Abstract
Many molecular epidemiological studies have been performed to explore the association between MTHFR C677T polymorphism and cancer risk in diverse populations. However, the results were inconsistent. Hence, we performed a meta-analysis to investigate the association between cancer risk and MTHFR C677T (150,086 cases and 200,699 controls from 446 studies) polymorphism. Overall, significantly increased cancer risk was found when all eligible studies were pooled into the meta-analysis. In the further stratified and sensitivity analyses, significantly increased breast cancer risk was found in Asians and Indians, significantly decreased colon cancer risk was found, significantly decreased colorectal cancer risk was found in male population, significantly increased gastric cancer risk was found in Caucasians and Asians, significantly increased hepatocellular cancer risk was found in Asians, significantly decreased adult acute lymphoblastic leukemia (AALL) risk was found in Caucasians, significantly decreased childhood acute lymphoblastic leukemia (CALL) risk was found in Asians, and significantly increased multiple myeloma and NHL risk was found in Caucasians. In summary, this meta-analysis suggests that MTHFR C677T polymorphism is associated with increased breast cancer, gastric cancer, and hepatocellular cancer risk in Asians, is associated with increased gastric cancer, multiple myeloma, and NHL risk in Caucasians, is associated with decreased AALL risk in Caucasians, is associated with decreased CALL risk in Asians, is associated with increased breast cancer risk in Asians, is associated with decreased colon cancer risk, and is associated with decreased colorectal cancer risk in male population. Moreover, this meta-analysis also points out the importance of new studies, such as Asians of HNC, Asians of lung cancer, and Indians of breast cancer, because they had high heterogeneity in this meta-analysis (I 2 > 75 %).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Folate deficiency has been associated with cardiovascular diseases and anemia, and more recently with a rapidly growing number of cancers [1, 2]. The methylenetetrahydrofolate reductase (MTHFR) gene codes for a key enzyme in the folate cycle, which affects DNA methylation and synthesis [3–6]. The MTHFR gene is located on short arm of chromosome 1 (1p36.3), and the total length of the gene cDNA is 2.2 kb [7]. MTHFR converts, irreversibly, 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, and the 5-methyltetrahydrofolate is the predominant circulatory form of folate that donates a methyl group for the remethylation of homocysteine (Hcy) to methionine [8]. A change of C to T at nucleotide 677 in MTHFR C677T (Ala 222 Val, rs 1801133) results in an amino acid substance change of an alanine to valine, and this substance is associated with reduced enzyme activity that leads to reduced plasma folate levels [9, 10]. MTHFR 677TT variant genotype have approximately 30 % enzyme activity in vitro compared with the 677CC wild-type [11], which has been shown to be associated with DNA hypomethylation, genomic instability, and derepression of proto-oncogenes [9].
To date, MTHFR C677T polymorphism has been reported to be associated with risk of several types of cancer, such as breast cancer, lung cancer, colorectal cancer, and so on (Supplementary reference [1–428]). However, the results were inconsistent or even contradictory. Some recent meta-analyses analyzed such an association only for single cancer such as head and neck, lung, breast, gastric, esophageal, colorectal cancer and so on [12–20]. In addition, one recent meta-analysis has studied the association between MTHFR C677T polymorphism and risk of cancer. However, many published studies were not included in the meta-analysis [21]. Hence, we performed a meta-analysis to obtain a more precise estimation of the association between MTHFR C677T polymorphism and risk of cancer.
Materials and methods
Identification and eligibility of relevant studies
We performed a comprehensive literature search using the databases PubMed, ISI, and EMBASE, and articles published before the end of December 2014 were identified with a combination of the following key words: “MTHFR,” or “methylenetetrahydrofolatereductase,” “polymorphism,” “mutation” or “variant,” and “cancer” or “carcinoma”. We used no lower date limit. All eligible studies were retrieved, and their bibliographies were checked for other relevant publications. The reference lists of reviews and retrieved articles were hand searched simultaneously.
Inclusion criteria
All studies selected have to fulfill the following three criteria: (1) case–control study of the MTHFR A1298C polymorphism and cancer risk; (2) the genotype distribution of the polymorphisms in cases and controls were described in details and the results were expressed as odds ratio (OR) and corresponding 95 % confidence interval (95 % CI); (3) When multiple publications reported on the same or overlapping data, only the largest or most recent publication was included, as recommended by Little er al. [22].
Data extraction
The following data were extracted from each included publication: the first author’s name, year of publication, country of origin, ethnicity, source of controls, sample size, cancer type, and numbers of different genotype in all subjects. Different ethnicity descents were categorized as “Caucasian,” “African,” (including African Americans), “Asian,” “Indian” (we considered the samples of studies from India and Pakistan as of “Indian”), “Middle Eastern” (we considered the samples of studies from Middle Eastern countries), and “mixed population” (When one study did not state which ethnic groups was included or if it was impossible to separate participants according to phenotype, the sample was termed as “mixed population’). When studies included subjects of more than one ethnicity and were able to separate, data were collected separately for each ethnic group.
