue [19]. In breast cancer tissue, moderate or high levels of MALAT-1 were also observed [26]. There is mounting evidence proposing that alterations in X chromosome inactivation are present in breast cancer cells [27]. In a recent study, expressions of six lncRNAs, HOTAIR, H19, KCNQ1OT1, Maternally Expressed 3 (MEG3), MALAT1, and ZFAS1, plus HER2 and MKI67 mRNAs, were investigated comprising normal epithelia, ductal carcinoma in situ (DCIS), and invasive carcinoma (IC) from 45 patients, and the results demonstrated that H19 and HOTAIR, specifically and possibly KCNQ1OT1, are potential biomarkers in breast cancer diagnosis [45].
Latest studies have demonstrated that ncRNAs such as microRNAs and lncRNAs are present in body fluids, and they are identified as feasible noninvasive biomarkers for diagnosis and prognosis of malignant tumors. For instance, in a recent study, circulating HOTAIR-derived fragment was measured in serum of breast cancer patients and healthy individuals, and the results showed that HOTAIR circulating DNA is a potential biomarker for breast tumor [46]. Lately, researchers have investigated circulating serum lncRNA RP11-445H22.4 in breast cancer patients. Their data represented lncRNA RP11-445H22.4 as a potential breast cancer biomarker [47].
Long noncoding RNAs as prognostic and predictive biomarkers
In a recent study, it was indicated that BC040587 is significantly downregulated in breast cancer tissues and breast cancer cell lines; therefore, BC040587 may represent a new prognostic marker in breast cancer [32]. HOTAIR and BC200 are upregulated in breast cancer and thus may be useful to predict outcome and tumor aggressiveness in breast cancer patients [48]. Lately, the expression of SPRY4-IT1 (PRY4 intronic transcript 1) was investigated in 48 breast cancer tumor tissues compared with normal tissues and showed a significant increase. Moreover, in the mentioned study, high expression level of SPRY4-IT1 displayed an association with larger tumor size and progressed pathological stage in breast tumor patients; therefore, it may be represented as a novel prognostic biomarker and a possible therapeutic target for breast cancer [33]. In another study, researchers identified that eosinophil granule ontogeny transcript (EGOT) is downregulated in breast cancer tissues and is strongly related to tumor stage and poor prognosis of breast cancer patients [49]. LINC00472 is a novel long intergenic noncoding RNA, and study results demonstrated that LINC00472 upregulation was related with less aggressive breast tumors and better prognosis [50].
Potential clinical applications of lncRNAs
Compared to protein-coding RNAs, lncRNAs are generally more tissue-specific; thus, this tissue-specific expression signature will allow personalized medicine to utilize lncRNAs as highly specific biomarkers in early detection of localized tumors from various biological fluids‚ the prediction of clinical outcome, and as gene therapy targets [51]. As time passes, more lncRNAs are found to be involved in breast tumor outcomes. For example, a study by Gupta et al. revealed that HOTAIR is overexpressed up to 2000-fold in breast cancer metastases and also showed that a high level of HOTAIR expression is a significant predictor of metastasis and mortality independent of other risk factors such as tumor volume, hormone receptor status, and tumor stage [19]. Colon cancer associated transcript 2 (CCAT2), a novel lncRNA in breast cancer, showed an overexpression in two out of three examined primary breast cancer patient sets and was represented as a valuable predictive marker for metastasis-free survival and overall survival in patients with local metastasis of lymph node who had received adjuvant chemotherapy [52].
Meng et al. have identified a set of four-lncRNA signature, which predicts the overall survival. The four-lncRNA signature (AK024118, U79277, AK000974, BC040204) may have clinical applications as molecular diagnosis markers and therapeutic targets, including the selection of high-risk patients for adjuvant therapy [3]. In a recent study, it is demonstrated that reduced GAS5 diminishes cell responses to apoptosis induction by conventional chemotherapeutic agents, and restoration of GAS5 expression may have implications in breast cancer chemotherapy [53].
One of lncRNA clinical approaches is the application of RNAi-mediated gene silencing to selectively silence oncogenic lncRNAs. Implementing the gene therapy is another therapeutic approach which can deliver tumor suppressor lncRNAs to breast cancer cells. However, it should be noted that there are still many technical challenges in implementing therapeutic RNAi and gene therapy [10, 54]. Another strategy is to target lncRNAs by antisense oligonucleotides (ASO), single-stranded DNA or RNA molecules which can bind to their target RNAs with high sequence specificity, and motivate their sequestration by RNAseH1 [55]. An example of this strategy application is mentioned in a recent study. In this investigation, it was demonstrated that ASO blocking MALAT1 prevents metastasis formation in lung cancer cells. ASOs could be designed to target other lncRNAs which function as oncogenic RNAs so as to restrict their expression [56].
