Abstract
Ribosomal protein s15a (RPS15A), a highly conserved cytoplasmic protein, promotes mRNA/ribosome interaction in translation. Recent evidence showed that RPS15A is essential for tumor growth. RPS15A expression level was measured in glioblastoma tissue samples and normal brain (NB) tissue samples. RPS15A RNAi stable cell line U87 and U251 was generated by the pLVTHM-GFP lentiviral RNAi expression system. The knockdown efficiency was confirmed by quantitative real-time PCR and western blot. Molecular mechanisms and the effect of RPS15A on cell growth and migration were investigated by using western blot, MTT assay, wound healing assay, transwell migration assay, and tumorigenesis in nude mice. Here, we report that RPS15A is overexpressed in human glioblastoma tumor tissues. RPS15A knockdown inhibits proliferation and migration of glioblastoma cells in vitro. Knocking down RPS15A leads to the level of p-Akt decrease and cell cycle arrested in G0/G1 phase in U87 and U251 cells. Furthermore, the growth of glioblastoma cell-transplanted tumors in nude mice is inhibited by transduction with Lv-shRPS15A. Our findings indicate that RPS15A promotes cell proliferation and migration in glioblastoma for the first time. RPS15A might play a distinct role in glioblastoma and serve as a potential target for therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiforme (GBM) is the most common form of malignant brain tumor with poor survival from 1 week to 3 years [1, 2]. Current treatment includes radiation, surgery, and chemotherapy. Concomitant use of temozolomide and radiotherapy in patients who were newly diagnosed with GBM prolongs survival and improves patients’ life quality [3]. Previously, researchers have found some prognostic markers of glioblastoma and its relevant mechanisms [4–7]. Despite the great advances in therapeutic strategies, the median survival time of GBM patients were still unfavorable.
Ribosomal protein S15A (RPS15A) encodes long 130 amino acids with a molecular weight of 14.3 kD, which belongs to the 40S ribosomal subunits. RPS15A was reported to be a responsive protein of transforming growth factor-beta 1 (TGF-beta 1) and promotes cell proliferation of the human lung carcinoma cell line A549 [8]. Lian et al. [9] reported that overexpression of RPS15A via HBxAg promotes the growth of HepG2 cells. Moreover, Xu et al. [10] found that depletion of RPS15A arrests HepG2 cells at the G0/G1 phase and thus suppresses cell growth. An increase in RPS15A has also been detected in many other tumors such as astrocytoma, prostate cancer, colorectal cancer, and breast cancer [11, 12]. However, the role of RPS15A in the development of glioblastoma is less understood.
In the present study, RPS15A expression was evaluated in GBM patients’ samples. A lentivirus-mediated RNA interference (RNAi) system was applied to examine the effect of targeted knockdown of RPS15A on cell proliferation and migration both in vitro and in vivo. Possible mechanisms of RPS15A’s role in cancer cell growth were also explored.
Results
The RPS15A is highly expressed in glioblastoma tissues compared with NB tissues
To evaluate the role of RPS15A in glioblastoma, quantitative real-time PCR was performed to detect the expression level of RPS15A mRNA in 20 freshly collected glioblastoma samples and 15 NB tissues. Glioblastoma tissues presented a much higher expression levels of RPS15A mRNA compared with normal brain (NB) tissues (P < 0.05) (Fig. 1a). Furthermore, RPS15A protein level in glioblastoma tissues was consistently higher than that in NB tissues (Fig. 1b, c) (P < 0.05). To measure the expression level of RPS15A in 123 glioblastoma samples and 15 NB tissues in archived embedded paraffin, we used immunohistochemical (IHC) staining assay. The result revealed that RPA15A was expressed low in normal brain tissues (Fig. 1d) in low-grade glioblastoma tissues (Fig. 1e), while highly expressed in high-grade glioblastoma tissues (Fig. 1f, g).The IHC assay showed the RPS15A protein was highly expressed in 73.2 % glioblastoma tissues while only 33.3 % of NB tissues (P = 0.001) (Table 1).
