Abstract
Human urinary bladder cancer (UBC) is the fourth most common cancer and the eighth most common cause of cancer death in the USA. High mobility group box 3 (HMGB3), a member of a family of proteins containing one or more high mobility group DNA binding motifs, was reported to be overexpressed in a variety of human cancers. However, the expression and role of HMGB3 in human UBC remains unclear. Here, we found that UBC patients had upregulated HMGB at both mRNA and protein levels. Immunochemistry (IHC) evaluation of HMGB3 expression in 113 UBC clinical specimens showed that high expression of HMGB3 had positive correlation with UBC tumor size (P = 0.019), tumor WHO grade (P = 0.031), stage (P = 0.028), and lymph node metastasis (P = 0.017). Moreover, patients with higher HMGB3 expression showed a poorer overall survival rate than those with relatively low HMGB3 (P = 0.0079, log-rank test). Multivariate analysis revealed that HMGB3 expression is an independent prognostic marker. The UBC cancer cell proliferation and migration ability were measured by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) and wound healing assays, respectively. RNA interference of HMGB3 in UBC cell lines inhibited cancer cell growth and migration, along with the downregulation of PCNA and MMP2 protein levels. In sum, our data suggests HMGB3 may serve as an important oncoprotein and indicate that overexpression of HMGB3 in UBC could be used as a potential prognostic marker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An estimated 386,300 new cases and 150,200 deaths from urinary bladder cancer (UBC) occurred in 2008 worldwide [1]. Although enormous improvements have been made in UBC biomarker prediction and detection tools [2–4], UBC is still the fourth most common cancer and the eighth most common cause of cancer death in the USA [5]. UBC can be divided into two main phenotypic variants based on the different biological behaviors and prognoses. The 5-year survival rate for advanced invasive UBC is just 6 %, which is extremely low compared with the rate of noninvasive UBC (90 %) [6]. Different molecular mechanisms are the causes for the different outcomes, and identification of the key genetic events may help develop therapeutic targets for invasive UBC [7].
High mobility group box 3 (HMGB3) is a member of a family of proteins containing one or more high mobility group DNA-binding motifs. HMGB3 plays an important role in DNA replication, recombination, and repair [8]. In a mice leukemia model, HMGB3 was proven to be one of the key components in leukemia stem cell renewal through activation of MLL/Hox/Meis. Furthermore, the enriched expression of HMGB3 is linked with poor leukemia prognosis because of aberrantly self-renewing progenitor-like cancer stem cells (CSCs) [9]. HMGB3 overexpression was demonstrated in high-grade breast cancer [10]. HMGB3 messenger RNA (mRNA) levels were also increased in metastatic breast cancers and knockdown of HMGB3 decreased breast cancer cell invasiveness [11]. Upregulation of HMGB3 in gastric adenocarcinoma is associated with poor prognosis, and HMGB3 is involved in gastric cancer cell line proliferation, migration, and apoptosis [12, 13]. Recently, Song et al. found that HMGB3 expression in nonsmall cell lung cancer (NSCLC) tissue had positive correlation with clinicopathological features, including the survival of NSCLC patients [14]. Thus, HMGB3 plays a key role in leukemia, breast cancer, gastric cancer, and NSCLC. Moreover, studies show that HMGB3 contributes to cancer progression, such as CSCs, invasion, and migration.
To the best of our knowledge, the expression and role of HMGB3 in UBC have remained unclear. Here, we determined the expression levels of HMGB3 in UBC and compared protein expression data with clinicopathological parameters. Our results suggest that HMGB3 might be a potentially important protein for UBC proliferation and progression.
Material and methods
Patients and tissue specimens
A total of 113 primary UBCs and partial corresponding normal tissues were collected by the Department of Urology Medicine of Huai’an, First People’s Hospital using appropriate informed consent obtained after institutional review board approval. No patient underwent radiotherapy or chemotherapy prior to surgery. The data included patient age, gender, lymph nodal status, TNM stage, and histological type. The postoperative histological type was determined according to the seventh edition of the TNM classification. Follow-up information was obtained by phone investigations. The date of the last follow-up was April 11, 2014. Overall survival rates were calculated as the period from surgery until the date of death or last contact.
