Abstract
Epigenetic mechanisms such as DNA methylation are being increasingly recognized to play an important role in cancer and may serve as a cancer biomarker. The aim of this study was to evaluate the promoter methylation status of MGMT (O6-methylguanine-DNA methyltransferase) and a possible correlation with the expression of MGMT and standard clinicopathological parameters in invasive ductal breast carcinoma patients (IDC) of Kashmir. Methylation-specific PCR was carried out to investigate the promoter methylation status of MGMT in breast tumors paired with the corresponding normal tissue samples from 128 breast cancer patients. The effect of promoter methylation on protein expression in the primary breast cancer and adjacent normal tissues was evaluated by immunohistochemistry (n = 128) and western blotting (n = 30). The frequency of tumor hypermethylation was 39.8 % and a significant difference in methylation frequency among breast tumors were found (p < 0.001) when compared with the corresponding normal tissue. Immunohistochemical analysis showed no detectable expression of MGMT in 68/128 (53.1 %) tumors. MGMT promoter methylation mediated gene silencing was associated with loss of its protein expression (rs = −0.285, p = 0.001, OR = 3.38, 95 % CI = 1.59–7.17). A significant correlation was seen between loss of MGMT and lymph node involvement (p = 0.030), tumor grade (p < 0.0001), loss of estrogen receptors (ER; p = 0.021) and progesterone receptors (PR) (p = 0.016). Also, MGMT methylation was found to be associated with tumor grade (p = 0.011), tumor stage (p = 0.009), and loss of ER (p = 0.003) and PR receptors (p = 0.009). To our knowledge, our findings, for the first time, in Kashmiri population, indicate that MGMT is aberrantly methylated in breast cancer and promoter hypermethylation could be attributed to silencing of MGMT gene expression in breast cancer. Our data suggests that MGMT promoter hypermethylation could have a potential function as molecular biomarker of breast oncogenesis. Also, based on their predictive value of response to therapy, the immunohistochemical evaluation and interpretation of MGMT may also help in future to establish therapeutic strategies for patients with breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common form of malignant disease in women worldwide and also the principal cause of death from cancer among women globally [1]. During the last decade, breast cancer has been rising steadily, and now, breast cancer is the most common cancer in women in India with an estimated 144,937 incidences and 70,218 mortalities due to breast cancer [1, 2]. Breast cancer is emerging as a major concern in women populations of the Kashmir valley with its incidence showing a rising trend in recent years [3]. In the year 2013, approximately 186 women were diagnosed with breast cancer, accounting for 13.5 % of all adult female cancers [4]. There are well-understood genetic alterations associated with breast tumorigenesis, including specific gene amplifications, chromosome rearrangements, deletions, point mutations, and aneuploidy. In addition to these highly characterized mutations in oncogenes and tumor suppressor genes, epigenetic alterations resulting in aberrant gene expression are key contributors to breast tumorigenesis [5, 6]. Hypermethylation of the CpG islands in promoter region is the best-studied epigenetic modification associated with condensed chromatin, delayed replication, inhibition of transcription initiation, and downregulation of genes [7]. The delineation of specific DNA methylation markers in breast carcinogenesis could impact the development of new approaches for diagnosis, clinical management and therapeutics that target methylation.
O6-methyguanine-DNA methyltransferase (MGMT) lies on chromosome 10, and the product of MGMT mediates DNA repair by removing mutagenic and cytotoxic adducts from the O6-guanine in DNA [8–10]. If not removed, O6-alkylguanine mediates toxicity by forming interstrand cross-links, which inhibit DNA replication and can cause the incorporation of incorrect base pairs, resulting in mutations. The MGMT prevents cells from these lesions by transferring the alkyl group from O6 position of guanine to an active-site cysteine residue in a stoichiometric, direct damage-reversal pathway restoring guanine in the DNA [11]. Previous studies have shown that loss of MGMT expression occurs rarely due to mutation, deletion, or rearrangement of MGMT but is rather associated with methylation of specific regions of MGMT CpG island [12–14]. However, the occurrence of MGMT epigenetic modifications has not been extensively investigated in breast cancer patients.
Recent studies suggest that methylation profiles of cancers depend on tumor type and are ethnicity specific [15–17]. There are several reports on methylation profile of MGMT in breast cancer patients from different populations [14, 18–20] and few studies on Indian population [12, 21]. However, to our knowledge hypermethylation of genes in Kashmiri breast cancer population, ethnically distinct from rest of India, is still not well-studied. Taking all these things into consideration, we found it rational to investigate the methylation status of MGMT and correlate our findings with gene expression at protein level. Finally, we aimed to analyze statistical correlations of clinicopathological patient characteristics with MGMT methylation and its expression data.