Quantitative real-time PCR, western blot, and immunohistochemical (IHC) staining assay were used to measure the expression of RPS15A in glioblastoma tissues compared with normal brain tissues. a The mRNA expression of RPS15A is increased in tumor tissues (20 clinical cases) compared with NB tissues (15 clinical cases) by qRT-PCR assay. GAPDH was used as loading control. b, c Western blot assay was used to test the RPS15A protein expression levels between NB tissues (15 clinical cases) and glioblastoma tissues (20 clinical cases) with GAPDH as loading control. Data are presented as mean ± SD for three independent experiments (*P < 0.05). d–g IHC images showed RPS15A expression (arrow) was decreased in 15 normal brain tissues compared with 123 primary glioma samples. d Staining of RPS15A in normal tissues. e Staining of RPS15A in low-grade (WHOII) glioma tissues. f, g Staining of RPS15A in high-grade glioma and glioblastoma tissues (WHOIII-IV). Original magnification ×400, ×100
RPS15A expression level was the independent prognostic factor to glioma
The clinicopathological characteristics of patients were shown in Table 2. There was no significant association between RPS15A expression and patients’ age, gender, or histologic type in the free-selected glioma cases. Nevertheless, the level of RPS15A expression was correlated with the grade of pathology classification in glioma patients (P < 0.05). To confirm whether RPS15A was performed as an independent prognostic factor to glioma, we conducted univariate and multivariate analysis of RPS15A expression that is adjusted for the same parameters. RPS15A expression levels were indicative of an independent prognostic factor to glioma patients (P < 0.000) (Table 3).
Patients with low level RPS15A expression have a positive survival time
In order to explore the prognostic value of RPS15A expression in glioma, the relationship between levels of RPS15A expression and patients’ survival time was assessed by GraphPad Prism 5.0 software (www.graphpad.com, USA). Our results of the glioma cases manifested that RPS15A protein expression was remarkably correlated with overall survival (P < 0.0001). Patients with low levels of RPS15A expression in glioma tissues have a better survival than those with high expression (P < 0.05)(Fig. 2).
Knockdown RPS15A inhibits proliferation and migration in U87 and U251 cell lines
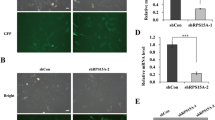
Lentiviral shRNA vector was used to knock down RPS15A expression in U87, U118, and U251 cells. RPS15A mRNA was measured by quantitative real-time PCR. The mRNA level was significantly reduced in shRPS15A transducing cells than in negative vector control [pLVTHMGFP-Control (PLV-Ctrl)] cells (P < 0.05) (Fig. 3a). Same was applied to protein expression level (P < 0.05) (Fig. 3b–d). To investigate possible functions of RPS15A in glioblastoma, we conducted series of experiments to determine the loss of function of RPS15A on glioblastoma cell growths in vitro. MTT assay showed that U87 and U251 cells with shRPS15A grew significantly slower than PLV-Ctr-U87 and PLV-Ctr-U251 cells (P < 0.05) (Fig. 4a, b). We adopted the wound healing and Boyden chamber assay to examine the effect of RPS15A on glioblastoma cell migration. As the wound healing assay showed that shRPS15A U87 and U251 cells displayed a lower speed into the gap compared with the shPLV-Ctrl-U87 and shPLV-Ctrl-U251cells (P < 0.05) (Fig. 4c).Moreover, in the Boyden chamber assay, there were less cells on the bottom surface of the shRPS15A cells compared with the shPLV-Ctrl-U87 and shPLV-Ctrl-U251 cells. The strained cells were counted with Image J 1.43r software (http://rsb.info.nih.gov/ij, Wayne Rasband National Institutes of Health, USA) (P < 0.05) (Fig. 4d). These results suggested that knockdown of RPS15A depressed motility of U87 and U251 cells.