Statistical analysis
Odds ratios (ORs) with 95 % CIs were used to assess the strength of association between the MTHFR C677T polymorphism and cancer risk. The pooled ORs were performed for dominant model (CT + TT vs. CC), recessive model (CC + CT vs. TT), homozygous model (TT vs. CC), and heterozygous model (CT vs. CC), respectively. Between-study heterogeneity was assessed by calculating Q-statistic (heterogeneity was considered statistically significant if P < 0.10) [23] and quantified using the I 2 value, a value that describes the percentage of variation across studies that are due to heterogeneity rather than chance, where I 2 = 0 % indicates no observed heterogeneity, with 25 % regarded as low, 50 % as moderate, and 75 % as high [24]. If results were not heterogeneous, the pooled ORs were calculated by the fixed-effect model (we used the Q-statistic, which represents the magnitude of heterogeneity between-studies) [25]. Otherwise, a random-effect model was used (when the heterogeneity between-studies were significant) [26]. In addition to the comparison among all subjects, we also performed stratification analyses by cancer type (if one cancer type contained less than three individual studies, it was combined into the “other cancers” group), source of control, and ethnicity. Moreover, the extent to which the combined risk estimate might be affected by individual studies was assessed by consecutively omitting every study from the meta-analysis (leave-one-out sensitivity analysis). This approach would also capture the effect of the oldest or first positive study (first study effect). In addition, we also ranked studies according to sample size, and then repeated this meta-analysis. Sample size was classified according to a minimum of 200 participants and those with fewer than 200 participants. The cite criteria were previously described [27]. Last, sensitivity analysis was also performed, excluding studies whose allele frequencies in controls exhibited significant deviation from the Hardy–Weinberg equilibrium (HWE), given that the deviation may denote bias. Deviation of HWE may reflect methodological problems such as genotyping errors, population stratification, or selection bias. HWE was calculated by using the goodness-of-fit test, and deviation was considered when P < 0.05. Begg’s funnel plots [28] and Egger’s linear regression test [29] were used to assess publication bias. A meta-regression analysis was carried out to identify the major sources of between-studies variation in the results, using the log of the ORs from each study as dependent variables, and cancer type, ethnicity, sample size, and source of controls as the possible sources of heterogeneity. All of the calculations were performed using STATA version 10.0 (STATA Corporation, College Station, TX).
Results
Eligible studies and meta-analysis databases
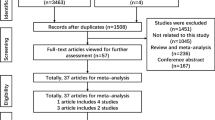
Figure 1 graphically illustrates the trial flow chart. A total of 1,948 publications regarding MTHFR C677T polymorphism with respect to cancer were identified. After screening the titles and abstracts, 1521 publications were excluded because they were duplicate articles, review articles, case reports, and other polymorphisms of MTHFR. In addition, of these published articles, 32 publications (Supplemental reference [1, 20, 53, 54, 61, 104, 125, 150, 157, 158, 164, 194, 202, 244, 251, 252, 272, 296, 298, 301, 302, 309, 321, 326, 331, 344, 350, 369, 376, 407, 419, 421]) were excluded because of their populations overlapped with another 28 included studies (Supplemental reference [4, 25, 28, 68, 83, 85, 105, 107, 134, 154, 165, 169, 171, 179, 184, 217, 235, 258, 309, 318, 352, 360, 363, 366, 391, 416, 423, 426]). As summarized in Supplemental Table 1, 395 publications with 446 case–control studies were selected in the meta-analysis, including 150,086 cases and 200,699 controls. Among these studies, there were 17 bladder cancer studies, 9 brain cancer studies, 68 breast cancer studies, 14 cervical cancer studies, 83 colorectal cancer studies, 3 endometrial cancer studies, 18 esophageal cancer studies, 27 gastric cancer studies, 12 hepatocellular cancer studies, 23 head and neck cancer studies, 62 leukemia studies, 20 lung cancer studies, 27 lymphoma studies, 8 multiple myeloma studies, 10 ovarian cancer studies, 7 pancreatic cancer studies, 21 prostate cancer studies, 3 renal cancer studies, 5 skin cancer studies, and 9 studies with the “other cancers.” All of the cases were pathologically confirmed.
Quantitative synthesis