Methodology
Tissue specimens
A total of 128 newly diagnosed breast cancer patients from Kashmiri ethnic population admitted to the Sher-I-Kashmir Institute of Medical Sciences (SKIMS), Srinagar during the period December 2011 to May 2013 were included in the study. Tissue samples consisting of tumor and adjacent matched normal breast tissues (taken 5–10 cm away from the site of tumor) were taken from all the breast cancer patients enrolled in the study from the Department of General Surgery, SKIMS. All the patients included in this study were diagnosed with invasive ductal carcinoma and all were females. The tissue samples were immediately shock-frozen after surgical resection and stored at −80 °C in deep freezer until further analysis. Each sample was histopathologically evaluated to ensure the presence of at least 80 % of tumoral cells. Clinical and pathological information was obtained from medical records and pathology reports. Disease staging was performed according to the tumor–node–metastasis (TNM) system of the American Joint Committee on Cancer/the Union Internationale Contre le Cancer (AJCC/UICC). Histopathological grading was carried out according to the Scarff-Bloom-Richardson classification as GI, GII, and GIII. All patients received a patient information sheet and signed a written consent form, approved by the SKIMS ethical committee.
DNA isolation
DNA was extracted from breast tumor and normal tissue using standard technique of digestion with proteinase K in the presence of SDS at 37 °C overnight, followed by phenol/chloroform extraction or genomic DNA extraction kit (Bioserve biotechnologies Pvt. Ltd., India). The quality and integrity of the DNA were checked by electrophoresis on 0.8 % agarose gel, quantitated spectrophotometrically and stored at −20 °C till further use.
Methylation-specific PCR (MSP)
The sensitive methylation-specific PCR (MSP) was used to detect promoter methylation. Bisulphite modification of the DNA (up to 2 μg) was carried out by EZ DNA Methylation™ Kit (Zymo Research, USA) according to manufactures protocol. Treatment of genomic DNA with sodium bisulfite converts unmethylated cytosines (but not methylated cytosines) to uracil, which are then converted to thymidine during the subsequent PCR step, giving sequence differences between methylated and unmethylated DNA. PCR primers that distinguish between these methylated and unmethylated DNA sequences were then used. Primer sequences of MGMT were as described previously in the literature [22] for the unmethylated reaction 5′-TTT GTG TTT TGA TGT TTG TAG GTT TTT GT-3′ (upper primer) and 5′-AAC TCC ACA CTC TTC CAA AAA CAA AAC A+3′ (lower primer) and for the methylated reaction 5′-TTT CGA CGT TCG TAG GTT TTC GC-3′ (upper primer) and 5′-GCA CTC TTC CGA AAA CGA AAC G-3′ (lower primer). The PCR amplification was carried in a total volume of 25 μl containing 2 μl of bisulfite-modified DNA, 1X PCR buffer, 2 mM MgCl2, 200 ng of each primer, 0.3 mM dNTPs (Fermentas life sciences, Inc. USA), and 1U of Taq polymerase (Fermentas life sciences, Inc. USA). Amplification was performed in Thermal cycler (Mastercycle, Ependroff) with an initial denaturation step at 95 °C for 5 min, followed by 35 cycles at 94 °C for 50 s, 59 °C for 50 s, and 72 °C for 50 s and a final extension at 72 °C for 10 min. Each PCR product was loaded onto a 3 % agarose gel, stained with ethidium bromide and visualized under UV illumination and photographed with Alpha Imager 1220 v5.5 Camera software. PCR generated a 91 bp product both for methylated and 98 bp for unmethylated, as shown in Fig. 1. The PCR for all samples demonstrating methylation were repeated to confirm these results.
Methylation specific polymerase chain reaction (PCR) assay. PCR product indicating a methylated O6-methylguaninemethyltransferase (MGMT) promoter is shown by arrows. U, unmethylated MGMT promoter; M, methylated MGMT promoter; L, 100-bp DNA marker ladder; In vitro SssI methyltransferase-treated and untreated DNA from normal lymphocytes were used as the positive controls (PC) for methylation and nonmethylation (NC), respectively
DNA from normal lymphocytes was used as negative control for methylated alleles of MGMT. Water with no DNA template as a control for contamination were included in each experiment. DNA from peripheral blood lymphocytes of healthy volunteers treated with SssI methyltransferase (New England Biolabs, Beverly, MA, USA) and then subjected to bisulfite modification was used as positive controls for methylated alleles.