Effect of shRNA to stably down-regulate RPS15A expression in human glioma cell lines U87, U251 and U118. a qRT-PCR assay shows transcriptional levels of the RPS15A gene with GAPDH used as loading control. b–d Western blot assay showing the RPS15A protein expression levels in sh-RPS15A and PLV-Ctrl treatments in vitro. GAPDH served as a loading control. c Data are presented as mean ± SD for three independent experiments (P < 0.05)
Stably knockdown the expression of RPS15A suppressed cell proliferation, migration. a, b MTT assay was used to measure the proliferation of U87 and U251 cell lines down-regulated by shRPS15A. Absorbance was read at 490 nm with averages from triplicate wells. c Stably knockdown RPS15A reduced the migration ability of U87 and U251 cell in vitro. d The Boyden chamber assay was used to measure the migration effect. Each experiment was repeated three times (P < 0.05, statistically significant difference)
Knockdown of RPS15A induces G0/G1 phase arrest and regulates cell cycle protein expression in U87 and U251 cell lines
Cell cycle analysis was employed to study the potential mechanisms that are involved in RPS15A knockdown-mediated cell growth inhibition in U87 and U251 cells. Our results showed a 69.52 and 72.44 % of G0/G1 phase cells in shRPS15A U87 and U251 cells while only 41.16 and 56.23 % of G0/G1 phase cells in PLV-Ctrl U87 and U251 cells (P < 0.05) (Fig. 5a–d). These data confirmed that the cell cycle was arrested in G0/G1 phase after knockdown of RPS15A expression in U87 and U251 cells. Moreover, cell cycle regulation protein CDK4 was expressed at a lower level in shRPS15A U87 and U251cells, whereas the protein p18INK4C expression was increased (Fig. 5e, f)
Knockdown of RPS15A in U87 and U251 cell lines could arrest the cell cycle at G0/G1 phase and regulate the CDK4, p18INK4C, and AKT pathway expression in vitro. a–d The cell cycle distribution of U87 and U251 cells transfected with sh-RPS15A and PLV-Ctrl were measured using propidium iodide (PI) staining and flow cytometry analyses. Down-regulated RPS15A in U87 and U251 cell lines could arrest the cell cycle at G0/G1 phase. e, f Suppressing endogenous RPS15A expression reduced the expression of CDK4. However, the p18INK4C was highly expressed. The GAPDH was used as loading control. g, h Down-regulating RPS15A expression depressed the expression of p-Akt, while the total protein and p-Erk, Erk, and Akt were not affected with GAPDH as loading control. Each experiment was repeated three times (P < 0.05, statistically significant difference)
Knockdown of RPS15A inhibits AKT activations
Since AKT and ERK pathways were reported to be important in cell cycle progress, the effect of RPS15A on AKT and ERK pathways were investigated. Results showed that reduced RPS15A could remarkably inhibit the phosphorylation of AKT but not the phosphorylation of ERK. Total AKT or ERK protein level were not affected (Fig. 5g, h). These results suggested that RPS15A regulated AKT activation in glioblastoma.
RPS15A and Akt immunoreactivity in glioblastoma samples and NB tissues
We examined the degree of co-expression of RPS15A and p-Akt in glioblastoma samples and NB tissues with fluorescent double-staining. The data revealed that the red fluorescence labeled the p-Akt was localized in the cytoplasm; the green fluorescence labeled the RPS15A also localized in cytoplasm; the blue fluorescence counterstained by DAPI was in the nucleus. Then, we built up the three fluorescence passages with the picture analysis software; the yellow fluorescence appeared in the position at which red and green fluorescence occurred simultaneously. This fact indicated that RPS15A and p-Akt were located in the same position in glioblastoma and normal brain tissues. Furthermore, this assay confirmed that RPS15A and p-Akt were overexpressed in glioblastoma samples compared with NB tissues (Fig. 6).
Immunofluorescence double-labeling assay was used to investigate the co-expression of phosphorylated p-Akt and RPS15A in glioblastoma and normal brain tissues. a Staining in normal tissues. b Staining in glioblastoma tissues. The blue fluorescence counterstained by DAPI was in the nucleus. The red fluorescence labeled the p-Akt was localized in the cytoplasm. The green fluorescence labeled the RPS15A also appeared in cytoplasm. Yellow fluorescence appeared in the position at which red and green occurred simultaneously (arrow). Original magnification ×400
Knockdown of RPS15A suppresses tumorigenesis in vivo
In order to confirm the effect of RPS15A on proliferation in vivo, nude mouse xenograft model was applied. As shown in Fig. 7a, the average weight of tumors from shPLV-Ctrl injecting mice was 2.970 g, whereas the weight of tumors from shRPS15A injecting mice was 1.564 g (Fig. 7a, b). Furthermore, IHC results showed that the RPS15A was located at the cytoplasm. The expression of RPS15A in the shRPS15A-xenografted tumors was significantly reduced compared to the PLV-Ctr-xenografted tumors (Fig. 7c, d).
Tumor weight from subcutaneously injected PLV-Ctrl-U87 and sh-RPS15A-U87 measured at 25 days post-injection. a, b Tumorigenicity of sh-RPS15A-U87 cells was markedly reduced in vivo compared with PLV-Ctrl. Ten mice were used for each treatment (*P < 0.05). c, d IHC staining of RPS15A expression in subcutaneous tumors of mice injected with shRPS15A and PLV-Ctrl cells (arrow). Original magnification ×400, ×100
Discussion
GBM is a malignant disease in brain with a poor survival. The development of molecular biological technologies allowed researchers to develop new methods to treat glioblastoma. Some biomarkers were identified during the progression of glioma, including PTEN mutations, EGFR amplification, loss of heterozygosity (LOH) 10q, and TP53 mutations, which may in turn help to improve the targeted therapy of glioma [13, 14].