The evaluations of the association of MTHFR C677T polymorphism with cancer risk are shown in Table 1. Overall, significantly increased cancer risk was observed in all genetic models (dominant model: odds ratio (OR) = 1.05, 95 % CI = 1.02–1.08, P value of heterogeneity test [P h] < 0.001, I 2 = 58.3 %; recessive model: OR = 1.06, 95 % CI = 1.02–1.10, P h < 0.001, I 2 = 50.7 %; homozygous model: OR = 1.08, 95 % CI = 1.04–1.13, P h < 0.001, I 2 = 57.3 %; heterozygous model: OR = 1.04, 95 % CI = 1.01–1.06, P h < 0.001, I 2 = 50.4 %) when all the eligible studies were pooled into meta-analysis. In the stratified analysis by cancer type, significant association was observed between MTHFR C677T polymorphism and breast cancer (recessive model: OR = 1.09, 95 % CI = 1.01–1.18, P h < 0.001, I 2 = 47.6 %; homozygous model: OR = 1.10, 95 % CI = 1.01–1.20, P h < 0.001, I 2 = 43.8 %), colorectal cancer (recessive model: OR = 0.92, 95 % CI = 0.85–0.99, P h < 0.001, I 2 = 50.5 %), esophageal cancer (heterozygous model: OR = 1.29, 95 % CI = 1.06–1.57, P h < 0.001, I 2 = 68.7 %), gastric cancer (recessive model: OR = 1.21, 95 % CI = 1.07–1.36, P h = 0.009, I 2 = 43.4 %; homozygous model: OR = 1.29, 95 % CI = 1.10–1.52, P h < 0.001, I 2 = 59.1 %), leukemia (dominant model: OR = 0.91, 95 % CI = 0.85–0.98, P h < 0.001, I 2 = 42.8 %; homozygous model: OR = 0.88, 95 % CI = 0.78–0.98, P h = 0.011, I 2 = 31.9 %; heterozygous model: OR = 0.92, 95 % CI = 0.86–0.99, P h = 0.002, I 2 = 37.9 %), lung cancer (recessive model: OR = 1.13, 95 % CI = 1.01–1.26, P h = 0.003, I 2 = 53.1 %; homozygous model: OR = 1.20, 95 % CI = 1.04–1.39, P h < 0.001, I 2 = 66.1 %), multiple myeloma (dominant model: OR = 1.26, 95 % CI = 1.06–1.50, P h = 0.207, I 2 = 27.7 %; heterozygous model: OR = 1.23, 95 % CI = 1.03–1.48, P h = 0.214, I 2 = 26.9 %; homozygous model: OR = 1.32, 95 % CI = 1.03–1.68, P h = 0.230, I 2 = 24.9 %), and renal cell cancer risk (dominant model: OR = 1.38, 95 % CI = 1.18–1.62, P h = 0.244, I 2 = 29.2 %; homozygous model: OR = 1.36, 95 % CI = 1.04–1.78, P h = 0.297, I 2 = 17.7 %; heterozygous model: OR = 1.40, 95 % CI = 1.19–1.65, P h = 0.368, I 2 = 0.0 %). In the stratified analysis by ethnicity, significantly increased cancer risk was observed in Asians (dominant model: OR = 1.08, 95 % CI = 1.03–1.14, P h < 0.001, I 2 = 70.1 %; recessive model: OR = 1.11, 95 % CI = 1.04–1.18, P h < 0.001, I 2 = 68.0 %; homozygous model: OR = 1.16, 95 % CI = 1.07–1.26, P h < 0.001, I 2 = 74.4 %; heterozygous model: OR = 1.06, 95 % CI = 1.01–1.11, P h < 0.001, I 2 = 58.8 %) and Indians (recessive model: OR = 1.44, 95 % CI = 1.12–1.86, P h = 0.455, I 2 = 0.5 %; homozygous model: OR = 1.42, 95 % CI = 1.09–1.83, P h = 0.427, I 2 = 2.5 %). We also found that there was a statistically significant link between the MTHFR C677T polymorphism and cancer risk in the hospital-based studies (dominant model: OR = 1.06, 95 % CI = 1.02–1.10, P h < 0.001, I 2 = 61.6 %; recessive model: OR = 1.10, 95 % CI = 1.04–1.15, P h < 0.001, I 2 = 47.2 %; homozygous model: OR = 1.12, 95 % CI = 1.06–1.19, P h < 0.001, I 2 = 55.1 %; heterozygous model: OR = 1.04, 95 % CI = 1.01–1.08, P h < 0.001, I 2 = 54.7 %), but not in population-based studies, as also shown in Table 1.
Ethnicity and cancer risk attributed to the MTHFR C677T polymorphism
We further examined the association of the MTHFR C677T polymorphism and cancer risk by cancer type and ethnicity (Table 2) because there was significant heterogeneity between studies. For samples of Caucasians, significant association was observed between MTHFR C677T polymorphism and gastric cancer (recessive model: OR = 1.23, 95 % CI = 1.03–1.48, P h = 0.550, I 2 = 0.0 %; homozygous model: OR = 1.34, 95 % CI = 1.03–1.75, P h = 0.094, I 2 = 39.5 %), leukemia (dominant model: OR = 0.87, 95 % CI = 0.79–0.95, P h = 0.021, I 2 = 36.7 %; recessive model: OR = 0.87, 95 % CI = 0.78–0.96, P h = 0.229, I 2 = 15.0 %; homozygous model: OR = 0.82, 95 % CI = 0.74–0.92, P h = 0.100, I 2 = 25.1 %; heterozygous model: OR = 0.92, 95 % CI = 0.86–0.99, P h = 0.106, I 2 = 24.5 %), lymphoma (homozygous model: OR = 1.16, 95 % CI = 1.01–1.34, P h = 0.696, I 2 = 0.0 %), multiple myeloma (dominant model: OR = 1.54, 95 % CI = 1.14–2.08, P h = 0.278, I 2 = 22.1 %; homozygous model: OR = 1.64, 95 % CI = 1.02–2.62, P h = 0.702, I 2 = 0.0 %; heterozygous model: OR = 1.50, 95 % CI = 1.10–2.06, P h = 0.237, I 2 = 29.1 %), and skin cancer risk (dominant model: OR = 1.56, 95 % CI = 1.05–2.34, P h = 0.038, I 2 = 69.5 %; recessive model: OR = 1.64, 95 % CI = 1.11–2.41, P h = 0.739, I 2 = 0.0 %; homozygous model: OR = 1.84, 95 % CI = 1.24–2.74, P h = 0.876, I 2 = 0.0 %). For samples of Asians, significant association was observed between MTHFR C677T polymorphism and esophageal cancer (heterozygous model: OR = 1.39, 95 % CI = 1.11–1.74, P h < 0.001, I 2 = 71.7 %), cervical cancer (homozygous model: OR = 1.34, 95 % CI = 1.01–1.77, P h = 0.389, I 2 = 0.0 %), gastric cancer (recessive model: OR = 1.23, 95 % CI = 1.03–1.47, P h = 0.002, I 2 = 60.6 %; homozygous model: OR = 1.30, 95 % CI = 1.02–1.65, P h < 0.001, I 2 = 71.2 %), and lymphoma risk (dominant model: OR = 0.73, 95 % CI = 0.62–0.85, P h = 0.238, I 2 = 30.4 %; recessive model: OR = 0.81, 95 % CI = 0.66–1.00, P h = 0.187, I 2 = 40.4 %; homozygous model: OR = 0.69, 95 % CI = 0.54–0.86, P h = 0.320, I 2 = 12.2 %; heterozygous model: OR = 0.74, 95 % CI = 0.63–0.88, P h = 0.108, I 2 = 55.1 %). For samples of Africans, significant association was only observed between MTHFR C677T polymorphism and bladder cancer risk (recessive model: OR = 0.59, 95 % CI = 0.37–0.95, P h = 0.940, I 2 = 0.0 %). For samples of Indians, significant association was observed between MTHFR C677T polymorphism leukemia risk (dominant model: OR = 1.31, 95 % CI = 1.05–1.63, P h = 0.115, I 2 = 46.2 %).
