Abstract
SET and MYND domain-containing protein 3 (SMYD3), a histone methyltransferase, plays a key function in the progression of human cancer. However, the role of SMYD3 in gastric carcinoma carcinogenesis has yet to be elucidated. This study aimed to determine the relationships of SMYD3 expression with clinicopathological characteristics and prognosis in gastric carcinoma. The expression of SMYD3 was detected by real-time quantitative reverse transcription PCR and Western blot in gastric carcinoma (GC) cell lines, normal gastric mucosa cell line, GC tissues, and adjacent non-tumor tissues. SMYD3 expression in tissue sections of 180 gastric carcinoma samples were evaluated using immunohistochemistry. The staining results were compared with clinicopathological characteristics and to the outcome of patients. The expression levels of SMYD3 messenger RNA (mRNA) and protein in GC tissues were both higher than those in adjacent non-tumor tissues (p < 0.05). SMYD3 mRNA and protein expression levels were higher in GC cell lines MKN28, SGC7901, and MGC803 than normal gastric mucosa cell line GES-1. SMYD3 expression in gastric carcinoma was significantly correlated with primary tumor size (p < 0.001), lymph node metastasis (p < 0.001), and TNM stage (p = 0.011). Degree of differentiation [hazard ratio (HR) = 5.113; p = 0.006], serosal invasion (HR = 2.074; p = 0.024), lymph node metastasis (HR = 1.354; p < 0.001), and SMYD3 expression (HR = 0.564; p = 0.004) were identified as the independent factors of the overall survival (OS) in all enrolled GC patients. For patients with positive lymph node metastasis, degree of differentiation (HR = 5.974; p = 0.015), lymph node metastasis (HR = 1.257; p < 0.001), and SMYD3 expression (HR = 0.529; p = 0.004) were the independent prognostic factors of the OS. SMYD3 performed an important function in the aggressiveness of gastric carcinoma and may act as a promising target for prognostic prediction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epigenetics, which is defined as heritable changes in gene expression that are not coded in the DNA sequence itself, is increasingly linked with tumorigenesis [1, 2]. Among epigenetic regulatory ways, histone methylation has demonstrated power of modifications over the DNA gene. Histone modifications occur at selected residues and multiple modifications that function in a combinational or sequential fashion, in single or multiple tails, and dictate “histone codes” that are closely linked to the biological consequences [3]. Recent studies identified the SET and MYND domain-containing protein 3 (SMYD3), which possessed histone methyltransferase activity responsible for catalyzing the methylation of histone H3 at K4 [4]. SMYD3 contained a SET domain crucial for HMT activity, as well as an MYND-type zinc-finger domain (zf-MYND) domain, which is common to developmental proteins [5, 6].
SMYD3 promotes dimethyltransferase and trimethyltransferase in histone H3-K4, eliciting the oncogenic effect of SMYD3 by activating the transcription of its downstream target genes [7]. SMYD3 interacts with its binding motif 5′-CCCTCC-3′ in the promoter region of its target genes. These downstream genes include several oncogenes (e.g., N-Myc, CrkL, Wnt10b, RIZ, and hTERT), as well as genes involved in the control of the cell cycle (e.g., cyclin G1 and CDK2) and signal transduction (e.g., STAT1, MAP3K11, and PIK3CB). The functions of such downstream genes are involved in numerous aspects of the process of cell growth and apoptosis [8]. Enhanced expression of SMYD3 gene is essential for the growth, adhesion, and migration of cancer cells, whereas suppression of SMYD3 by RNAi or other reagents inhibit cell proliferation and migration [9, 10]. SMYD3 was found to be overexpressed in various cancers [11–13]. In addition, SMYD3 expression was shown to be enhanced in hepatocellular carcinoma (HCC) and involved in the growth of HCC cells. Conversely, depletion of SMYD3 reduced cell growth and migration and induced cell apoptosis [11]. Overexpressed levels of SMYD3 had been observed in breast cancer tissues and cell lines [12]. Wang et al. [13] revealed that downregulation of SMYD3 expression resulted in inhibition of cell growth, colony formation, migration, invasion, and induction of apoptosis. In addition, these researchers confirmed that SMYD3 was essential for the proliferation and migration in HeLa cells.