Immunohistochemistry
The IHC for HMGB3 (1:100; Abnova, Taipei, Taiwan) was performed on formalin-fixed, paraffin-embedded tissue sections using steam heat-induced epitope retrieval and the DAB chromogen (Boster, Wuhan, China). Nuclear HMGB3 expression was quantified using a ten-value intensity score (0–9) and the percentage (0–100 %) of the extent of reactivity by two pathologists. The whole field in each slide was examined for final quantification. An immunohistochemical expression score was obtained by multiplying the intensity and reactivity extension values, and these expression scores were used to determine expression levels.
Real-time reverse-transcription polymerase chain reaction analysis
Total RNA was extracted with Trizol® (Invitrogen Inc., Carlsbad, CA, USA). The expression levels of hmgb3 were detected by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). The primers are listed as follows: the forward primer 5′-GACAGGGACCTCTGGGTTTT-3′ and the reverse primer 5′-TGCCCAGACTAGCGAACAAT-3′; β-actin was used as an internal control. The primers for β-actin were 5′-AGCGAGCATCCCCCAAAGTT-3′ and 5′-GGGCACGAAGGCTCATCATT-3′. qPCR was performed using the SYBR® Green (TaKaRa Biotechnology Co. Ltd, Dalian, China) dye detection method on ABI StepOne Sequence Detection System under default conditions: 95 °C for 10 min, and 40 cycles of 95 °C for 5 s and 55 °C for 31 s. Comparative Ct method was used for quantification of the transcripts.
Western blotting assay
Twenty micrograms of total protein was electrophoresed on a 10 % SDS-PAGE gel, transferred onto polyvinylidene difluoride membrane, blocked, and then incubated overnight with the antibodies against HMGB3 (Abnova, Taipei, Taiwan), PCNA (Santa Cruz, CA), MMP2 and β-actin (Bioworld, China). Corresponding horseradish peroxidase-conjugated secondary antibody was then used on them at room temperature for 2 h. Protein bands were detected on a G:Box iChem Imager (Syngene, Cambridge, UK) using Pierce West Pico chemiluminescent detection system (Thermo Fisher Scientific Inc., Rockford, IL, USA).
Cell lines and siRNA transfection
Human UBC cell lines, 5637 and UMUC3 were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI1640 medium containing 10 % FBS (Hyclone). The small interfering RNAs (siRNAs) for HMGB3 and one negative control siRNA were synthesized by RiboBio Co., Ltd (Guangzhou, China). The sequence of the 23-nucleotide siRNA targeting the human HMGB3 sequence (5′-ugAGAAGGAUGUUGCUGACUAua-3′) was designed by online software provided by KRIBB siRNA project (http://sysbio.kribb.re.kr:8080/AsiDesigner/menuDesigner.jsf). Transfection was carried out using Lipofectamine 2000 (Invitrogen) as described before [15].
MTT assay
Cells were plated at a density of 2 × 103 cells/well in 96-well plates in 100-μL medium. After siRNA was transfected for 0, 24, 48, and 72 h, 10 μL MTT was added to each well. The mixture was incubated at 37 °C for 4 h in the dark, remove media and MTT, and 100 μL dimethyl sulfoxide (DMSO) added to each well. The absorption value at 490 nm was recorded.
Wound healing assay
Transfected cells in six-well plates were cultured until monolayer formed. Cell layers were wounded using a 200-μL pipette tip and cultured for another 24 h. Photographs were taken at time 0 and 24 h. Analysis of the wound closure was performed by NIH Image program and calculated by the percentage of the uncovered wound area at 24 h.
Statistical analysis
Results were expressed as mean ± SE. Chi-squared analysis was done to evaluate the significance of differences between the experimental groups. For a single comparison of two groups, Student’s t test was used. Survival analyses were performed using the log-rank test and Kaplan–Meier plots approach. For all analyses, the level of significance was set at P < 0.05.