Source of controls and cancer risk attributed to the MTHFR C677T polymorphism
We also examined the association between the MTHFR C677T polymorphism and cancer risk by cancer type and source of controls (Table 3). For the population-based studies, significant association was found between MTHFR C677T polymorphism and cervical cancer (dominant model: OR = 0.67, 95 % CI = 0.53–0.84; homozygous model: OR = 0.66, 95 % CI = 0.43–0.99; heterozygous model: OR = 0.67, 95 % CI = 0.53–0.85) and colorectal cancer risk (recessive model: OR = 0.88, 95 % CI = 0.81–0.95, P h = 0.001, I 2 = 47.2 %; homozygous model: OR = 0.88, 95 % CI = 0.82–0.96, P h = 0.013, I 2 = 38.2 %). For the hospital-based studies, significant association was observed between MTHFR C677T polymorphism and brain tumor (recessive model: OR = 1.48, 95 % CI = 1.19–1.84, P h = 0.122, I 2 = 38.6 %), breast cancer (recessive model: OR = 1.17, 95 % CI = 1.04–1.31, P h = 0.002, I 2 = 41.6 %; homozygous model: OR = 1.17, 95 % CI = 1.04–1.32, P h = 0.004, I 2 = 39.1 %), gastric cancer (recessive model: OR = 1.27, 95 % CI = 1.13–1.44, P h = 0.125, I 2 = 29.2 %; homozygous model: OR = 1.38, 95 % CI = 1.12–1.72, P h = 0.016, I 2 = 47.4 %), leukemia (dominant model: OR = 0.89, 95 % CI = 0.81–0.96, P h < 0.001, I 2 = 45.0 %; recessive model: OR = 0.88, 95 % CI = 0.79–0.98, P h = 0.084, I 2 = 21.6 %; homozygous model: OR = 0.84, 95 % CI = 0.74–0.95, P h = 0.016, I 2 = 31.5 %; heterozygous model: OR = 0.90, 95 % CI = 0.83–0.98, P h = 0.001, I 2 = 40.2 %), lung cancer (homozygous model: OR = 1.24, 95 % CI = 1.02–1.52, P h < 0.001, I 2 = 70.7 %), multiple myeloma (dominant model: OR = 1.41, 95 % CI = 1.15–1.72, P h = 0.492, I 2 = 0.0 %; homozygous model: OR = 1.42, 95 % CI = 1.06–1.91, P h = 0.209, I 2 = 28.8 %; heterozygous model: OR = 1.39, 95 % CI = 1.13–1.72, P h = 0.601, I 2 = 0.0 %), prostate cancer (recessive model: OR = 0.65, 95 % CI = 0.51–0.82, P h = 0.289, I 2 = 15.5 %; homozygous model: OR = 0.63, 95 % CI = 0.48–0.82, P h = 0.277, I 2 = 26.6 %), renal cell cancer (dominant model: OR = 1.38, 95 % CI = 1.18–1.62, P h = 0.244, I 2 = 29.2 %; homozygous model: OR = 1.36, 95 % CI = 1.04–1.78, P h = 0.297, I 2 = 17.7 %; heterozygous model: OR = 1.40, 95 % CI = 1.19–1.65, P h = 0.368, I 2 = 0.0 %), and skin cancer risk (homozygous model: OR = 1.45, 95 % CI = 1.04–2.02, P h = 0.202, I 2 = 35.1 %).
Anatomical site, histological type, and association of the MTHFR C677T polymorphism with cancer risk
We next completed a subgroup analysis by tumor site and histological type or anatomical location (Table 4). Regarding colorectal cancer, the MTHFR C677T polymorphism was associated with cancer risk for colon cancer (recessive model: OR = 0.84, 95 % CI = 0.74–0.96, P h = 0.057, I 2 = 38.0 %) and rectal cancer (recessive model: OR = 0.87, 95 % CI = 0.77–0.98, P h = 0.373, I 2 = 7.3 %). For leukemia, the MTHFR C677T polymorphism was associated with cancer risk for children acute lymphoblastic leukemia (CALL; homozygous model: OR = 0.87, 95 % CI = 0.77–0.98, P h = 0.139, I 2 = 21.7 %) and children acute myeloid leukemia (CAML; heterozygous model: OR = 0.71, 95 % CI = 0.51–0.98, P h = 0.068, I 2 = 46.9 %). In the stratified analysis by ethnicity, for CALL, significant association was observed in Asians (dominant model: OR = 0.86, 95 % CI = 0.74–0.99, P h = 0.195, I 2 = 30.6 %) and Indians (heterozygous model: OR = 1.45, 95 % CI = 1.10–1.91, P h = 0.115, I 2 = 49.4 %). For esophageal cancer, there was significant association between the MTHFR C677T polymorphism and esophageal squamous cell carcinoma (ESCC) risk (recessive model: OR = 1.43, 95 % CI = 1.13–1.81, P h = 0.002, I 2 = 64.9 %; homozygous model: OR = 1.83, 95 % CI = 1.28–2.63, P h < 0.001, I 2 = 75.0 %; heterozygous model: OR = 1.37, 95 % CI = 1.04–1.81, P h < 0.001, I 2 = 72.2 %). In the stratified analysis by ethnicity, significant association was observed between the MTHFR C677T polymorphism and ESCC risk in Asians (dominant model: OR = 1.68, 95 % CI = 1.24–2.28, P h < 0.001, I 2 = 73.9 %; recessive model: OR = 1.50, 95 % CI = 1.16–1.94, P h = 0.002, I 2 = 69.4 %; heterozygous model: OR = 1.53, 95 % CI = 1.13–2.06, P h = 0.002, I 2 = 69.4 %). For gastric cancer, significant association was observed in cardia gastric cancer (recessive model: OR = 1.44, 95 % CI = 1.21–1.72, P h = 0.105, I 2 = 39.4 %) and intestinal gastric cancer (recessive model: OR = 1.43, 95 % CI = 1.10–1.86, P h = 0.964, I 2 = 0.0 %). In the stratified analysis by ethnicity, significant association was observed between the MTHFR C677T polymorphism and cardia gastric cancer risk in Asians (recessive model: OR = 1.53, 95 % CI = 1.26–1.87, P h = 0.255, I 2 = 23.8 %; homozygous model: OR = 1.45, 95 % CI = 1.15–1.81, P h = 0.114, I 2 = 41.5 %). For lymphoma, the results suggested a differential contribution of the MTHFR T allele, with a significant association in Hodgkin lymphoma (HL) (dominant model: OR = 0.75, 95 % CI = 0.60–0.94, P h = 0.518, I 2 = 0.0 %; heterozygous model: OR = 0.74, 95 % CI = 0.58–0.95, P h = 0.474, I 2 = 0.0 %). In the stratified analysis by ethnicity, significant association was observed between the MTHFR C677T polymorphism and non-Hodgkin lymphoma (NHL) risk in Asians (dominant model: OR = 0.73, 95 % CI = 0.62–0.85, P h = 0.241, I 2 = 29.8 %; recessive model: OR = 0.71, 95 % CI = 0.55–0.93, P h = 0.363, I 2 = 0.0 %; homozygous model: OR = 0.64, 95 % CI = 0.48–0.85, P h = 0.216, I 2 = 34.8 %) and Caucasians (dominant model: OR = 1.10, 95 % CI = 1.02–1.20, P h = 0.585, I 2 = 0.0 %; recessive model: OR = 1.16, 95 % CI = 1.02–1.32, P h = 0.954, I 2 = 0.0 %; homozygous model: OR = 1.21, 95 % CI = 1.06–1.38, P h = 0.885, I 2 = 0.0 %).