RPS15A encodes a conserved ribosomal protein which belongs to the 40S subunits of ribosomes catalyzing protein synthesis [15–17]. It is reported previously that in response to the induction of Myc transcription factor or transforming growth factor-β, RPS15A was up-regulated [8, 18]. In actively dividing tissues of oilseed like flower buds, apical meristem, and new leaves, RPS15A was highly expressed, but was expressed low in the expanded leaves and mature stem [19, 20]. Moreover, recent studies confirmed that RPS15A took roles in some kinds of human cancers. RPS15A expressed differently in poor differentiated ductal carcinoma in situ and in invasive grade 3 breast cancer, which suggested that RPS15A might play roles in the development of breast cancer [12]. RPS15A was highly expressed in HBxAg-positive cells, indicating that RPS15A might contribute to the progression of hepatocellular cancer [9]. Meta-analysis also showed an RPS15A over-expression in colorectal cancer, astrocytoma, as well as prostate cancer [11]. Despite the evidence that RPS15A could promote cell proliferation in many kinds of cells, its biological effect in human glioblastoma remains unknown.
In this study, we have shown that both RPS15A mRNA and protein levels were over-expressed in 20 glioblastoma tissues compared to those in 15 NB tissues (Fig. 1a, b). Then, we used IHC to measure the expression level of RPSS15A in 123 glioma and 15 NB tissues. The result inferred that RPS15A was highly expressed in 73.2 % of glioma patients and only in 33.3 % of normal tissues (Fig. 1c, Table 1). Furthermore, to investigate the possible relationship between clinicopathologic characteristics and RPS15A in glioma patients, we conducted survival analysis experiments. The results shown in Tables 2 and 3 suggest that RPS15A expression could be considered as a prognostic factor of patients with glioma. The survival analysis indicated that patients with a higher level of RPS15A suffered a worse survival than those with a lower level of RPS15A (Fig. 2).
Previous studies indicated that shRNA-targeted MAP4K4 or COMMD7 significantly inhibited U87 and U251 growth [21, 22], and lentivirus-mediated shRNA to knockdown RPS15A in U87 and U251 cells could also inhibit cell proliferation [10]. We, therefore, successfully established lentivirus-mediated shRNA knockdown of RPS15A in U87 and U251cells to investigate the biological functions of RPS15A in glioma cells (Fig. 3). When infected with shRPS15A, both of the two cells presented an inhibited cell proliferation in vitro (Fig. 4a, b) and decreased tumorigenesis in vivo (Fig. 7) compared to the PLVM-Ctrl groups. Besides, cell migration was also inhibited when U87 and U251 cells were infected with shRPS15A (Fig. 4 c, d).
Previous studies reported that RPS15A could promote interactions between mRNA and ribosome in translation and facilitate the translation via interacting with eukaryotic initiation factor 4F (elF-4F) coded by CDC33 in yeast. CDC33 mutation led to cell growth arrest during the G1-S transition, which could be reversed by abundant expression of RPS15A [23]. This suggests that RPS15A could conquer G1 arrest and further promote cell cycle process. Reduced RPS15A expression induced G0/G1 phase arrest and strongly suppressed cell growth in human hepatic carcinoma [10]. Results from FACS flow cytometry showed that increased number of U87 and U251 cells infected with shRPS15A was mainly arrested at G0/G1 phase compared with that of shPLV-Ctrl cells (Fig. 5a). According to previous research, p18INK4C and CDK4 were both important cell cycle regulation protein. They played significant role in the cell cycle process [24]. Moreover, the p18INK4C also was an inhibitor of CDK4 [25]. We found that the expression of CDK4 was decreased in shRPS15A U87 and U251 cells, whereas the expression of p18INK4C was increased (Fig. 5e, f).
Previous studies have showed that AKT pathways were involved in the regulation of cell cycle progression [26, 27], cell growth, proliferation, and migration [28–31]. We therefore tested its activity in our scenario. We found that knockdown of RPS15A significantly inhibited the phosphorylation of AKT, whereas it had no effect on phosphorylation of ERK (Fig. 5g, h). Therefore, the function of RPS15A on tumor progression was mediated by AKT pathway. Moreover, fluorescent double assay confirmed that RPS15A and p-Ark were highly expressed compared with NB tissues, and they were co-expressed in cytoplasm (Fig. 6). We confirmed that RPS15A may contribute to glioblastoma growth, proliferation, migration, and cell cycle via the AKT pathway.