Immunohistochemistry
Sections of formalin-fixed, paraffin-embedded breast were obtained on polyl-lysine-coated slides. Sections were deparaffinized in xylene, then rehydrated through a graded alcohol series. Antigen retrieval was performed by incubating slides in citrate buffer (pH 6.0) (10 mM) at 95 °C for 20 min. Endogenous peroxidase activity was blocked with 3 % H2O2 for 30 min. Thereafter, slides were incubated with specific primary antibody, ERα (sc-543, dilution 1:100; Santacruz Biotechnology Inc., USA), PR (1:100 dilution, clone 1A6, Biocare Medical, USA), Her2/neu (sc-08, dilution 1:100; Santacruz Biotechnology Inc., USA), MGMT (sc-28241, dilution 1:100; Santacruz Biotechnology Inc., USA) for 16 h at 4 °C and washed with Tris buffered saline (TBS). Next day, the slides were washed three times in Tris buffers (pH 7.6) and bound primary antibody was detected by MACH1 Universal HRP-Polymer (Biocare medical, USA) for 30 min at room temperature. After washing in Tris buffer, the immunostaining reaction product was developed using 3,3-diaminobenzidine (Betazoid DAB Plus substrate, Biocare Medical, USA). After immunoreactivity, slides were dipped in distilled water, counterstained with Harris hematoxylin and finally the sections were dehydrated in xylene, mounted with DPX and coverslipped. In all cases, adjacent normal surrounding tissue served as an internal positive control. In negative controls, the primary antibody was replaced by non-immune mouse IgG of the same isotype to ensure specificity.
In case of ERα, PR, and MGMT only nuclear staining was considered as immunopositivity. The slides were scored as follows: negative: <10 % tumor cells showing immunoreactivity and tumors showing >10 % cells with nuclear immunopositivity were considered as positive [23–26]. For Her2/neu protein expression, membrane immunostaining was considered as positive. Briefly, the scoring system was as follows: no staining or membrane staining in fewer than 10 % of tumor cells, 0; faint, barely perceptible membrane staining in more than 10 % of tumor cells, the cells are stained only in part of the membrane, 1+; weak to moderate complete membrane staining observed in more than 10 % of tumor cells, 2+; and strong, complete membrane staining in more than 10 % of tumor cells, 3+ [27].
Western blotting
Total protein extracts from 30 frozen breast cancer and adjacent normal samples were prepared with lysis buffer (50 mM Tris–HCl, 0.3 M NaCl, 1 mM EDTA and 1 mM dithiothreitol) supplemented with a cocktail of protease inhibitors (Sigma Aldrich, USA). Equal amounts (100 μg) of protein extracts were resolved on SDS-polyacrylamide gels and subsequently electrotransferred onto nitrocellulose membranes (Invitrogen, USA).) in trans-buffer (25 mM Tris; 129 mM glycine; 10 % methanol; 0.05 % SDS). Membranes were blocked in 5 % non-fat milk in 1 % TBST for 1 h at room temperature, and incubated overnight at 4 °C with a rabbit anti-MGMT monoclonal antibody (Santa Cruz Biotechology, USA) diluted in the same blocking buffer. After washing, membranes were incubated with goat antirabbit (Abcam, USA) antibodies conjugated to horseradish peroxidase. Blots were developed with SuperSignal chemiluminescence reagent (Pierce). For loading control, membranes were stripped and reprobed with a human-specific antibody against β-actin (Sigma Aldrich, USA).
Statistical analysis
Comparisons of MGMT methylation and expression with clinicopathological characteristics were made using Pearson Chi-squared test. The logistic regression model was used to assess univariate association between MGMT methylation (methylated vs. unmethylated) and MGMT expression (low vs. normal) with clinicopathological variables. The variables that were significantly associated with MGMT expression in univariate analysis were then simultaneously put in multivariate logistic regression model to assess the association while adjusting for other significantly associated variables. All p values are two-sided and considered statistically significant at the 0.05 level. All statistical analyses were performed using SPSS (version 16; USA).
Results
Clinicopathological findings
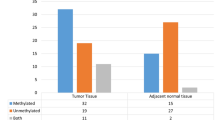
Relevant clinicopathological characteristics of the patients included in this study are summarized in Table 1. A total of 128 cases of IDC were included in this study. The mean age of patients was 51.85 ± 1.02 years, median age was 53 (range 26–80). The mean body mass index (BMI) was 23.64 ± 0.21. Age was grouped into two categories with 56.2 % of cases lying in the group >50 years of age. Approximately 68.0 % were rural. The stage distribution was typical with 38.3, 31.3, 23.4, and 7.0 % presenting with stages I, II, III, and IV, respectively. Stages III and IV were combined for all statistical analysis. Most of the tumors were estrogen receptors (ER) positive (69.5 %) and progesterone receptors (PR) positive (66.4 %). Her-2-neu immunostaining was detected in only 25 % of breast tumors (Fig. 2).