Interaction of MTHFR C677T and other risk factors for cancer susceptibility
Finally, we tested if the MTHFR C677T polymorphism could interact with other known risk factors (Table 5). For lung cancer, significant association was observed in smokers (dominant model: OR = 0.84, 95 % CI = 0.75–0.93, P h = 0.372, I 2 = 6.0 %, homozygous model: OR = 0.86, 95 % CI = 0.74–0.99, P h = 0.346, I 2 = 10.5 %; heterozygous model: OR = 0.83, 95 % CI = 0.70–0.98, P h = 0.295, I 2 = 18.8 %). For colorectal cancer, significant association was observed in male (homozygous model: OR = 0.80, 95 % CI = 0.70–0.92, P h = 0.183, I 2 = 29.5 %) and female (homozygous model: OR = 0.84, 95 % CI = 0.72–0.99, P h = 0.376, I 2 = 7.2 %).
Heterogeneity analysis
In this meta-analysis, high between-study heterogeneity was observed in endometrial cancer, esophageal cancer, pancreatic cancer, Caucasians of cervical cancer, Caucasians of hepatocellular cancer, Caucasians of HNC, Caucasians of prostate cancer, Caucasians of skin cancer, Asians of bladder cancer, Asians of HNC, Asians of lung cancer, Asians of multiple myeloma, Africans of lymphoma, Asians of esophageal cancer, Indians of breast cancer, Indians of cervical cancer, population-based of esophageal cancer, hospital-based of pancreatic cancer, population-based of prostate cancer, hospital-based of skin cancer, proximal colorectal cancer, meningioma, adult acute lymphoblastic leukemia (AALL), Asians of AALL, Caucasians of AALL, Caucasians of chronic myeloid leukemia, esophageal squamous cell caicinoma, Asians of ESCC, and Africans of non-hodgkin lymphoma. However, when we excluded the studies of HWE deviation, high between-study heterogeneity was eliminated in Caucasians of hepatocellular cancer, Caucasians of HNC, Caucasians of prostate cancer, Caucasians of skin cancer, Africans of lymphoma, and hospital-based of skin cancer, as shown in Table 6. When we excluded the small sample studies, high between-study heterogeneity was eliminated in endometrial cancer and Asians of multiple myeloma, as shown in Table 7. In addition, when the study of Qu et al. (Supplemental reference [7]) was excluded, high between-study heterogeneity was eliminated in esophageal cancer. When the study of Wang et al. (Supplemental reference [318]) was excluded, high between-study heterogeneity was eliminated in pancreatic cancer and hospital-based studies of pancreatic cancer. When the study of Zoodsma et al. (Supplemental reference [353]) was excluded, high between-study heterogeneity was eliminated in Caucasians of cervical cancer. When the study of Chung et al. (Supplemental reference [122]) was excluded, high between-study heterogeneity was eliminated between MTHFR C677T polymorphism and bladder cancer risk in Asians. When the study of Shekari et al. (Supplemental reference [209]) was excluded, high between-study heterogeneity was eliminated between MTHFR C677T polymorphism and cervical cancer risk in Indians. When the study of Song et al. (Supplemental reference [224]) was excluded, high between-study heterogeneity was eliminated in the population-based of esophageal cancer. When the study of Naghibalhossaini et al. Supplemental reference [134]) was excluded, high between-studies heterogeneity was deleted in proximal colorectal cancer. When the study of Li et al. (Supplemental reference [18]) was excluded, high between-studies heterogeneity was deleted in meningioma. When the study of Kim et al. (Supplemental reference [147]) was excluded, high between-studies heterogeneity was deleted in Asians of AALL. When the study of Deligezer et al. (Supplemental reference [383]) was excluded, high between-studies heterogeneity was deleted in Caucasians of AALL and Caucasians of chronic myeloid leukemia. When the study of Song et al. (Supplemental reference [436]) was excluded, high between-studies heterogeneity was deleted in ESCC and Asians of ESCC. When the study of Nasr et al. (Supplemental reference [22]) was excluded, high between-studies heterogeneity was deleted in Africans of non-hodgkin lymphoma.
There was significant heterogeneity in any genetic model when all the eligible studies were pooled into the meta-analysis (P h < 0.001). Then, we assessed the source of heterogeneity by ethnicity, cancer type, source of controls, HWE, and sample size. The results of meta-regression indicated that sample size (recessive model: P = 0.030) but not ethnicity (dominant model: P = 0.494; recessive model: P = 0.230; homozygous model: P = 0.197; heterozygous model: P = 0.822), source of controls (dominant model: P = 0.586; recessive model: P = 0.371; homozygous model: P = 0.429; heterozygous model: P = 0.731), cancer type (dominant model: P = 0.886; recessive model: P = 0.379; homozygous model: P = 0.439; heterozygous model: P = 0.770), and HWE (dominant model: P = 0.275; recessive model: P = 0.476; homozygous model: P = 0.897; heterozygous model: P = 0.104) contributed to substantial heterogeneity in the meta-analysis.