Materials and methods
Reagents and antibodies
Antibodies of p-Akt (Cat 4060p), P-p44/42(Cat 4370S), p44/42(Cat 7695S), anti-rabbit antibody (Cat 7074P2), CDK4 (Cat12790P), and p18INK4C (Cat 2896P) antibodies were obtained from Cell Signaling Technology, Inc. (Beverly, MA, USA). Anti-RPS15A (ab175054, Cat GR169656-1, UK), anti-Akt (Cat 10176-2-AP, US), and anti-GAPDH (Cat 60004-1-lg, US) were obtained from Proteintech company. Dulbecco’s modified Eagles medium (DMEM, Hyclone, Cat no.SH30022.01B) together with fetal bovine serum (FBS) were purchased from GIBCO® (lot 1616964, Life Technologies, USA). All of the other reagents used were in analytic grade.
Patients and tissue samples
All of the 138 archived paraffin-embedded samples were obtained from operations performed between 2004 and 2009 in the Department of Neurosurgery of the First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning, China. There were 123 glioma tissues (including 9 oligodendroglioma, 14 astrocytic tumors, 60 anaplastic astrocytoma, and 40 glioblastoma) and 15 normal brain samples (obtained from normal brain tissue that was excised during the glioblastoma operation). The average and median age of these patients was 52.58 and 56.86 years (5–81 years). The glioma patients group contained 50 females and 73 males. This work was performed with ethical approval of the Human Ethics Committee of the First Affiliated Hospital of Dalian Medical University (approval number KY2014-50) in accordance with institutional medical requirements, and written informed consents were obtained from all of the enrolled subjects.
Cell culture
The human glioma U87, U251, and U118 cells were donated by YZ, Wang lab, Institute of Neuroscience, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences. All glioma cell lines were cultured in the Dulbecco’s modified Eagles medium (Cat SH30022.01B, Thermo, US) with 10 % FBS. Cells were cultured at 37 °C in 5 % CO2.
Animals
Animal care and experimental procedures were approved by the ethics committee according to the guidelines about animal care and experiments to laboratory animals. Nude female mice (BALB/c-nu/nu) of 4–6 weeks old were purchased and raised in the Center of Experimental Animals, Dalian Medical University (China).
Immunohistochemistry
Parts of the tumor tissues were cut, fixed in 10 % formaldehyde, and then embedded in paraffin. Paraffin section were soaked in pure xylene for dewaxing, and then re-hydrated in diluted ethanol series with water. Tissues were blocked with serum, followed by incubation in rabbit anti-human RPS15A antibody overnight at 4 °C. The tissues were washed twice, incubated in biotin-labeled goat anti-rabbit antibody for 10 min at room temperature, and followed with incubation of horseradish peroxidase (HRP). Peroxidase reaction was performed using 3, 3-diaminobenzidine (DAB, Cat 15111A03, Zhong Shan Jin Qiao, China). Samples were counterstained with hematoxylin and analyzed using a microscope (Original magnification ×400 and ×100).
Evaluation of staining
IHC-stained tissues were analyzed using two pathologists regardless of clinical parameters. RPS15A expression was observed in the cytoplasm. For the cytoplasm, the staining score was analyzed according to the cytoplasm staining intensity and the amount of positive cytoplasm staining of RPS15A expression. The results of percentage of positive staining in cytoplasm were regarded as follows: 0, <10 %, 1, 10–25 %, 2, 26–75 %, and 3, >76 %. Staining of 0–1 and 2–3 in cytoplasm were defined to be low and high expression of RPS15A, respectively.
Immunofluorescence double-labeling method
We used the immunofluorescence double-labeling method assay to investigate expression levels of p-Akt and RPS15A in glioblastoma samples and NB tissues following previous protocol [32, 33]. Antibody included anti-RPS15A (1:50), anti-p-Akt (1:50), goat anti rabbit (1:50) and goat anti-mouse (1:50).