Prevalence of MGMT promoter gene methylation in breast cancer
Representative results for MSP analysis are demonstrated in Fig. 1. When the promoter methylation status of MGMT in 128 invasive breast cancer tissues and corresponding normal tissue from the same patient was analyzed, MSP products were obtained successfully in all cases. MS-PCR results were interpreted data using appropriate statistical tests.
MGMT gene methylation was present in 39.8 % (51 of 128) of breast cancer tissues. Methylation profile of these genes in normal adjacent tissues from the same patient was also investigated, and we found very low methylation (2.6 %). Patients who showed methylation of this gene in normal tissue were found to be methylated in tumor tissue as well.
Correlation between methylation of the MGMT promoter and Clinicopathological Features using univariate and multivariate analysis
Table 2 summarizes the various demographic, clinical, and pathologic factors that were included in the univariate statistical analysis to explore their relationship with MGMT hypermethylation. We did not observe a relationship between MGMT hypermethylation and clinical factors such as age, menopausal status, dwelling, nodal status, and BMI. A significant association was observed between MGMT methylation and tumor grade (p (2) = 0.011). Univariate analysis for MGMT methylation between tumor grade types demonstrated grade II tumors with an OR = 0.99 (CI 0.40–2.42, p = 0.976) and tumor grade III with an OR = 4.43 (CI 1.30–15.09, p = 0.017) compared to grade I tumors. Similarly, there was a good correlation between increase in MGMT methylation and tumor stage (p (2) = 0.009) with OR = 2.28 (CI 0.92–5.65, p = 0.074) for stage II and OR = 3.99 (CI 1.61–9.89, p = 0.003) for stages III and IV tumors compared to stage I tumors. We did not find an association between MGMT promoter methylation status and the tumor HER2 status. Estrogen-receptor negative tumors (OR = 3.13, CI 1.44–6.83, p = 0.003) and progesterone receptor negative tumors (OR = 2.71, CI 1.27–5.78, p = 0.009) were associated with higher frequency of MGMT methylation compared to estrogen and progesterone-receptor positive tumors.
Immunohistochemical assessment of MGMT expression and correlation with promoter methylation
To further explore the functional consequence of MGMT promoter hypermethylation, MGMT expression was examined in primary breast tumors. Normal breast tissue exhibited positive immunostaining for MGMT protein in ductal epithelial cells. Loss or markedly reduced expression of MGMT protein was observed in 68 of 128 IDC of breast (Fig. 3) Comparison of methylation and Immunohistochemistry data revealed that out of the 51 tumors harboring promoter methylation, 36 (70.6 %) showed loss or weak immunoreactivity for MGMT protein. Among the remaining cases, 15 of 51(29.4 %) showed positive immunostaining and were interpreted as immunohistochemistry-positive. In contrast, in the unmethylated group, 45 of 77 (58.4 %) were heterogeneous or homogeneous immunohistochemistry-positive for MGMT while 32 (41.6 %) showed reduced or loss of expression. The inverse correlation between immunohistochemistry reactivity for MGMT protein and methylation status for MGMT gene was statistically significant. Collectively, evidence from promoter methylation and protein expression studies strongly suggest that MGMT promoter hypermethylation correlates significantly with reduction or loss of MGMT expression (rs = −0.285, p < 0.05; Table 3). Figure 4 gives the graphical representation of the correlation between MGMT promoter hypermethylation and MGMT expression.