SMYD3 promoted the progression of cancer via two mechanisms. On the one hand, SMYD3 catalyzed histone methylation, mainly bi/tri-methylation of histone H3 at lysine 4 (H3-K4), and rendered chromatin to be more accessible. On the other hand, SMYD3 interacted with its binding motif, CCCTCC or GGAGGG, in the promoter of the target genes and initiated transcription through association with RNA polymerase II and RNA helicase [14]. However, the carcinogenic role of SMYD3 in gastric carcinoma (GC) remained unknown. In this study, the expression of SMYD3 in GC was estimated using real-time quantitative reverse transcription PCR (qRT-PCR), Western blot analysis, and immunohistochemistry. In addition, we identified the relationships between SMYD3 expression and clinicopathological characteristics, as well as its relation with the overall survival (OS) of patients.
Materials and methods
Ethics statement
This research was approved by the Ethics Committee of the Tianjin Medical University, and written informed consent was obtained from each patient involved in the study.
Specimens
Specimens of 180 GC patients after potentially curative gastrectomy procedures at the Tianjin Medical University Cancer Hospital from January 2003 to September 2007 were included in this study. Eligibility criteria for this study included the following: (1) histologically proven adenocarcinoma, (2) no history of gastrectomy or other malignancy, (3) no distant metastasis or peritoneal dissemination, (4) the number of dissected lymph nodes greater than or equal to 15, (5) no patients died during the initial hospital stay or for 1 month after surgery, and (6) availability of complete follow-up data. Of these patients, 109 (60.6 %) were male and 71 (39.4 %) were female. Ages ranged between 23 and 79 years, with a mean age of 57.74 ± 11.32 years. According to 7th Union for International Cancer Control/American Joint Committee on Cancer TNM classification for GC, 130 patients (72.2 %) present lymph node metastasis. The mean number of dissected lymph nodes was 23.53 ± 8.19 (range, 15–66), and the mean number of the metastatic lymph nodes was 4.70 ± 5.30 (range, 0–35).
To investigate messenger RNA (mRNA) and protein expression of SMYD3 in gastric carcinoma, fresh GC tissues and adjacent non-tumor tissues were collected from 40 gastric carcinoma patients between January 2013 and July 2013. After surgical resection, the fresh tissue samples were immediately frozen at −80 °C until RNA and protein extraction. Both tumor tissue and adjacent non-tumor tissue, which was located more than 5 cm away from the gastric carcinoma, were sampled and verified by pathological examination. Table 1 shows the clinicopathological characteristics of 40 GC patients.
Gastric carcinoma cell lines, normal gastric mucosa cell line
GC cell lines MKN28, SGC7901, and MGC803 were purchased from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human normal gastric mucosa cell line GES-1 was purchased from Biowit Technologies, Ltd. (Shenzhen, China). SGC7901, MKN28, MGC803, and GES-1 cell lines were maintained at 37 °C in a humidified atmosphere at 5 % CO2 and 95 % air in RPMI 1640 (Thermo Electron Corporation, Beijing, China) with 10 % (v/v) FBS (Life Tech, Mulgrave Victoria, Australia) and penicillin–streptomycin (10,000 IU/mL penicillin and 20 mg/mL streptomycin; Roche, Swiss).
Extraction of total RNA and real-time quantitative PCR
We evaluated the mRNA expression of SMYD3 in GC cell lines, normal gastric mucosa cell line, and 40 paired gastric carcinoma tissues and adjacent non-tumor tissues by qRT-PCR analysis. Total RNA was extracted from frozen tissue using TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s protocol. Reverse transcription was performed in a 25 mL reaction volume with 2 mg total RNA treated with 0.5 mg of Oligo (dt), 200 U M-MLV reverse transcriptase, 25 U of RNase inhibitor, and 2.5 mM of dNTP to synthesize first-strand complementary DNA (cDNA) (Promega, USA), according to the manufacturer’s recommendations. The reaction system was incubated at 70 °C for 5 min (primer annealing) and 42 °C for 1 h (synthesis). Resulting cDNA was stored at −20 °C. The resulting cDNA was then subjected to real-time quantitative PCR for evaluation of the relative mRNA levels of SMYD3 and GAPDG (as internal control) with the following primers: SMYD3 forward (5′-CCCAGTATGTCTTTGCTGAATCAC-3′) and reverse (5′-ACTTCCAGTGCGCCTTCAGCTC-3′); and human GAPDH forward (5-ATTCAACGGCACAGTCAAGG-3′) and reverse (5′-GCAGAAGGGGCGGAGATGA-3′). Gene special amplification was performed in an ABI PRISM 7900HT real-time PCR system (Life Technologies, USA) with a 20 μL PCR mix containing 2 μL of cDNA, 10 μL of 2× SYBR Green PCR Master Mix (Invitrogen, USA), and 200 nM of the appropriate primers. The mixture was preheated for 5 min at 95 °C, followed by 30 cycles of amplification (30 s at 94 °C, 30 s at 48 °C, and 1 min at 72 °C) and a final elongation step of 72 °C for 10 min. All experiments were performed in triplicate, after which the average was calculated. The relative quantification of SMYD3 mRNA expression was normalized to GAPDH value (2−ΔΔCT method).