Results
HMGB3 is overexpressed in UBC
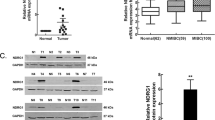
In this study, the HMGB3 mRNA level difference between normal bladder tissues (seven normal urothelium and five normal mucosa) and UBC cases (46 specimens) was investigated in public GEO profile dataset (GDS1479) [16]. As shown in Fig. 1a, HMGB3 expression was significantly lower in the normal group compared with the tumor group (P < 0.0001). We next performed quantitative RT-PCR to evaluate HMGB3 expression in 30 pairs of collected UBC samples and adjacent normal tissues. The results showed that HMGB3 expression was significantly upregulated in UBC cases compared with normal tissues (P < 0.0001) (Fig. 1b). Consistent with the mRNA level results, the Western blot showed that HMGB3 protein was similarly overexpressed in UBC cases (Fig. 1c). We performed IHC staining of HMGB3 in 63 normal bladder tissues and 113 UBC samples. Three representative UBC staining results are shown in Fig. 1d. Analysis of the IHC scores showed that HMGB3 was significantly upregulated in UBC cases (P < 0.0001, Fig. 1e). When we divided the UBC group into invasive and noninvasive subgroups, IHC scores showed that HMGB3 was obviously upregulated in the invasive subgroup compared with the noninvasive subgroup (P = 0.0177, Fig. 1f). Together, our data suggest that the overexpression of HMGB3 may play an important role in UBC initiation and progression.
HMGB3 is overexpressed in UBC samples. a HMGB3 mRNA levels in normal bladder tissues (seven normal urothelium and five normal mucosa) and 46 UBC tumors. b Quantitative RT-PCR evaluation of HMGB3 expression in 30 pairs of collected UBC samples and adjacent normal tissues. c Representative Western blot of HMGB3 protein expression from clinical specimens. N normal bladder tissue, T UBC tumor. d Representative HMGB3 staining in UBC specimens. Bar, 100 μm. e Box–whisker plots presenting the mean ± SEM value of HMGB staining values in a cohort of UBC tissues (n = 113) and normal bladder tissues (n = 63). f Box–whisker plots presenting the mean ± SEM value of HMGB staining values between noninvasive (n = 87) and invasive UBC (n = 26)
The overexpression of HMGB3 is related with clinicopathological and UBC patient survival
To identify the potential association between HMGB3 expression and tumor clinicopathological features in UBC, the patients were classified as low HMGB3 and high HMGB3 groups according to the HMGB3 immunoreactive intensity. A total of 46 (40.7 %) patients were placed in the high HMGB3 group, with over 20 % of tumor nuclei were positively stained (Fig. 1d, the right panel). As summarized in Table 1, HMGB3 expression was significantly correlated with tumor size (P = 0.019), tumor grade (P = 0.031), pN (P = 0.017), and pT (P = 0.028) status. Furthermore, overall survival (OS) of UBC patients was evaluated by Kaplan–Meier survival analysis as performed in previous reports [17, 18]. Log-rank test showed that high HMGB3 groups had significantly lower OS than the low HMGB3 group (P = 0.0079; Fig. 2). These results suggest that HMGB3 overexpression could be a promising prognostic marker for UBC patients. Multivariate analysis showed that HMGB3 (P = 0.008), N stages (P = 0.023), and TNM stages (P = 0.005) were independent prognostic factors in UBC patients (Table 2).
Downregulation of HMGB3 decreases UBC cell proliferation
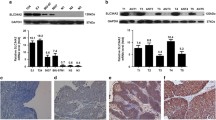
To evaluate the impact of HMGB3 on UBC cell proliferation, we knocked down HMGB3 by RNA interference in UBC cancer cell lines, 5637 and UMUC-3, which express HMGB3 (Fig. 3). RNA interference reduced HMGB3 mRNA levels to 27.6 and 31.0 % in 5637 and UMUC-3 cells, respectively, compared with cells transfected with control sequences (Fig. 3a, b). MTT assay showed that the HMGB3 gene silencing inhibited the proliferation of both 5637 and UMUC-3 cells at 48 and 72 h after transfection (Fig. 3a, b). Moreover, we examined the changes of the proliferation-related molecule, PCNA. We found reduced protein levels of PCNA when HMGB3 was knocked down in both cancer cell lines (Fig. 3c, d).