Sensitivity analysis
Examining genotype frequencies in the controls, significant deviation from HWE was detected in 51 studies (Supplemental reference [4, 5, 9, 10, 12, 22, 25, 30, 48, 51, 52, 67, 68, 74, 75, 76, 82, 108, 112, 114, 121, 137, 139, 143, 165, 195, 198, 219, 228, 229, 232, 235, 241, 242, 253, 257, 262, 271, 273, 275, 283, 284, 287, 288, 300, 312, 322, 343, 352, 366, 423]). When these studies were excluded, the results were changed in colorectal cancer (recessive model: OR = 0.94, 95 % CI = 0.87–1.02), esophageal cancer (heterozygous model: OR = 1.21, 95 % CI = 0.97–1.52), hepatocellular cancer (recessive model: OR = 1.17, 95 % CI = 1.01–1.37), Asians of breast cancer (recessive model: OR = 1.16, 95 % CI = 1.00–1.35), Asians of hepatocellular cancer (recessive model: OR = 1.20, 95 % CI = 1.00–1.44; homozygous model: OR = 1.26, 95 % CI = 1.01–1.58), Africans of lymphoma (dominant model: OR = 0.65, 95 % CI = 0.45–0.94; heterozygous model: OR = 0.63, 95 % CI = 0.43–0.93), Caucasians of lymphoma (homozygous model: OR = 1.14, 95 % CI = 0.97–1.33), hospital-based of esophageal cancer (heterozygous model: OR = 1.13, 95 % CI = 0.88–1.45), hospital-based of lung cancer (homozygous model: OR = 1.25, 95 % CI = 0.99–1.59), and hospital-based of skin cancer (homozygous model: OR = 1.41, 95 % CI = 0.98–2.02), as shown in Table 6. In addition, when the meta-analysis was performed excluding studies with small sample sizes, the results were changed in brain tumor (recessive model: OR = 1.39, 95 % CI = 1.00–1.92), hepatocellular cancer (recessive model: OR = 1.17, 95 % CI = 1.00–1.36), leukemia (dominant model: OR = 0.94, 95 % CI = 0.87–1.01; homozygous model: OR = 0.90, 95 % CI = 0.80–1.02; heterozygous model: OR = 0.95, 95 % CI = 0.88–1.02), multiple myeloma (dominant model: OR = 1.19, 95 % CI = 0.98–1.45; homozygous model: OR = 1.18, 95 % CI = 0.89–1.56; heterozygous model: OR = 1.24, 95 % CI = 0.92–1.69), African of bladder cancer (recessive model: OR = 0.62, 95 % CI = 0.36–1.09), Indians of breast cancer (recessive model: OR = 2.38, 95 % CI = 1.16–4.89), Asians of cervical cancer (homozygous model: OR = 1.29, 95 % CI = 0.96–1.74), Africans of lymphoma (dominant model: OR = 0.63, 95 % CI = 0.43–0.93; heterozygous model: OR = 0.63, 95 % CI = 0.42–0.95), and hospital-based of lung cancer (homozygous model: OR = 1.08, 95 % CI = 0.96–1.22), as shown in Table 7.
When the study of Hussain et al. (Supplemental reference [45]) was excluded, the results were changed in Indians of leukemia (dominant model: OR = 1.34, 95 % CI = 0.88–2.04). When the study of Semmler (Supplemental reference [280]) was excluded, the results were changed in the hospital-based studies of brain tumor (recessive model: OR = 1.32, 95 % CI = 0.96–1.81). When the study of Ulvik et al. (Supplemental reference [356]) was excluded, the result was changed between colorectal cancer risk and MTHFR C677T polymorphism in female subgroup analysis (homozygous model: OR = 0.88, 95 % CI = 0.74–1.05). When the study of Kim (Supplemental reference [52]) was excluded, the results were changed between MTHFR C677T polymorphism and rectal cancer risk. When the study of Reddy et al. (Supplemental reference [290]) was excluded, the result was changed in Indians of CALL (heterozygous model: OR = 1.23, 95 % CI = 0.88–1.72). When the study of Deligezer (Supplemental reference [396]) was excluded, the result was changed in Hodgkin lymphoma (dominant model: OR = 0.81, 95 % CI = 0.63–1.03; heterozygous model: OR = 0.79, 95 % CI = 0.61–1.04). When the study of Timuragaoglu et al. (Supplemental reference [287]) was excluded, the result was changed adult ALL (AALL) risk in Caucasian (recessive model: OR = 0.51, 95 % CI = 0.36–0.72; dominant model: OR = 0.75, 95 % CI = 0.59–0.95; TT vs. CC: OR = 0.45, 95 % CI = 0.31–0.66). When the study of Zheng et al. (Supplemental reference [214]) was excluded, the result was changed between MTHFR C677T polymorphism and CAML risk (heterozygous model: OR = 0.72, 95 % CI = 0.41–1.28).
Publication bias
We performed Begg’s funnel plot and Egger’s test to assess the publication bias of literatures. Begg’s funnel plots and Egger’s test suggested that there might be publication bias between MTHFR C677T polymorphism and cancer risk (homozygous model: P = 0.005; recessive model: P = 0.029; dominant model: P = 0.003). This might be a limitation for the meta-analysis because studies with null findings, especially those with small sample size, are less likely to be published. Adjusting for possible publication bias using the Duval and Tweedie nonparametric “trim and fill” method for overall studies, the results did not change between MTHFR C677T polymorphism and cancer risk. Figure 2 lists the Duval and Tweedie nonparametric “trim and fill” methods funnel plot.