Establishment of U87 cell lines with the stable expression of RPS15A short hairpin RNA
In order to prepare lentivirus that expressed human RPS15A short hairpin RNA (top: GATCCGCCATCGGCGCCATCCTGCAATCTAATTCAAGAGATTAGATTGCAGGATGGCGCC GATGGTTTTTTC; bottom: AATTGAAAAAACCATCGGCGCCATCCTGCAATCTAATCTCTTGAATTAGATTGCAGGATGGCGCCGATGGCG), pLVTHM-GFP lentiviral RNAi expression system was employed. Lentiviral particles that contained specific or negative control vectors were infected into U87 and U251 cells. FACS flow cytometry (BD FACSAria II, US) was used to select polyclonal cells with GFP signals. Total RNA was extracted, and levels of RPS15A mRNA were detected using quantitative real-time PCR analysis (Applied Biosystems, 7500 Real-Time PCR System, Thermo, US).
Cell proliferation assay
Cell proliferation was determined by MTT (5 mg/ml, Sigma, St. Louis, MO) assay as described previously [12]. Cells were plated at a certain density (1 × 103cells per well) in 96-well plates. Cells were then incubated in complete media for another 1–6 days. MTT was added and incubated for 4 h. At the end of the incubation, supernatants were removed, and DMSO (100 μl, Amresco, Cat: D8370, USA) was added, and absorbance of each well was measured at 490 nm in a Bio-Rad microplate reader (Bio-Rad, San Diego, CA, USA).
Wound-healing assay
Cell proliferation was analyzed with wound-healing assay in vitro. The steadily transfected with shRNA U87 and U251 cells were cultured in 6-well plates. Until U87 and U251 cells became confluent, we made a cell-free scratched gap with a pipette tip about 500 μm wide at the bottom of the plates and then incubated it for another 6 and 12 h. We compared the length of the healed wound with the length of the initial wound. The lengths from two different wounded regions were measured with Image J software (public domain software, http://rsb.info.nih.gov/ij/).
Boyden chamber migration assay
For the cell Boyden chamber assay, U87 and U251 were collected, diluted at the density of 1 × 105 cells/ml in FBS-free DMEM medium, and took out 100 μl seeded on the transwell apparatus (Costar, MA) with Boyden chamber(0.8 μm,Corning,USA). DMEM (600 μl) with 10 % FBS was added outside the transwell. Cells were incubated for another 4 h at 37 °C, washed twice with PBS, and then removed the cells on the top surface of the transwell. The lower surface cells were fixed with methanol, then stained with Giemsa solution, and counted using a microscope with Image J software.
Tumorigenesis in nude mice in vivo
U87 cells grown in vitro were collected and implanted 200 μl 1 × 107 cells sc under the shoulder of the 4–6-week-old female BALB/c nu/nu mice. The mice were divided randomly into two groups (ten mice per group) and were administered an intratumoral injection of sh-PLV-Ctl-U87 cells sh-RPS15A-U87 (1 × 107pfu). After 25 days of breeding, all mice were sacrificed, and the tumor tissues were excised and weighed. All animal studies were conducted under the principles and procedures given by the National Institute of Health Guide.
Flow cytometry analysis of the cell cycle
Cells were harvested at the logarithmic growth phase and seeded in the 6-well plates. After 48 h of growth, cells were digested with trypsin and harvested with ice-cold PBS. Then, suspended cells were centrifuged at 1000 rpm, for 5 min, at 4 °C. Cells were suspended with 300 μl ice-cold PBS and recompensed in 70 % ethanol for 30 min at 4 °C. Lastly, 1 ml propidium iodide (PI, lot 1685935, Life Technologies, USA) solution was added and analyzed on the FACScan flow cytometry (Becton Dickinson, CA, USA).
Quantitative real-time PCR
For U87 and U251 cells, they were harvested and washed twice with ice-cold PBS. Cell total RNA was scraped using RNA isoPlus® Reagent Kit (Cat RR820A, Takara Biotechnology) according to the protocols. For glioblastoma and NB tissues, their RNA were homogenized using RNA isoPlus® Reagent Kit. Certain quality of total RNA was taken to produce cDNA using PrimeScript® RT Reagent Kit (Cat: RR037A). cDNA was augmented using SYBR® Premix Ex Taq™ Kit with 7500 Real-Time PCR System. Data was analyzed by comparative ΔΔCt method. Target genes were normalized to GAPDH (forward 5′-GCACCGTCAAGGCTGAGAAC-3′, reverse: 5′-TGGTGAAGACGCCAGTGGA-3′).