Representative immunohistochemical detection of MGMT in mammary gland. Positive staining was identified by the presence of brown staining in the nuclei and in the cytoplasm. Nuclear MGMT was observed in normal breast (a), with variable expression in IDCs ranging from strong (b), weak (c), or completely absent (d), negative control (e) (original magnification—×20)
Correlation between MGMT expression status and clinicopathological features
The relationships between the expression levels of MGMT and clinicopathological parameters were assessed to determine their clinical significance (Table 4). The relationships between the expression level of MGMT and clinicopatholigical characteristics were almost comparable to those observed in tumors with MGMT methylation which again strengthens our finding that promoter methylation and expression of MGMT are highly correlated with each other. A very significant correlation was observed between MGMT expression and tumor grade with MGMT expression reduced in 8 (27.6 %) of 29 grade I, 43 (54.4 %) of 79 grade II tumors, and 17 (85 %) of 60 grade III tumors (p (2) = <0.0001). Univariate analysis for MGMT expression between tumor grades demonstrated grade II with an OR = 3.14 (CI 1.24–7.92, p = 0.016) and grade III with an OR = 14.88 (CI 3.41–64.88, p < 0.0001) in comparison to grade I tumors. Low MGMT expression was also found to be associated with lymph node involvement (OR = 2.25, CI 1.08–4.70, p = 0.03). However, no significance was found with tumor stage and other demographic or pathologic features. Estrogen-receptor- and progesterone-receptor negative tumors were associated with low MGMT expression compared to estrogen-receptor-positive (OR = 3.12, CI 1.38–7.03, p = 0.005) and progesterone-receptor positive (OR = 2.44, CI 1.13–5.26, p = 0.021) tumors, respectively. In the multivariate analysis (Table 5), low MGMT expression was found associated with ER negativity (p = 0.015), PR negativity (p = 0.007) and advanced tumor grade (p = 0.004) only and not nodal involvement.
Expression of MGMT in breast tissues using western blotting
We examined MGMT protein expression by Western blot analysis in 30 tumor tissues and 30 adjacent normal tissues to confirm the results obtained by immunohistochemistry showing association between promoter methylation and its impaired synthesis. Among the 30 tumor tissues 12 were methylated and 18 were unmethylated whereas all the adjacent normal tissues were without any promoter methylation. Loss or marked reduction of MGMT expression was observed in 8 of the 12 hypermethylated tumors whereas the remaining 4 tissues showed normal MGMT expression. In contrast, 15 of the 18 unmethylated tumors expressed almost normal amounts of MGMT whereas only three unmethylated tumor tissues showed loss of expression of MGMT. Representative results are shown in Fig. 5, methylated tumor tissues (lane 6 and 8) showed loss of expression of MGMT compared to their adjacent normal tissues (lane 5 and 7). These results confirmed that hypermethylation of the CpG islands located in the promoter region of the MGMT gene is associated with loss of MGMT expression of protein in breast cancer tissues.
The representative western blot shows the expression of MGMT protein using the MGMT antibody and corresponding loading control (β-actin). Lane 1: empty vector (pB513B1) transfected HEK cell lysate (25 μg), lane 2: pB513B1-MGMT transfected HEK cell lysate (10 μg), lane 3: pB513B1-MGMTtransfected HEK cell lysate (40 μg), lane 4:pB513B1-MGMT transfected HEK cell lysate (60 μg), lane 5: (N1) normal breast tissue lysate (25 μg), lane 6: (T1) breast tumor tissue lysate (25 μg), lane 7: (N2) normal breast tissue lysate (25 μg), lane 8: (T2) breast tumor tissue lysate (25 μg)
Discussion
Methylation is the main epigenetic event in humans involved in cancer initiation and progression. Early detection is critical for the successful treatment of numerous types of cancer, including breast cancer. As aberrant methylation is frequently observed during early tumorigenesis even before cancer development, their profiling can be exploited for the detection of potentially premalignant disease and prediction of human malignancies [28–32].
The main aim of this study was to determine the methylation level in the promoter region of MGMT gene in breast cancer patients and to investigate the association between the methylation status of the examined gene promoter and the available demographic, clinicopathological characteristics, i.e., age, tumor grade, lymph node status, stage, ER/PR and Her-2-neu status in primary breast tumors from 128 patients. Aberrant promoter DNA methylation has been examined with a variety of methodologies having different sensitivities, including SSCP, COBRA, MSP, and sequencing [33]. MSP is currently the most commonly used method due to its claimed efficiency in heterogeneous cancer cell populations [34]. In the present study, we used MSP approach in order to determine methylation status of MGMT, a DNA repair gene. From each patient, a breast cancer sample and the adjacent normal breast tissue were evaluated. With this design, pair of cancer and normal samples was available for each case. In our cohort MGMT promoter methylation was found in 39.8 % of the cases as against 2.6 % of the corresponding normal breast samples. In accordance with our findings, many earlier studies have found that the frequency of MGMT methylation fluctuates between 22 and 32 % [12, 18, 19]. Our data, however, are at variance with the findings of Fumagalli et al. who reported a very high frequency of MGMT methylation [20] and several other studies reporting either rare or total absence of methylation of MGMT gene in breast cancer [21, 24, 30]. The reason for this discrepancy remains unknown, but may be due to sample size differences in individual studies, differences in ethnic origins of patients, well-known heterogeneity of breast tumors and analytical methods used to determine methylation status.