Suppression of HMGB3 inhibits UBC cell proliferation. qRT-PCR and MTT assay were used to detect effect of RNA interference against HMGB3 on cancer cell proliferation inhibition in 5637 (a) and UMUC-3 cells (b). *P < 0.05, ***P < 0.0001 compared with control siRNA. Western blotting assay analyzes the changes of HMGB3 and PCNA protein levels in c 5637and d UMUC-3 cells. siNC control siRNA, siHMGB3 HMGB3 siRNA
Knockdown of HMGB3 reduces UBC migration
The overexpression of HMGB3 showed a positive correlation with UBC lymph metastatic status and cancer stage (Table 1). Combined with the observation that invasive UBC has relatively higher HMGB3 expression compared with noninvasive (Fig. 1f), we proposed that HMGB3 was possibly be involved in UBC progression. As cell motility is a measurement of the metastatic potential of cancer [19], we performed wound healing assays to examine the role of HMGB3 in UBC cells’ migration. Confluent monolayers of cells were scratched and cultured for 24 h. As shown in Fig. 4a, b, 5637 and UMUC-3 cells transfected with negative control siRNA (siNC) spread into 85.1 and 83.0 % of wound area, respectively, while cells downregulated for HMGB3 spread into 49.7 and 58.7 % of the wound area. These results indicate that knockdown of HMGB3 significantly decreased migration of both UBC cell lines (Fig. 4a, b). To analyze the potential mechanism underlying HMGB3 abrogation-induced migration block, protein levels of MMP-2, an extracellular matrix enzyme involved in migration, was examined in HMGB3 silenced cancer cell lines. As shown in Fig. 4c, d, MMP2 protein levels were downregulated in both cancer cell lines with downregulation of HMGB3.
Effect of downregulation of HMGB3 on UBC cells migration activity in vitro. a 5637 cells and b UMU-3 transfected with siNC and siHMGB3 were wounded by a plastic tip. Cells were monitored under a microscope equipped with a camera at 0 and 24 h, and the wound healing was quantified by NIH Image program. HMGB3 and MMP2 protein levels changes were assessed by Western blotting in c 5637 and d UMUC-3 cells. siNC control siRNA, siHMGB3 HMGB3 siRNA
Discussion
As one of the most common cancers worldwide, the incidence of UBC has been increasing steadily [1]. Like other malignant tumors, the mechanism for carcinogenesis and progression of UBC remains unclear. Previous studies have suggested HMGB3, one of the HMGB family members, as an oncoprotein. HMGB3 has been studied in some types of cancers, including leukemia, breast cancer, gastric cancer, and NSCLC [13, 14, 20, 21], but little is known about the diversification of HMGB3 expression in UBC and its relation to clinicopathological parameters. In this study, we investigated HMGB3 expression in UBC. Compared with the normal group, the HMGB3 mRNA level in UBC was relatively high in both public data and our qRT-PCR data. We also detected the protein level of HMGB3 by IHC and Western blot. Data showed that HMGB3 was mainly localized in the nuclei of cancer cells, and the expression level of HMGB3 was higher in UBC than in normal tissues. Of note, HMGB3 expression has significantly higher expression in invasive UBC than noninvasive UBC. This is the first report demonstrating the overexpression of HMGB3 in UBC.
To assess the function of HMGB3 in UBC, we knocked down its gene expression in UBC cell lines, 5637 and UMUC-3. Our experiments showed that the downregulation of HMGB3 caused a decreased proliferation rate and migration ability. In UBC, PCNA was well documented to promote cancer cell proliferation [14, 22]. We detected downregulation of PCNA in 5637 and UMUC-3 cells knocked down for HMGB3. MMP2, which can degrade collagen IV, the major extracellular matrix component of the basement membrane, is implicated in nonphysiological tissue invasion and migration [23]. We found that the protein level of MMP2 was decreased in HMGB3-silenced 5637 and UMUC-3 cells. These results suggest that HMGB3 may influence cell proliferation and migration via the regulation of PCNA and MMP2 expression levels. However, the exact mechanism by which HMGB3 modulates these two proteins needs to be elucidated in future studies.
To further examine the role of HMGB3 in the development and progression of UBC, we analyzed the expression of HMGB3 in 113 UBC patients and found that HMGB3 overexpression was significantly associated with tumor size (P = 0.019), tumor grade (P = 0.031), pN (P = 0.017), and pT (P = 0.028) status. Combined with UBC cell data, overexpressed HMGB3 in UBC may accelerate tumor growth and migration. Our results indicate that HMGB3 may play significant roles in UBC progression, including tumor proliferation and migration. Moreover, HMGB3 expression in the invasive UBC group was higher than in the noninvasive group and the downregulation of HMGB3 decreases UBC cell migration. This data suggests that HMGB3 enhances UBC local cell invasion and metastasis.
We divided 113 UBC patients into high and low HMGB3 subgroups based on the median of IHC scores. We found that HMGB3 expression was associated with overall survival after surgery. The high HMGB3 patients had significantly low survival rate compared with the low HMGB3 group. Multivariate analyses of HMGB3 expression and other clinical prognostic factors in UBC patients found that HMGB3 expression could be a significant prognostic factor. In sum, HMGB3 expression might be a potentially useful prognostic marker in UBC.