Western blot analysis
Western blot analysis was conducted to measure the protein expression. Glioblastoma and NB tissues were homogenized and cracked in lysis buffer, shaken for 30 min at 4 °C. Protein were determined by BCA method (Cat PC0020, Solarbio, China) and then denatured. Same qualities of protein were loaded and separated by SDS-PAGE, transferred to PVDF (Cat IPVH00010, EMD Millipore Corporation Billerica Ma01821, USA) membrane, blocked with blocking solution, and immunoblotted with first antibodies overnight at 4 °C. On the second day, membranes were washed using TBST and incubated in second antibodies for 2 h at room temperature. Membranes were detected by ECL reagent with Bio-Spectrum Gel Imaging System (UVP, USA).
Statistical analysis
All data obtained were given as mean ± SD of at least three experiments. GraphPad Prism 5.0 and SPSS 17.0 software (SPSS, Chicago, USA) were used for statistical analysis. Two-tailed Student’s t test or one-way ANOVA test were taken for comparisons between groups. Chi-square test or Fischer’s test were used to analyze the difference between classified variables. Survival analysis was conducted by GraphPad Prism 5.0 software. The relationship between the variables and patient’s survival time was performed using univariate and multivariate cox proportional hazards method. Differences were considered to be statistically significant only when values of P were <0.05.
Conclusion
In conclusion, the expression of RPS15A correlated with disease progression in patients with GBM. Attenuated RPS15A expression led to suppressed cell growth as well as cell migration via AKT pathway in U87 and U251 glioma cells.
References
Chen T et al. Dasatinib reverses the multidrug resistance of breast cancer MCF-7 cells to doxorubicin by downregulating P-gp expression via inhibiting the activation of ERK signaling pathway. Cancer Biol Ther. 2015;16:106–14.
Chen X, Meng Q, Wang C, Liu Q, Sun H, Huo X, et al. Protective effects of calycosin against CCl4-induced liver injury with activation of FXR and STAT3 in mice. Pharm Res. 2015;32:538–48.
Lanzetta G, Campanella C, Rozzi A, Nappa M, Costa A, Fedele F. Temozolomide in radio-chemotherapy combined treatment for newly-diagnosed glioblastoma multiforme: phase II clinical trial. Anticancer Res. 2003;23:5159–64.
Sano T, Lin H, Chen X, Langford LA, Koul D, Bondy ML, et al. Differential expression of MMAC/PTEN in glioblastoma multiforme: relationship to localization and prognosis. Cancer Res. 1999;59:1820–4.
Arshad H, Ahmad Z, Hasan SH. Gliomas: correlation of histologic grade, Ki67 and p53 expression with patient survival. Asian Pacific J Cancer Prev: APJCP. 2010;11:1637–40.
Dellaretti M, Reyns N, Touzet G, Dubois F, Gusmao S, Pereira JL, et al. Diffuse brainstem glioma: prognostic factors. J Neurosurg. 2012;117:810–4.
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73.
Akiyama N, Matsuo Y, Sai H, Noda M, Kizaka-Kondoh S. Identification of a series of transforming growth factor beta-responsive genes by retrovirus-mediated gene trap screening. Mol Cell Biol. 2000;20:3266–73.
Lian Z, Liu J, Li L, Li X, Tufan NL, Wu MC, et al. Human S15a expression is upregulated by hepatitis B virus X protein. Mol Carcinog. 2004;40:34–46.
Xu M, Wang Y, Chen L, Pan B, Chen F, Fang Y, et al. Down-regulation of ribosomal protein S15A mRNA with a short hairpin RNA inhibits human hepatic cancer cell growth in vitro. Gene. 2014;536:84–9.
Xinmin Zhao, Lei Shen, Yu Feng, Hui Yu, Xianghua Xu, Jianhua Chang, Xuxia Shen, Jie Qiao, Jialei Wang. Decreased expression of RPS15A suppresses proliferation of lung cancer cells, Tumor Biol (2015); doi: 10.1007/s13277-015-3371-9
Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res: Bcr. 2006;8:R61.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110.
Yan W, Zhang W, Jiang T. Oncogene addiction in gliomas: implications for molecular targeted therapy. J Exp Clin Cancer Res: Cr. 2011;30:58.
Chan YL, Olvera J, Paz V, Wool IG. The primary structure of rat ribosomal protein S15a. Biochem Biophys Res Commun. 1994;200:1498–504.
Kenmochi N, Kawaguchi T, Rozen S, Davis E, Goodman N, Hudson TJ, et al. A map of 75 human ribosomal protein genes. Genome Res. 1998;8:509–23.