In this article, we reported the identification of MGMT gene as an epigenetic target in breast cancer patients of Kashmir. A significant correlation was observed between aberrant MGMT methylation and its gene silencing, suggesting that DNA methylation may be the possible mechanism involved in loss of MGMT expression in breast tumors. However, negative immunostaining in 41.6 % of unmethylated cases could be due to histone modifications—binding of methyl-CpG binding proteins, demethylation of histone H3 lysine 9 and MBD, which have also been identified to be the regulators of MGMT expression in breast cancer in other populations [12].
Several studies have found that mean MGMT activity are higher in tumors as compared to their normal tissue counterparts [35–40]. For example, Silber et al. assessed levels of MGMT in histologically normal brain tissue adjacent to primary brain tumors and demonstrated absence of detectable MGMT activity in normal brain [41]. However, we identified reduced expression of MGMT in 53 % of the breast cancer tumors. Our results are in agreement with others, which demonstrate significantly lower amounts of MGMT in tumors ranging from 19 to 93.3 % [14, 19, 42, 43].
We also investigated the association of promoter hypermethylation and loss of expression with various clinicopathological parameters of breast cancer. The prevalence of MGMT methylation was essentially dependent on tumor histological stage and grade, thus indicating that this event occurs at a later stage. In the present study, loss of expression of MGMT also tended to be associated with higher malignant nuclear grade of cancer cells which suggests that expression of MGMT may be a sign of low biologic aggressiveness. Therefore, our finding, in agreement with an earlier studies [14, 18, 42], supports the hypothesis that loss of MGMT function may be related to a more aggressive phenotype with a higher risk of metastatic disease. Our study also demonstrated the association of loss of MGMT expression with lymph node metastasis in breast cancer suggesting that the breast tumors with reduced expression of MGMT may possess a biologically aggressive phenotype or we can say that MGMT may have a metastasis suppressing function. Recently reported data also has indicated significantly higher MGMT protein expression in the tumors with no lymph node metastasis compared with the tumors positive for lymph node metastasis [44]. However, our finding is inconsistent with the earlier studies who did not find any association between loss of MGMT expression and nodal status in breast cancer [12, 14, 26, 43]. While lymph node involvement appeared to positively trend with loss of MGMT expression in univariate analysis, this association could not be confirmed on the multivariate analysis and may require a larger dataset to evaluate this.
In addition, we compared the methylation status and expression of MGMT with the standard Immunohistochemical parameters (Her-2, ER, and PR, Fig. 2). Both the promoter hypermethylation and the loss of MGMT expression were significantly correlated with negative ER and PR status supporting the hypothesis that disruption of the maintenance mechanisms of the genomic methylation pattern in cells could result in discrete expression profiles of primary breast cancer because all three genes are known to be epigenetically silenced [45, 46]. Similar to our findings, early studies have also demonstrated simultaneous loss of all the three proteins [14, 43]. Our data is also in agreement with the observations of Neto JC et al., who found significantly greater MGMT protein levels in luminal-type compared with basal-like tumors [42]. However, correlations with any other clinicopathological features were not attained, apart from a marginal correlation of MGMT methylation with age.
The reduced expression of MGMT in tumors than in normal tissue is a remarkable finding. Tumors that express little or no MGMT are more likely to show better response to alkylating agents; conversely, tumors with high MGMT levels are more likely to be drug-resistant [36]. The strong correlation of promoter methylation in MGMT with the lack of its expression, a favorable prognosis, and sensitiveness to alkylating drugs has been described in nervous system-related tumors. The presence of methylation on MGMT is currently used as an indicator of the susceptibility to the chemotherapeutic alkylating drugs in these tumors [47]. Our results indicate that immunohistochemical evaluation of MGMT could offer a new adjunct in predicting tumor response to alkylating drugs in breast cancer.
Conclusion
In conclusion, we found MGMT methylation frequently in invasive ductal carcinoma of breast and that MGMT methylation is associated with the loss of MGMT protein expression. Moreover, based on our results, we hypothesize that the immunohistochemical assessment of this marker predicts tumor progression and could be extended in future to provide molecular staging, and predictive information with regards to response to therapy. The clinical significance of our preliminary findings should be further confirmed in large cohorts that will undoubtedly lead to a greater understanding of breast cancer progression and may help in future to establish therapeutic strategies for patients with breast cancer.