Taken together, here, we first report overexpression of HMGB3 in UBC, especially in invasive UBC. The high expression of HMGB3 had positive correlation with clinicopathological characteristics and UBC patient survival. Reduced HMGB3 affected UBC cell proliferation and migration capabilities. These data suggest that HMGB3 could be a promising UBC predictive biomarker and potent therapeutic target for advanced invasive UBC.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Galustian C. Tools to investigate biomarker expression in bladder cancer progression. BJU Int. 2013;112(3):404–6.
Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374(9685):239–49.
Prasad SM, Decastro GJ, Steinberg GD, Medscape. Urothelial carcinoma of the bladder: definition, treatment and future efforts. Nat Rev Urol. 2011;8(11):631–42.
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29.
Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5(9):713–25.
Vishnu P, Mathew J, Tan WW. Current therapeutic strategies for invasive and metastatic bladder cancer. Onco Targets Ther. 2011;4:97–113.
Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799(1–2):101–13.
Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–40.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507.
Elgamal OA, Park JK, Gusev Y, Azevedo-Pouly AC, Jiang J, Roopra A, et al. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS One. 2013;8(10):e76402.
Tang HR, Luo XQ, Xu G, Wang Y, Feng ZJ, Xu H, et al. High mobility group-box 3 overexpression is associated with poor prognosis of resected gastric adenocarcinoma. World J Gastroenterol. 2012;18(48):7319–26.
Gong Y, Cao Y, Song L, Zhou J, Wang C, Wu B. HMGB3 characterization in gastric cancer. Genet Mol Res. 2013;12(4):6032–9.
Song N, Liu B, Wu JL, Zhang RF, Duan L, He WS, et al. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34(5):2599–603.
Zhou J, Bi H, Zhan P, Chang C, Xu C, Huang X, et al. Overexpression of HP1gamma is associated with poor prognosis in non-small cell lung cancer cell through promoting cell survival. Tumour Biol. 2014;35(10):9777–85.
Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64(11):4040–8.
Lu DY, Leung YM, Cheung CW, Chen YR, Wong KL. Glial cell line-derived neurotrophic factor induces cell migration and matrix metalloproteinase-13 expression in glioma cells. Biochem Pharmacol. 2010;80(8):1201–9.
Tong ZT, Wei JH, Zhang JX, Liang CZ, Liao B, Lu J, et al. AIB1 predicts bladder cancer outcome and promotes bladder cancer cell proliferation through AKT and E2F1. Br J Cancer. 2013;108(7):1470–9.
Zhu F, Zhang Z, Wu G, Li Z, Zhang R, Ren J, et al. Rho kinase inhibitor fasudil suppresses migration and invasion though down-regulating the expression of VEGF in lung cancer cell line A549. Med Oncol. 2011;28(2):565–71.
Petit A, Ragu C, Della-Valle V, Mozziconacci MJ, Lafage-Pochitaloff M, Soler G, et al. NUP98-HMGB3: a novel oncogenic fusion. Leukemia. 2010;24(3):654–8.
Nemeth MJ, Kirby MR, Bodine DM. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci U S A. 2006;103(37):13783–8.
Malmstrom PU, Wester K, Vasko J, Busch C. Expression of proliferative cell nuclear antigen (PCNA) in urinary bladder carcinoma. Evaluation of antigen retrieval methods. APMIS. 1992;100(11):988–92.
Gao X, Mi Y, Ma Y, Jin W. LEF1 regulates glioblastoma cell proliferation, migration, invasion, and cancer stem-like cell self-renewal. Tumour Biol. 2014;35(11):11505–11.
Acknowledgments
The study was supported by a grant from the “Twelve-Five Plan” of the Major Program of Jiangsu Medical Science and Technique Development Foundation and Young Professionals Foundation of Huai’an First People’s Hospital.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Minghui Li and Yong Cai contributed equally to this work.
Rights and permissions
About this article
Cite this article
Li, M., Cai, Y., Zhao, H. et al. Overexpression of HMGB3 protein promotes cell proliferation, migration and is associated with poor prognosis in urinary bladder cancer patients. Tumor Biol. 36, 4785–4792 (2015). https://doi.org/10.1007/s13277-015-3130-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3130-y