Vladimirov SN, Ivanov AV, Karpova GG, Musolyamov AK, Egorov TA, Thiede B, et al. Characterization of the human small-ribosomal-subunit proteins by N-terminal and internal sequencing, and mass spectrometry. Europ J Biochem/Febs. 1996;239:144–9.
Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69.
Bonham-Smith PC, Moloney MM. Nucleotide and protein sequences of a cytoplasmic ribosomal protein S15a gene from Arabidopsis thaliana. Plant Physiol. 1994;106:401–2.
Bonham-Smith PC, Oancia TL, Moloney MM. Cytoplasmic ribosomal protein S15a from Brassica napus: molecular cloning and developmental expression in mitotically active tissues. Plant Mol Biol. 1992;18:909–19.
Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, et al. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res: An Official J Am Assoc Cancer Res. 2011;17:710–20.
Zheng L, Liang P, Li J, Huang XB, Liu SC, Zhao HZ, et al. ShRNA-targeted COMMD7 suppresses hepatocellular carcinoma growth. PLoS One. 2012;7:e45412.
Lavoie C, Tam R, Clark M, Lee H, Sonenberg N, Lasko P. Suppression of a temperature-sensitive cdc33 mutation of yeast by a multicopy plasmid expressing a Drosophila ribosomal protein. J Biol Chem. 1994;269:14625–30.
Schreiber M, Muller WJ, Singh G, Graham FL. Comparison of the effectiveness of adenovirus vectors expressing cyclin kinase inhibitors p16INK4A, p18INK4C, p19INK4D, p21(WAF1/CIP1) and p27KIP1 in inducing cell cycle arrest, apoptosis and inhibition of tumorigenicity. Oncogene. 1999;18:1663–76.
Swarbrick A, Lee CS, Sutherland RL, Musgrove EA. Cooperation of p27(Kip1) and p18(INK4c) in progestin-mediated cell cycle arrest in T-47D breast cancer cells. Mol Cell Biol. 2000;20:2581–91.
Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, Blalock WL, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603.
Chang F, Steelman LS, Shelton JG, Lee JT, Navolanic PM, Blalock WL, et al. Regulation of cell cycle progression and apoptosis by the Ras/Raf/MEK/ERK pathway (Review). Int J Oncol. 2003;22:469–80.
Zhang, X., Shi, H., Tang, H., Fang, Z., Wang, J., and Cui, S. miR-218 inhibits the invasion and migration of colon cancer cells by targeting the PI3K/Akt/mTOR signaling pathway. Int J Mol Med (2015).
Yuan Y, Du W, Wang Y, Xu C, Wang J, Zhang Y, et al. Suppression of AKT expression by miR-153 produced anti-tumor activity in lung cancer. International Journal of Cancer. J Int Du Cancer. 2015;136:1333–40.
Yi, X. P., Han, T., Li, Y. X., Long, X. Y., and Li, W. Z. Simultaneous silencing of XIAP and survivin causes partial mesenchymal-epithelial transition of human pancreatic cancer cells via the PTEN/PI3K/Akt pathway. Mol Med Rep (2015).
Geng J, Li X, Zhou Z, Wu CL, Dai M, Bai X. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015;359:275–87.
Singh M, Savitz SI, Hoque R, Gupta G, Roth S, Rosenbaum PS, et al. Cell-specific caspase expression by different neuronal phenotypes in transient retinal ischemia. J Neurochem. 2001;77:466–75.
Wang ST, Liu JJ, Wang CZ, Lin B, Hao YY, Wang YF, et al. Expression and correlation of Lewis y antigen and TGF-beta1 in ovarian epithelial carcinoma. Oncol Rep. 2012;27:1065–71.
Acknowledgments
The study was sponsored by the Natural Science Foundation of China (NSFC) (No. 81172180). Hospital IRB approval has been signed for the use of human samples. The authors also thank YZ Wang lab for donating the cell lines.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Hospital IRB approval has been signed for the use of human samples. Informed consent was obtained from all individual participants included in the study. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Conflicts of interest
None
Rights and permissions
About this article
Cite this article
Yao, Y., Liu, Y., Lv, X. et al. Down-regulation of ribosomal protein S15A inhibits proliferation of human glioblastoma cells in vivo and in vitro via AKT pathway. Tumor Biol. 37, 4979–4990 (2016). https://doi.org/10.1007/s13277-015-4323-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4323-0