References
Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin D, Forman D, Bray F: GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://globocan.iarc.fr, accessed on 13/February/2014. In, 2013.
Indian Council of Medical Research ND, 2013.: National Cancer registry Programme: Consolidated Report of the Population Based Cancer Registries (PBCR) 2007–2011. In, 2013.
Wani SQ, Khan T, Wani SY, Koka AH, Arshad S, Rafiq L, et al. Clinicoepidemiological analysis of female breast cancer patients in Kashmir. J Cancer Res Therapeut. 2012;8:389.
RCC: Sheri-Kashmir Institute of Medical Sciences. Retreived on 04/07/2014 2013.
Dworkin AM, Huang THM, Toland AE: Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. In Seminars in Cancer Biology. Elsevier, 2009:165–171.
Visvanathan K, Sukumar S, Davidson NE. Epigenetic biomarkers and breast cancer: cause for optimism. Clin Cancer Res. 2006;12:6591–3.
Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59.
Jacinto FV, Esteller M. MGMT hypermethylation: a prognostic foe, a predictive friend. DNA Repair. 2007;6:1155–60.
Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–29.
Pegg AE, Dolan ME, Moschel RC. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog Nucleic Acid Res Mol Biol. 1995;51:167–223.
Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, et al. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol. 2004;11:714–20.
Sharma G, Mirza S, Parshad R, Srivastava A, Gupta SD, Pandya P, et al. Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci. 2010;87:83–91.
Danam RP, Qian XC, Howell SR, Brent TP. Methylation of selected CpGs in the human O6-methylguanine-DNA methyltransferase promoter region as a marker of gene silencing. Mol Carcinog. 1999;24:85–9.
Munot K, Bell SM, Lane S, Horgan K, Hanby AM, Speirs V. Pattern of expression of genes linked to epigenetic silencing in human breast cancer. Hum Pathol. 2006;37:989–99.
Mehrotra J, Ganpat MM, Kanaan Y, Fackler MJ, McVeigh M, Lahti-Domenici J, et al. Estrogen receptor/progesterone receptor-negative breast cancers of young African-American women have a higher frequency of methylation of multiple genes than those of Caucasian women1. Clin Cancer Res. 2004;10:2052–7.
Bae YK, Brown A, Garrett E, Bornman D, Fackler MJ, Sukumar S, et al. Hypermethylation in histologically distinct classes of breast cancer. Clin Cancer Res. 2004;10:5998–6005.
Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Pookot D, et al. Ethnic groupâ-related differences in CpG hypermethylation of the GSTP1 gene promoter among Africanâ-American, Caucasian and Asian patients with prostate cancer. Int J Cancer. 2005;116:174–81.
Tserga A, Michalopoulos NV, Levidou G, Korkolopoulou P, Zografos G, Patsouris E, et al. Association of aberrant DNA methylation with clinicopathological features in breast cancer. Oncol Rep. 2012;27:1630–8.
Alkam Y, Mitomi H, Nakai K, Himuro T, Saito T, Takahashi M, et al. Protein expression and methylation of DNA repair genes hMLH1, hMSH2, MGMT and BRCA1 and their correlation with clinicopathological parameters and prognosis in basal-like breast cancer. Histopathology. 2013;63:713–25.
Fumagalli C, Pruneri G, Possanzini P, Manzotti M, Barile M, Feroce I, et al. Methylation of O 6-methylguanine-DNA methyltransferase (MGMT) promoter gene in triple-negative breast cancer patients. Breast Cancer Res Treat. 2012;134:131–7.
Viswanathan M, Solomon SPR, Tsuchida N, Selvam GS, Shanmugam G. Methylation of E-cadherin and hMLH1 genes in Indian sporadic breast carcinomas. Indian J Exp Biol. 2006;44:115–9.
Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–71.
Pertschuk LP, Kim DS, Nayer K, Feldman JG, Eisenberg KB, Carter AC, et al. Immunocytochemical estrogen and progestin receptor assays in breast cancer with monoclonal antibodies. Histopathologic, demographic, and biochemical correlations and relationship to endocrine response and survival. Cancer. 1990;66:1663–70.
Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–7.
Belanich M, Randall T, Pastor MA, Kibitel JT, Alas LG, Dolan ME, et al. Intracellular localization and intercellular heterogeneity of the human DNA repair protein O6-methylguanine-DNA methyltransferase. Cancer Chemother Pharmacol. 1996;37:547–55.
Matsukura S, Harimaya K. Expression and prognostic significance of O6-methylguanine-DNA methyltransferase in hepatocellular, gastric, and breast cancers. Ann Surg Oncol. 2001;8:807–16.