Discussion
Gene variants of key enzymes in the folate metabolism, such as the gene MTHFR, were suggested to be responsible for differences in folate levels and DNA methylation [30, 31]. The MTHFR C677T polymorphism influences genomic DNA methylation status through an interaction with the folate status in the peripheral blood mononuclear cells [10]. In addition, MTHFR C677T polymorphism may cause uncontrolled gene expression, genomic instability, and induce carcinogenesis [32, 33]. To date, a lot of studies have performed the association of MTHFR C677T polymorphism with risk of cancer, however, the results remained controversial. In order to resolve this conflict, we performed a meta-analysis to derive a more precise estimation of the association between MTHFR C677T polymorphism polymorphism and risk of cancer.
Overall, significantly increased cancer risk was found when all the eligible studies were pooled into meta-analysis. In the stratified analysis by ethnicity, significantly increased cancer risk was observed in Asians and Indians, but not in Caucasians, Africans, and Middle East population. However, when we performed the further stratified and sensitivity analyses by ethnicity and cancer type, significantly increased breast cancer risk was found in Asians and Indians, significantly decreased colon cancer risk was found, significantly increased gastric cancer risk was found in Caucasians and Asians, significantly increased hepatocellular cancer risk was found in Asians, significantly decreased adult acute lymphoblastic leukemia (AALL) risk was found in Caucasians, significantly decreased childhood acute lymphoblastic leukemia (CALL) risk was found in Asians, and significantly increased multiple myeloma and NHL risk was found in Caucasians. It should be considered that the apparent inconsistency of these results may underlie differences in ethnicity, lifestyle and disease prevalence as well as possible limitations due to the relatively small sample size. The current knowledge of carcinogenesis indicates a multi-factorial and multistep process that involves various genetic alterations and several biological pathways. Thus, it is unlikely that risk factors of cancer work in isolation from each other. One important property of the gene polymorphisms is that their incidence can vary substantially between different racial or ethnic populations. We also found a wide variation of the T allele frequencies of control resources in Asians (0.396), Indians (0.132), Caucasians (0.326), Middle East (0.201), and Africans (0.196), and this different allele frequency might account for the association between the C677T polymorphism and cancer risk in different ethnicity. In the subgroup analysis by source of control, significantly increased cancer risk was found in the hospital-based studies, but not the population-based studies. The hospital-based studies may have certain biases for such controls and may only represent a sample of an ill-defined reference population, and may not be representative of the general population or it may be that numerous subjects in the population-based controls were susceptible individuals.
Based on biochemical properties described for MTHFR C677T polymorphism, we expected that MTHFR C677T polymorphism was associated with risk of all types of cancer. However, significant association was only observed among breast cancer, colon cancer, gastric cancer, hepatocellular cancer, AALL, CCLL, and NHL, suggesting that other factors may be modulating the MTHFR C677T polymorphism functionality. The same polymorphisms may play different roles in different cancer susceptibility, because cancer is a complicated multi-genetic disease, and different genetic backgrounds may contribute to the discrepancy [34]. However, the exact mechanism for association between different tumor sites and MTHFR C677T polymorphism was not clear, carcinogenetic mechanism may differ by different tumor sites and the MTHFR genetic variants may exert varying effects in different cancers. The results of meta-regression indicated that sample size contributed to substantial heterogeneity among the meta-analysis. The small number of studies may hinder the ability of drawing more definite conclusions for some results. For these cases, the interpretation of the results should be taken carefully. There was some evidence of publication bias, which may also have contributed to the high heterogeneity observed. However, such limitations highlight the need for further studies in specific tumor sites and different ethnicities. Our meta-analysis results indicated that a significantly increased risk was observed to be associated with MTHFR C677T polymorphism and lung cancer risk in smokers. There is a consensus that tobacco smoking is the major cause of lung cancer and tobacco smoke contains some carcinogens that induce MTHFR C677T may contribute to the development of lung cancer.
We noticed with great interest that one previous meta-analysis had been reported on the risk of all cancer with MTHFR C677T polymorphism [21]. The study of Tang et al. [21] only had 134 case–control studies, in which a total of 46,207 cancer cases and 69,160 controls were included. Their meta-analysis supported an association between the MTHFR C677T polymorphism and increased risk of esophageal and stomach cancer, especially in Asians. However, many published studies were not included in their meta-analysis [21]. By analyzing a larger number of studies than the previous meta-analysis [21], our meta-analysis included 150,086 cases and 200,699 controls (from 446 studies) to perform the MTHFR C677T polymorphism and risk of cancer. Our meta-analysis seems to confirm and establish the trend in the meta-analysis of the MTHFR C677T polymorphism by the previous meta-analysis [21].
Some limitations of this meta-analysis should be acknowledged. First, the controls of some studies were not uniformly defined and some were hospital-based normal individuals or patients of other diseases which were not representative enough. Second, some studies with small sample size may not have enough statistical power to explore the real association. Third, the effect of gene-environment interactions was not addressed in the analysis. Fourth, study quality was not evaluated for included studies. However, the current meta-analysis has also some strength compared with individual studies and previous meta-analyses. First, differently from previous meta-analyses [12–20], we explored the association of MTHFR C677T polymorphism with risk of cancer risk, allowing for a general view of its influence on cancer susceptibility. Second, we explored the interaction of MTHFR C677T polymorphism and other risk factors, such as smoking, age, and menopausal status. Third, our meta-analysis explores and analyzes the sources of heterogeneity between MTHFR C677T polymorphism and cancer risk.
In summary, this meta-analysis suggests that MTHFR C677T polymorphism is associated with increased breast cancer, gastric cancer, and hepatocellular cancer risk in Asians, is associated with increased gastric cancer, multiple myeloma and NHL risk in Caucasians, is associated with decreased AALL risk in Caucasians, is associated with decreased CALL risk in Asians, is associated with increased breast cancer risk in Asians, and is associated with decreased colon cancer risk. Moreover, this meta-analysis also points out the importance of new studies, such as Asians of HNC, Asians of lung cancer, Indians of breast cancer, because they had high heterogeneity in this meta-analysis (I 2 > 75 %).
References
Kwasniewska A, Tukendorf A, Semczuk M. Folate deficiency and cervical intraepithelial neoplasia. Eur J Gynaecol Oncol. 1997;18:526–30.