Conclusions and future perspectives
LncRNAs as significant regulators of gene expression have demonstrated a wide spectrum of functions which enable them to modify breast cancer progressions via diverse pathways. Due to aggressive entity of most breast tumors, it is necessary to find noninvasive biomarkers with high sensitivity and specificity to detect breast cancer at primary stages and monitor the response to therapy.
Although some of the lncRNAs have shown the potential to serve as diagnostic biomarkers, to predict stages, the survival rate, and metastasis of breast cancer patients, there are still notable obstacles to the transfer of lncRNAs into clinical application that should be overcome. Desirable lncRNA biomarker requires stability and robust detection in biological fluids such as plasma and urine, but the stability of circulating lncRNAs remains largely unknown. Furthermore, the origin of circulating lncRNAs is not easily detectable. The lncRNA transcripts and their post-transcriptional modification levels are fluctuating or hard to determine during various disease states. Finally, the lncRNA biomarkers are still in need of further analytic and clinical validation for implementing in clinical practice [57].
New advances in RNA-based therapeutics introduce lncRNAs as novel targets for therapy and may provide new hope for safer and more effective treatment of breast tumors. Novel technologies for detection of circulating tumor cells may allow the investigations in metastasis stage [58]. The application of new genome editing tools such as the CRISPR/Cas9 system may pave a way for lncRNA research [54]. The utilization of synthetic oligonucleotides, e.g., locked nucleic acid modified oligonucleotides (LNA) that have high affinity and specificity to certain lncRNAs, may represent a new approach for targeted modification of lncRNA expression [59]. High-throughput screening of small-molecule modulators of lncRNA–protein interactions may introduce potential candidates to interrupt lncRNA–protein interaction or make alterations in lncRNA with its loading region target genome. Targeting lncRNA–protein interactions would increase compound specificity and reduce off-target effects to achieve more selective therapeutic effect [60, 61].
However, many common challenges remain in the development of RNA therapeutics, including the lack of secure, specific, and effective vector types for delivery methods, undetermined optimal dosage regimen, and off-target effects [62]. In addition, it is difficult to target lncRNA transcripts using RNAi technologies, such as siRNA and ASO, which may be due to their large size and extensive secondary structures [63].
In conclusion, although there is much to know about the functional role of the plenty of new characterized lncRNAs, they provide a new gold mine of cancer therapies and biomarkers. More studies are required in the context of lncRNAs to mirror their potential in discriminating normal and tumor tissue or even the different stages of breast cancer, which makes them clinically beneficial as possible biomarkers in the diagnosis and prognosis of breast tumors. In this brief review, some of well-described as well as novel lncRNAs which have the potential to serve as diagnostic, prognostic, and predictive biomarkers were discussed. Although they may have therapeutic applications, there is still a great need for further researches before they can be applied in clinical practice and there are multiple challenges that should be overcome for a wider use of lncRNAs.
References
International Agency for Research on Cancer (IARC). Cancer Facts and Figures. [ONLINE]. 2013. Available at: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
Koboldt DCFR, McLellan MD, Schmidt H, Kalicki-Veizer J, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi:10.1038/nature11412.
Meng J, Li P, Zhang Q, Yang Z, Fu S. A four-long non-coding RNA signature in predicting breast cancer survival. J Exp Clin Cancer Res. 2014;33(1):84. doi:10.1186/s13046-014-0084-7.
Hansji H, Leung EY, Baguley BC, FInlay G, Askarian-Amiri ME. Keeping Abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014;5. doi:10.3389/fgene.2014.00379.
Vikram R, Ramachandran R, Abdul K. Functional significance of long non-coding RNAs in breast cancer. Breast Cancer. 2014;21(5):515–21. doi:10.1007/s12282-014-0554-y.
Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5(20):9864–76.
Ye N, Wang B, Quan Z-F, Cao S-J, Wen X-T, Huang Y, et al. Functional roles of long non-coding RNA in human breast cancer. Asian Pac J Cancer Prev. 2014;15(15):5993.
Merry CR, Niland C, Khalil AM. Diverse functions and mechanisms of mammalian long noncoding RNAs. Methods Mol Biol. 2015;1206:1–14. doi:10.1007/978-1-4939-1369-5_1.
Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10(1):103. doi:10.1186/1479-5876-10-103.