Rhodes A, Jasani B, Anderson E, Dodson AR, Balaton AJ. Evaluation of HER-2/neu immunohistochemical assay sensitivity and scoring on formalin-fixed and paraffin-processed cell lines and breast tumors a comparative study involving results from laboratories in 21 countries. Am J Clin Pathol. 2002;118:408–17.
Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–9.
Kang S, Kim JW, Kang GH, Lee S, Park NH, Song YS, et al. Comparison of DNA hypermethylation patterns in different types of uterine cancer: cervical squamous cell carcinoma, cervical adenocarcinoma and endometrial adenocarcinoma. Int J Cancer. 2006;118:2168–71.
Muggerud AA, Ronneberg JA, Warrnberg F, Botling J, Busato F, Jovanovic J, et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010;12:R3.
Lo P-K, Lee JS, Liang X, Han L, Mori T, Fackler MJ, et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70:6047–58.
Catto JWF, Azzouzi A-R, Rehman I, Feeley KM, Cross SS, Amira N, et al. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol. 2005;23:2903–10.
Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41.
Dulaimi E, Hillinck J, de Caceres II, Al-Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin Cancer Res. 2004;10:6189–93.
Sabharwal A, Middleton MR. Exploiting the role of O6-methylguanine-DNA-methyltransferase (MGMT) in cancer therapy. Curr Opin Pharmacol. 2006;6:355–63.
Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307.
Sharma S, Salehi F, Scheithauer BW, Rotondo F, Syro LV, Kovacs K. Role of MGMT in tumor development, progression, diagnosis, treatment and prognosis. Anticancer Res. 2009;29:3759–68.
Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair. 2007;6:1079–1099.
Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204.
Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem Cell Biol. 2005;83:429–37.
Silber JR, Blank A, Bobola MS, Mueller BA, Kolstoe DD, Ojemann GA, et al. Lack of the DNA repair protein O6-methylguanine-DNA methyltransferase in histologically normal brain adjacent to primary human brain tumors. Proc Natl Acad Sci. 1996;93:6941–6.
Neto JC, Ikoma MM, Carvalho KC, Vassallo J, De Brot M, Gobbi H, et al. MGMT and PTEN as potential prognostic markers in breast cancer. Exp Mol Pathol. 2012;92:20–6.
Osanai T, Takagi Y, Toriya Y, Nakagawa T, Aruga T, Iida S, et al. Inverse correlation between the expression of O6-methylguanine-DNA methyl transferase (MGMT) and p53 in breast cancer. Jpn J Clin Oncol. 2005;35:121–5.
Isono S, Fujishima M, Azumi T, Hashimoto Y, Komoike Y, Yukawa M, et al. O6-methylguanine-DNA methyltransferase as a prognostic and predictive marker for basal- like breast cancer treated with cyclophosphamide- based chemotherapy. Oncol Lett. 2014;7:1778–84.
Lapidus RG, Ferguson AT, Ottaviano YL, Parl FF, Smith HS, Weitzman SA, et al. Methylation of estrogen and progesterone receptor gene 5′CpG islands correlates with lack of estrogen and progesterone receptor gene expression in breast tumors. Clin Cancer Res. 1996;2:805–10.
Skliris GP, Munot K, Bell SM, Carder PJ, Lane S, Horgan K, et al. Reduced expression of oestrogen receptor β in invasive breast cancer and its re- expression using DNA methyl transferase inhibitors in a cell line model. J Pathol. 2003;201:213–20.
Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003.
Acknowledgments
The authors are thankful to the University Grants Commission (UGC), New Delhi, India, for providing the funds that allowed us to carry out this research.
Conflicts of interest
The contributing authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
The upper panel of the representative images show HEK293 cells after 48 h of transient empty vector transfection (pB513B1) without human MGMT gene. These cells did not express MGMT as seen by the absence of MGMT antibody immunofluorescence. Whereas the human MGMT transfected (pB513B1-MGMT)HEK293 cells show strong nuclear immunofluorescence (green) as seen in the lower panel. (GIF 3384 kb)
ESM 2
(DOCX 11 kb)
Rights and permissions
About this article
Cite this article
Asiaf, A., Ahmad, S.T., Malik, A.A. et al. Protein expression and methylation of MGMT, a DNA repair gene and their correlation with clinicopathological parameters in invasive ductal carcinoma of the breast. Tumor Biol. 36, 6485–6496 (2015). https://doi.org/10.1007/s13277-015-3339-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3339-9