Eichholzer M, Luthy J, Moser U, Fowler B. Folate and the risk of colorectal, breast and cervix cancer: the epidemiological evidence. Swiss Med Wkly. 2001;131:539–49.
Fodinger M, Horl WH, Sunder-Plassmann G. Molecularbiology of 5,10-methylenetetrahydrofolate reductase. J Nephrol. 2000;13:20–33.
Lucock M. Folic acid: nutritional biochemistry, molecular biology, and role in disease processes. Mol Genet Metab. 2000;71:121–38.
Lucock M. Is folic acid the ultimate functional food component for disease prevention? BMJ. 2004;328:211–4.
Cicek MS, Nock NL, Li L, Conti DV, Casey G, Witte JS. Relationship between methylenetetrahydrofolate reductase C677T and A1298C genotypes and haplotypes and prostate cancer risk and aggressiveness. Cancer Epidemiol Biomarkers Prev. 2004;13:1331–6.
García-Closas R, García-Closas M, Kogevinas M, Malats N, Silverman D, Serra C, et al. Food, nutrient and heterocyclic amine intake and the risk of bladder cancer. Eur J Cancer. 2007;43:1731–40.
Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–3.
Jacques PF, Bostom AG, Williams RR, Ellison RC, Eckfeldt JH, Rosenberg IH, et al. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation. 1996;93:7–9.
Friso S, Choi SW. Gene-nutrient interactions in one-carbon metabolism. Curr Drug Metab. 2005;6:37–46.
Rozen R. Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR). Thromb Haemost. 1997;78:523–6.
Niu YM, Deng MH, Chen W, Zeng XT, Luo J. MTHFR C677T gene polymorphism and head and neck cancer risk: a meta-analysis based on 23 publications. Dis Markers. 2015;2015:681313.
Macis D, Maisonneuve P, Johansson H, Bonanni B, Botteri E, Iodice S, et al. Methylenetetrahydrofolate reductase (MTHFR) and breast cancer risk: a nested-case–control study and a pooled meta-analysis. Breast Cancer Res Treat. 2007;106:263–71.
Li K, Li W, Dong X. Association of 677 C>T (rs1801133) and 1298 A>C (rs1801131) polymorphisms in the MTHFR gene and breast cancer susceptibility: a meta-analysis based on 57 individual studies. PLoS One. 2014;9, e71290.
Liu YX, Wang B, Wan MH, Tang WF, Huang FK, Li C. Meta-analysis of the relationship between the metholenetetrahydrofolate reductase C677T genetic polymorphism, folate intake and esophageal cancer. Asian Pac J Cancer Prev. 2011;12:247–52.
Fang Y, Xiao F, An Z, Hao L. Systematic review on the relationship between genetic polymorphisms of methylenetetrahydrofolate reductase and esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2011;12:1861–6.
Xia LZ, Liu Y, Xu XZ, Jiang PC, Ma G, Bu XF, et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and gastric cancer susceptibility. World J Gastroenterol. 2014;20:11429–38.
Ramsey SD, Holmes RS, McDermott CL, Blough DK, Petrin KL, Poole EM, et al. A comparison of approaches for association studies of polymorphisms and colorectal cancer risk. Colorectal Dis. 2012;14:e573–86.
Hubner RA, Houlston RS. MTHFR C677T and colorectal cancer risk: a meta-analysis of 25 populations. Int J Cancer. 2007;120:1027–35.
Wang X, Yue K, Hao L. Meta-analysis of methylenetetrahydrofolate reductase polymorphism and lung cancer risk in Chinese. Int J Clin Exp Med. 2015;8:1521–5.
Tang M, Wang SQ, Liu BJ, Cao Q, Li BJ, Li PC, et al. The methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and tumor risk: evidence from 134 case–control studies. Mol Biol Rep. 2014;41:4659–73.
Little J, Bradley L, Bray MS, Clyne M, Dorman J, Ellsworth DL, et al. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156:300–10.
Davey SG, Egger M. Meta-analyses of randomized controlled trials. Lancet. 1997;350:1182.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analysis. Br Med J. 2003;327:557–60.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Natl Cancer Inst. 1959;22:719–48.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Klug SJ, Ressing M, Koenig J, Abba MC, Agorastos T, Brenna SM, et al. TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies. Lancet Oncol. 2009;10:772–84.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101.
Egger M, Smith DG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315:629–34.
Friedman G, Goldschmidt N, Friedlander Y, Ben-Yehuda A, Selhub J, Babaey S, et al. A common mutation A1298C in human methylenetetrahydrofolate reductase gene: association with plasma total homocysteine and folate concentrations. J Nutr. 1999;129:1656–61.
Parle-McDermott A, Mills JL, Molloy AM, Carroll N, Kirke PN, Cox C, et al. The MTHFR 1298CC and 677TT genotypes have opposite associations with red cell folate levels. Mol Genet Metab. 2006;88:290–4.
Duthie SJ. Folic acid deficiency and cancer: mechanisms of DNA instability. Br Med Bull. 1999;55:578–92.
Ames BN. DNA damage from micronutrient deficiencies is likely to be a major cause of cancer. Mutat Res. 2001;475:7–20.
Hirschhorn JN, Lohmueller K, Byrne E. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Shu-Zhe Xie, Zhi-Zhong Liu, Jun-hua Yu, Li Liu, Wei Wang, Dao-Lin Xie, and Jiang-Bo Qin are co-first authors.
Shu-Zhe Xie, Zhi-Zhong Liu, Jun-hua Yu, Li Liu, Wei Wang, Dao-Lin Xie and Jiang-Bo Qin contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOC 719 kb)
ESM 1
(DOC 325 kb)
Rights and permissions
About this article
Cite this article
Xie, SZ., Liu, ZZ., Yu, Jh. et al. Association between the MTHFR C677T polymorphism and risk of cancer: evidence from 446 case–control studies. Tumor Biol. 36, 8953–8972 (2015). https://doi.org/10.1007/s13277-015-3648-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3648-z