Western blot analysis
Frozen tissue samples from patients with gastric carcinoma, including the tumor and non-tumor tissue, were homogenized in RIPA lysis buffer for 30 min, and the lysates were cleared by centrifugation (12,000 rpm) at 4 °C for 15 min. Total protein extracts were separated on a 10 % sodium dodecyl sulfate polyacrylamide gel electrophoresis and were electrotransferred to PVDF membranes. After blocking nonspecific binding sites for 60 min with 5 % nonfat milk, the membranes were incubated overnight at 4 °C with a primary rabbit antihuman SMYD3 polyclonal antibody (Cell Signaling, #12859, 1:1,000 dilution). The membranes were washed three times for 15 min with phosphate-buffered saline (PBS)-T and probed with a horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin antibodies (ZhongShan Biotechnology, SP-9001, 1:2,000 dilution) for 60 min at room temperature. The membranes were then washed three times with PBS-T for 10 min. Immunocomplexes were visualized by enhanced chemiluminescence system.
Immunohistochemistry analysis
Formalin-fixed tissues were embedded in paraffin, sectioned at 4 μm, and mounted on silane-coated slides for immunohistochemistry analysis. The sections were deparaffinized with dimethylbenzene and rehydrated through 100, 95, 90, 80, and 75 % ethanol. Antigen retrieval treatment was conducted at 95 °C for 20 min in 1 × 10−5 mol/mL sodium citrate buffer (pH 6.0), and endogenous peroxidases were blocked using 3 % hydrogen peroxide for 15 min at room temperature. The sections were washed in PBS and blocked with 10 % goat serum (ZhongShan Biotechnology) for 30 min and then incubated with rabbit anti-human SMYD3 polyclonal antibody (Cell Signaling, #12859, 1:50 dilution) in a humidified chamber at 4 °C overnight. Following three additional washes using PBS, the sections were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. Finally, the visualization signal was developed with 3,3′-diaminobenzidine solution, and all slides were counterstained with 20 % hematoxylin. The slides were dehydrated and mounted on cover slips. For negative controls, PBS was used in place of the primary antibody.
All immunostained sections were estimated in a blinded manner by two independent pathologists. Cells were positive for SMYD3 protein when cell cytoplasm was stained. Each slide was evaluated under two fields at 400× magnification, and 100 cells were counted. We used a scoring standard for SMYD3 protein expression, and both distribution and intensity were considered. The staining distribution of SMYD3 was evaluated with the percentage of stained cells, which was scored as follows: 0, <5 %; 1, 5–25 %; 2, 26–50 %; 3, 51–75 %; and 4, 76–100 %. Staining intensity was scored as follows: 0, no staining; 1, buff; 2, buffy; and 3, puce. When the multiplication product of the two scores was ≥3, the samples were considered positively stained.
Follow-up
After surgery, all patients were followed up every 6 months for 2 years, then every year or until death. The median follow-up for the entire cohort was 52 (range, 3–89) months. Ultrasound, computed tomographic scans, chest X-ray, and endoscopy were performed at every visit.
Statistical analysis
All statistical analyses were carried out using SPSS software 17.0. A paired sample t-test was used to compare SMYD3 mRNA and protein levels in gastric carcinoma tissue samples with those of adjacent non-tumor tissue samples. Associations between SMYD3 expression and clinicopathological characteristics were analyzed using chi-square test. Survival analysis was performed by Kaplan–Meier method and compared by log-rank test. Multivariate analysis by Cox proportional hazard regression model was performed to find the potential independent prognostic factors in gastric carcinoma. p < 0.05 was regarded as statistically significant.
Result
Expression of SMYD3 mRNA in GC and adjacent non-tumor tissues and cell lines
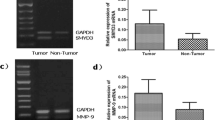
SMYD3 mRNA expression levels were respectively detected in tumor tissue specimens of GC and adjacent non-tumor tissue specimens from 40 patients. The relative mRNA expression values of SMYD3 in GC tissues were significantly higher than those in adjacent non-tumor tissues (0.12 ± 0.65 vs 0.05 ± 0.27, p < 0.001) (Fig. 1).