Juracek J, Iliev R, Svoboda M, Slaby O. Long noncoding RNAs in breast cancer: implications for pathogenesis, diagnosis, and therapy. In: Barh D, editor. Omics approaches in breast cancer. India: Springer; 2014. p. 153–70.
Serviss JT, Johnsson P, Grandér D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. 2014;5:234. doi:10.3389/fgene.2014.00234.
Qiu M-T, Hu J-W, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumor Biol. 2013;34(2):613–20. doi:10.1007/s13277-013-0658-6.
Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–66. doi:10.1016/j.canlet.2013.06.013.
Shen X-h, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol. 2015;28(1):4–13. doi:10.1038/modpathol.2014.75.
Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed). 2015;7:94–108.
Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32(6):473–80. doi:10.1002/bies.200900170.
Subramanian M, Jones MF, Lal A. Long non-coding RNAs embedded in the Rb and p53 pathways. Cancers (Basel). 2013;5(4):1655–75. doi:10.3390/cancers5041655.
Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330–7. doi:10.1158/0008-5472.can-06-0037.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6. doi:10.1038/nature08975.
Wang L, Zeng X, Chen S, Ding L, Zhong J, Zhao JC, et al. BRCA1 is a negative modulator of the PRC2 complex. EMBO J. 2013;32(11):1584–97. doi:10.1038/emboj.2013.95.
Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2008;28(2):195–208. doi:10.1038/onc.2008.373.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA Gas5 is a growth arrest– and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi:10.1126/scisignal.2000568.
Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J Cell Sci. 2008;121(7):939–46. doi:10.1242/jcs.024646.
Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin [beta]4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41.
Gutschner T, Hämmerle M, Diederichs S. MALAT1—a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl). 2013;91(7):791–801. doi:10.1007/s00109-013-1028-y.
Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2006;26(6):851–8.
Froberg JE, Yang L, Lee JT. Guided by RNAs: X-inactivation as a model for lncRNA function. J Mol Biol. 2013;425(19):3698–706. doi:10.1016/j.jmb.2013.06.031.
Ganesan S, Silver DP, Drapkin R, Greenberg R, Feunteun J, Livingston DM. Association of BRCA1 with the inactive X chromosome and XIST RNA. Philos Trans R Soc Lond B Biol Sci. 2004;359(1441):123–8.
Vincent-Salomon A, Ganem-Elbaz C, Manié E, Raynal V, Sastre-Garau X, Stoppa-Lyonnet D, et al. X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer Res. 2007;67(11):5134–40. doi:10.1158/0008-5472.can-07-0465.
Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35(8):1751–6. doi:10.1007/s00268-010-0951-0.
Cayre A, Rossignol F, Clottes E, Penault-Llorca F. aHIF but not HIF-1α transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 2003;5(6):R223–30.
Chi Y, Huang S, Yuan L, Liu M, Huang N, Zhou S, et al. Role of BC040587 as a predictor of poor outcome in breast cancer. Cancer Cell Int. 2014;14. doi:10.1186/s12935-014-0123-7.
Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, et al. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14(1):51. doi:10.1186/s12943-015-0318-0.
Liu J, Shen L, Yao J, Li Y, Wang Y, Chen H, et al. Forkhead box C1 promoter upstream transcript, a novel long non-coding RNA, regulates proliferation and migration in basal-like breast cancer. Mol Med Rep. 2015;11(4):3155–9. doi:10.3892/mmr.2014.3089.
Khoshhali M, Moslemi A, Saidijam M, Poorolajal J, Mahjub H. Predicting the categories of colon cancer using microarray data and nearest shrunken centroid. J Biostat Epidemiol. 2014;1(1).
Xu N, Wang F, Lv M, Cheng L. Microarray expression profile analysis of long non-coding RNAs in human breast cancer: a study of Chinese women. Biomed Pharmacother. 2015;69:221–7. doi:10.1016/j.biopha.2014.12.002.
Yadegarazari R, Saidijam M. Using RT-PCR and qRT-PCR to assay RNA markers in detection of peripheral colorectal circulating cells: a systemic review. Clin Biochem. 2011;44(13):S196. doi:10.1016/j.clinbiochem.2011.08.479.
Karimi S, Mohamadnia A, Nadji SA, Yadegarazari R, Khosravi A, Bahrami N, et al. Expression of two basic mRNA biomarkers in peripheral blood of patients with non-small cell lung cancer detected by real-time rt-PCR, individually and simultaneously. Iran Biomed J. 2015;19(1):17.