In cell lines, SMYD3 mRNA expression levels were higher in GC cell lines MKN28, SGC7901, and MGC803 than in normal gastric mucosa cell line GES-1 (0.13, 0.26, and 0.37 vs 0.06).
Western blot analysis for SMYD3 protein expression in GC and adjacent non-tumor tissues and cell lines
Protein expression levels of SMYD3 were detected in tumor tissue specimens of GC and adjacent non-tumor tissues from 40 patients by Western blot, simultaneously. The results show a SMYD3 band at the expected size of 42 kDa (Fig. 2a). The relative protein expression values of SMYD3 in GC tissues were significantly higher than those in adjacent non-tumor tissues (1.23 ± 0.64 vs 0.39 ± 0.20, p < 0.001) (Fig. 2b).
a Western blot analysis of SMYD3 protein expression in gastric carcinoma tissue and adjacent non-tumor tissue. b Relative protein expression of SMYD3 in gastric carcinoma tissues and adjacent non-tumor tissues assessed by Western blotting. Differences were analyzed with a paired sample t-test (p < 0.001). c Western blot analysis of SMYD3 protein expression in human normal gastric mucosa cell line GES-1 and gastric carcinoma cell lines MKN28, SGC7901, and MGC803
In cell lines, the protein expression levels were higher in GC cell lines MKN28, SGC7901, and MGC803 than in normal gastric mucosa cell line GES-1 (0.62, 1.07, and 1.34 vs 0.39) (Fig. 2c).
Relationship between SMYD3 expression in GC tissues and various clinicopathological variables
To elucidate the biological significance of SMYD3 in gastric carcinoma, we examined the immunohistochemical expression of SMYD3 in GC tissues. SMYD3 protein was mainly localized in the cytoplasm of GC cells, and positive staining was not seen in the smooth muscles, vessels, and stromal fibroblasts (Fig. 3). According to immunohistochemical results, we correlated SMYD3 expression with clinicopathologic characteristics (Table 2). Our analyses showed that positive SMYD3 expression was significantly correlated with larger size of the primary tumor (p < 0.001), greater lymph node metastasis (p < 0.001), and advanced TNM stage (p = 0.011). By contrast, positive SMYD3 expression was not associated with sex, age at surgery, location of primary tumor, degree of differentiation, Lauren’s classification, and serosal invasion (p > 0.05) (Table 2).
Survival analysis for gastric carcinoma patients
Table 3 shows the results of survival analysis of 180 GC patients. Univariate analysis showed significant relationships between the OS and location of primary tumor, degree of differentiation, serosal invasion, lymph node metastasis, TNM stage, and SMYD3 expression but not with sex, age at surgery, size of primary tumor, and Lauren’s classification of tumor. Degree of differentiation [hazard ratio (HR) = 5.113; p = 0.006], serosal invasion (HR = 2.074; p = 0.024), lymph node metastasis (HR = 1.354; p < 0.001), and SMYD3 expression (HR = 0.564; p = 0.004) were identified as the independent factors of OS in all enrolled GC patients following multivariate analysis (Cox proportional hazards model) (Fig. 4). Clinically, GC patients with negative SMYD3 expression presented significantly better 5-year survival rate (5-YSR) than those with positive SMYD3 expression.
Furthermore, we examined the prognostic value of SMYD3 expression in patients with positive lymph node metastasis. Univariate analysis showed significant relationships between OS and location of the primary tumor, degree of differentiation, lymph node metastasis, and SMYD3 expression. Multivariate Cox regression analysis showed that degree of differentiation (HR = 5.974; p = 0.015), lymph node metastasis (HR = 1.257; p < 0.001), and SMYD3 expression (HR = 0.529; p = 0.004) were the independent prognostic factors for OS (Fig. 5, Table 4).
Discussion
Although the rate of GC has declined over the last 50 years, GC remains to be the fourth most common cancer worldwide, with a total of 989,600 new cases and 738,000 deaths estimated in 2008 [15]. Recent progress in early diagnosis, surgical techniques, and perioperative management has improved patient satisfaction and outcomes; however, GC remains a major clinical challenge because of its high prevalence, poor prognosis, and limited treatment options [16, 17]. The etiology of GC is a complex process that involves activation of oncogenes and inactivation of tumor suppressor genes at different stages, but its exact pathogenesis remains unclear. Thus, identifying marker genes that will aid GC prognosis evaluation is a critically imminent issue.
SET-domain-containing protein, a class of lysine histone methyltransferases, has been regarded as an important factor in carcinogenesis [18]. SMYD3 is a novel SET-domain-containing protein. Several studies demonstrated that SMYD3 specifically methylates histone H3 at lysine 4 and activates the transcription of a set of downstream genes containing a “5′-CCCTCC-3′” sequence in the promoter region [8]. The biological function of SMYD3 depends to a large extent on the activity of downstream genes [19]. Enhanced expression of SMYD3 is essential for the growth of numerous cancer cells [20]. Overexpression of the SMYD3 gene affects cell viability, adhesion, migration, and invasion [13], whereas knocking down SMYD3 gene expression in cervical carcinoma cells inhibited cell proliferation, migration, and invasion [21]. Jia et al. [22] found that low levels of SMYD3 expression in tumor cells reduced the biological function of HGF and inhibited cancer cell migration and invasion. Wang et al. [23] analyzed 200 HCC patients and 261 healthy controls and found that SMYD3 polymorphism was not associated with the occurrence and metastasis of HCC in the Chinese population. In this study, we investigated SMYD3 expression in GC and its correlation with clinicopathological characteristics of patients, including OS.
We first investigated the SMYD3 mRNA and protein expression in GC specimens and cell lines by qRT-PCR and Western blot analysis, respectively. The expression levels of SMYD3 mRNA and protein in GC tissues and cell lines were both significantly higher than those in adjacent non-tumor tissues and normal gastric mucosa cell line. These observations suggested that SMYD3 may function as an oncogene in GC and suggested that SMYD3 may perform an important function in GC tumorigenesis.
Besides the results mentioned above, the SMYD3 expression data obtained from immunohistochemistry detection were analyzed for correlation with clinicopathological characteristics. We found that positive SMYD3 expression was significantly associated with larger size of primary tumor, greater lymph node metastasis, and advanced TNM stage, indicating that SMYD3 overexpression may affect the invasion, metastasis, and progression of GC. In addition, the results imply that this overexpression could be used to indicate the aggressive behavior of carcinomas.
To assess the prognostic value of SMYD3 expression in GC, we analyzed the expression of the protein in patients by using OS and Cox regression analysis. Our results identified the location of primary tumor, degree of differentiation, serosal invasion, lymph node metastasis, TNM stage, and SMYD3 expression as independent factors of OS. Patients with positive SMYD3 expression presented lower 5-YSR than those with negative SMYD3 expression. These findings suggested that GC patients with SMYD3 overexpression may be a high-risk group with poor survival and may require more aggressive additional postoperative systemic therapy.
More than 50 % of GC patients were accompanied by lymph node metastases at diagnosis, which led 5-YSR of GC to be less than 30 % [24]. Survival rates markedly decreased with increasing number of metastatic lymph nodes [25]. Hochwald et al. [26] reported that nodal status was the most powerful prognostic factor of outcome for GC. Deng et al. [27] analyzed 196 lymph node-positive GC patients and found that the number of metastatic lymph nodes and ratio of metastatic lymph nodes showed significant correlations with OS. In this study, a positive correlation was found between SMYD3 expression and lymph node metastasis. Meanwhile, we also found that lymph node metastasis and SMYD3 expression were the independent prognostic factors for the OS for patients with positive lymph node metastasis by multivariate Cox regression analysis. We deduced that SMYD3 expression may be a promising prognostic factor of survival in patients with positive lymph node metastasis.
Our study provided evidence that the expression of SMYD3 is elevated in GC and is related to tumor invasion and metastasis. The expression of the protein may serve as a potential prognostic biomarker in gastric carcinogenesis. However, the molecular mechanisms involved in the regulation of SMYD3 expression in GC are still not fully understood. Future studies in this field are necessary because greater understanding of SMYD3 function in malignant transformation presents the potential to improve prognosis in GC patients.
References
Jaskelioff M, Peterson CL. Chromatin and transcription: histones continue to make their marks. Nat Cell Biol. 2003;5:395–9.
Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83.
Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5.
Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–40.
Xu S, Wu J, Sun B, Zhong C, Ding J. Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding. Nucleic Acids Res. 2011;39:4438–49.
Jakowlew SB, Breathnach R, Jeltsch JM, Masiakowski P. Chambon P sequence of the pS2 mRNA induced by estrogen in the human breast cancer cell line MCF-7. Nucleic Acids Res. 1984;12:2861–78.