Yadegarazari R, Hassanzadeh T, Majlesi A, Keshvari A, Monsef Esfahani A, Tootoonchi A, et al. Improved real-time RT-PCR assays of two colorectal cancer peripheral blood mRNA biomarkers: a pilot study. Iran Biomed J. 2013;17(1):15–21. doi:10.6091/IBJ.1104.2012.
Yan B, Wang Z-H, Guo J-T. The research strategies for probing the function of long noncoding RNAs. Genomics. 2012;99(2):76–80. doi:10.1016/j.ygeno.2011.12.002.
Feng Y, Hu X, Zhang Y, Zhang D, Li C, Zhang L. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol Biol. 2014;1165:115–43. doi:10.1007/978-1-4939-0856-1_10.
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–19. doi:10.4161/rna.20481.
Klecka J, Holubec L, Pesta M, Pesta M, Topolcan O, Topolcan O, et al. Differential display code 3 (DD3/PCA3) in prostate cancer diagnosis. Anticancer Res. 2010;30(2):665–70.
Moradi Sardareh H, Goodarzi MT, Yadegar-Azari R, Poorolajal J, Mousavi-Bahar SH, Saidijam M. Prostate cancer antigen 3 gene expression in peripheral blood and urine sediments from prostate cancer and benign prostatic hyperplasia patients versus healthy individuals. Urol J. 2014;11(6):1952–8.
Zhang Z, Peng Z, Olsen D, de Kay J, Weaver DL, Evans MF. Abstract 1498: long non-coding RNA in situ hybridization signal patterns correlate with breast tumor pathology. Cancer Res. 2014;74(19 Supplement):1498. doi:10.1158/1538-7445.am2014-1498.
Zhang L, Song X, Wang X, Xie Y, Wang Z, Xu Y, et al. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res Treat. 2015;152(1):199–208.
Xu N, Chen F, Wang F, Lu X, Wang X, Lv M, et al. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumor Biol. 2015;1–7. doi:10.1007/s13277-015-3469-0.
Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25(11):2125–33. doi:10.1093/carcin/bgh228.
Xu S-P, Zhang J-F, Sui S-Y, Bai N-X, Gao S, Zhang G-W, et al. Downregulation of the long noncoding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumor Biol. 2015;1–6. doi:10.1007/s13277-015-3746-y.
Shen Y, Katsaros D, Loo LWM, Hernandez BY, Chong C, Canuto EM, et al. Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget. 2015;6(11):8579–92.
Nguyen Q, Carninci P. Expression specificity of disease-associated lncRNAs: toward personalized medicine. Current topics in microbiology and immunology. Berlin: Springer; 2015. p. 1–22.
Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4(10):1748–62.
Pickard M, Williams G. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat. 2014;145(2):359–70. doi:10.1007/s10549-014-2974-y.
Haemmerle M, Gutschner T. Long non-coding RNAs in cancer and development: where do we go from here? Int J Mol Sci. 2015;16(1):1395–405.
Zong X, Huang L, Tripathi V, Peralta R, Freier SM, Guo S et al. Knockdown of nuclear-retained long noncoding RNAs using modified DNA antisense oligonucleotides. Methods Mol Biol. 2015;1262:321–31. doi:10.1007/978-1-4939-2253-6_20.
Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–9. doi:10.1158/0008-5472.can-12-2850.
Engstrom PF, Bloom MG, Demetri GD, Febbo PG, Goeckeler W, Ladanyi M, et al. NCCN molecular testing white paper: effectiveness, efficiency, and reimbursement. J Natl Compr Cancer Netw. 2011;9 Suppl 6:S-1–16.
Ramskold D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–82. http://www.nature.com/nbt/journal/v30/n8/abs/nbt.2282.html#supplementary-information.
Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34(39):5003–11. doi:10.1038/onc.2014.456.
Fatemi RP, Velmeshev D, Faghihi MA. De-repressing LncRNA-targeted genes to upregulate gene expression: focus on small molecule therapeutics. Mol Ther Nucleic Acids. 2014;3:e196. doi:10.1038/mtna.2014.45.
Pedram Fatemi R, Salah-Uddin S, Modarresi F, Khoury N, Wahlestedt C, Faghihi MA. Screening for small-molecule modulators of long noncoding RNA-protein interactions using AlphaScreen. J Biomol Screen. 2015;20(9):1132–41. doi:10.1177/1087057115594187.
Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–38.
Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–65.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Rights and permissions
About this article
Cite this article
Malih, S., Saidijam, M. & Malih, N. A brief review on long noncoding RNAs: a new paradigm in breast cancer pathogenesis, diagnosis and therapy. Tumor Biol. 37, 1479–1485 (2016). https://doi.org/10.1007/s13277-015-4572-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4572-y