Kunizaki M, Hamamoto R, Silva FP, Yamaguchi K, Nagayasu T, Shibuya M, Nakamura Y, Furukawa Y. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 67:10759–10765.
Luo XG, Xi T, Guo S, Liu ZP, Wang N, Jiang Y, et al. Effects of SMYD3 overexpression on transformation, serum dependence, and apoptosis sensitivity in NIH3T3 cells. IUBMB Life. 2009;61:679–84.
Dong SW, Zhang H, Wang BL, Sun P, Wang YG, Zhang P. Effect of the downregulation of SMYD3 expression by RNAi on RIZ1 expression and proliferation of esophageal squamous cell carcinoma. Oncol Rep. 2014;32:1064–70.
Sponziello M, Durante C, Boichard A, Dima M, Puppin C, Verrienti A, et al. Epigenetic-related gene expression profile in medullary thyroid cancer revealed the overexpression of the histone methyltransferases EZH2 and SMYD3 in aggressive tumours. Mol Cell Endocrinol. 2014;392:8–13.
Chen LB, Xu JY, Yang Z, Wang GB. Silencing SMYD3 in hepatoma demethylates RIZI promoter induces apoptosis and inhibits cell proliferation and migration. World J Gastroenterol. 2007;13:5718–24.
Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–8.
Wang SZ, Luo XG, Shen J, Zou JN, Lu YH, Xi T. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cell growth and invasion in vitro. BMB Rep. 2008;41:294–9.
Kim H, Heo K, Kim JH, Kim K, Choi J, An W. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J Biol Chem. 2009;284:19867–77.
Yuan Y. A survey and evaluation of population-based screening for gastric cancer. Cancer Biol Med. 2013;10:72–80.
Yang L. Incidence and mortality of gastric carcinoma in China. World J Gastroenterol. 2006;12:17–20.
Zhang XM, Zhou C, Gu H, Yan L, Zhang GY. Correlation of RKIP, STAT3 and cyclin D1 expression in pathogenesis of gastric cancer. Int J Clin Exp Pathol. 2014;7:5902–8.
He C, Xu J, Zhang J, Xie D, Ye H, Xiao Z, et al. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum Pathol. 2012;43:1425–35.
Cock-Rada AM, Medjkane S, Janski N, Yousfi N, Perichon M, Chaussepied M, et al. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 2012;72:810–20.
Ren TN, Wang JS, He YM, Xu CL, Wang SZ, Xi T. Effects of SMYD3 over-expression on cell cycle acceleration and cell proliferation in MDA-MB-231 human breast cancer cells. Med Oncol. 2011;28:S91–8.
Wang SZ, Luo XG, Shen J, Zou JN, Lu YH, Xi T. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cells growth and invasion in vitro. BMB Rep. 2008;41:294–9.
Zou JN, Wang SZ, Yang JS, Luo XG, Xie JH, Xi T. Knockdown of SMYD3 by RNA interference down-regulates c-Met expression and inhibits cells migration and invasion induced by HGF. Cancer Lett. 2009;280:78–85.
Wang XQ, Miao X, Cai Q, Garcia-Barcelo MM. Fan STSMYD3 tandem repeats polymorphism is not associated with the occurrence and metastasis of hepatocellular carcinoma in a Chinese population. Exp Oncol. 2007;29:71–3.
Hundahl SA, Phillips JL, Menck HR. The national cancer data base report on poor survival of U.S. gastric carcinoma patients treated with gastrectomy: Fifth edition American joint committee on cancer staging, proximal disease, and the “different disease” hypothesis. Cancer. 2000;88:921–32.
Abe N, Watanabe T, Suzuki K, et al. Risk factors predictive of lymph node metastasis in depressed early gastric carcinoma. Am J Surg. 2002;183:168–72.
Hochwald SN, Kim S, Klimstra DS, Brennan MF, Karpeh MS. Analysis of 154 actual 5-year survivors of gastric carcinoma. J Gastrointest Surg. 2000;4:520–5.
Deng J, Liang H, Sun D, Pan Y. The prognostic analysis of lymph node-positive gastric carcinoma patients following curative resection. J Surg Res. 2010;161:47–53.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 31470816).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, Y., Luo, X., Deng, J. et al. SMYD3 overexpression was a risk factor in the biological behavior and prognosis of gastric carcinoma. Tumor Biol. 36, 2685–2694 (2015). https://doi.org/10.1007/s13277-014-2891-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2891-z