Abstract
In the world, hepatocellular carcinoma (HCC) is one of the most common and most lethal cancers. Currently, standard therapy for unresectable HCC is a local–regional therapy with transarterial chemoembolisation (TACE). In this study, we sought to assess whether plasma circulating microRNAs (miRNAs) can be used to predict the prognosis of HCC patients receiving the TACE treatment. Firstly, we systematically examined TACE therapeutic effectiveness-related circulating miRNAs through miRNA Profiling Chips. As a result, we identified 19 circulating miRNAs to be significantly differentially expressed between the TACE-response group and the TACE-nonresponse group. In the second stage, we performed quantitative analyses of these candidate miRNAs in additional HCC patients treated with TACE and validated two of the aforementioned 19 miRNAs (miR-1285-3p and miR-4741) as candidate biomarkers for predicting prognosis of TACE. Interestingly, we found that miR-1285-3p could directly repress JUN oncogene expression in HCC cells, indicating miR-1285-3p could act as a potential tumor suppressor. In conclusion, our data indicate that circulating miR-1285-3p and miR-4741 was predictive of response to TACE therapy in HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the world, hepatocellular carcinoma (HCC) is one of the most common and most lethal cancers [1, 2]. When diagnosed, only about 30 % HCC patients are eligible for receiving surgery. Currently, standard therapy for unresectable HCC is local–regional therapy with transarterial chemoembolisation (TACE) [1–4]. While blocking the primary artery from feeding the tumor cells, TACE also concentrates on chemotherapeutic agents at the tumor site. Therefore, TACE is an effective and widely used approach to prolong the survival of HCC patients. Multiple baseline clinical factors, including liver function, performance status, and tumor stage could affect treatment outcomes of TACE among HCC patients [3, 4]. There were few studies on the identification of biomarkers for the prediction of HCC patient outcomes and prognosis after TACE treatment. Novel biomarkers are warrants to be discovered to improve patient clinical outcomes and tailor the treatments based on the individual profile of each patient.
As a class of regulatory small noncoding RNA, microRNAs (miRNAs) could bind to target messenger RNA (mRNA) and regulate posttranscriptional gene expression [5, 6]. During this regulation process, miRNAs can pair to complementary binding sites within the 3′ untranslated region (3′-UTR) of hundreds of target mRNAs [5, 6]. As a result, miRNAs impair the translation or promote the degradation of their target mRNAs and, thus, are involved in crucial processes [5–7]. Genetic changes interrupting regulation of miRNAs on their target genes (oncogenes or tumor suppressors) are associated with multiple malignants [8–11]. Previous studies have reported important roles of miRNAs during HCC development and progression [12–16]. Most previous studies on miRNA expression have been performed on tissue samples. However, accumulated evidences have shown diagnostic and prognostic potential for circulating miRNAs [17–21] because tumor-derived miRNAs can be present and exist stably in blood [17–21].
In this study, we sought to assess whether plasma circulating miRNAs can be used to predict the prognosis of HCC patients receiving the TACE treatment. To the best of our knowledge, this is the first study to investigate the role of circulating miRNAs in predicting HCC prognosis. We systematically examined TACE therapeutic effectiveness-related circulating miRNAs in a two-stage way. In the first stage, we determined differentially expressed plasma circulating miRNAs in TACE-treated HCC patients with different therapeutic effectiveness through miRNA microarrays. In the second stage, we then performed quantitative analyses of these candidate miRNAs in additional HCC patients treated with TACE. Moreover, we investigated the potential role of miR-1285-3p in HCC cells through a series of biochemistry assays.
Materials and methods
Study population
A total of 97 unresectable HCC patients treated with TACE were recruited into the current study between October 2012 and May 2013 at the Department of Intervention Surgery, Shandong Cancer Hospital, Shandong Academy of Medical Sciences (Jinan, Shandong Province, China). All HCC patients had no history of other cancers or cancer-related therapy. There was no restriction on age, sex, or disease stage for patient recruitment. All patients received TACE as the first-line treatment and were Han Chinese. During TACE treatment, the chemo regimens include 30 mg epirubicin, 30 mg cyano-camptothecin, and 40 mg cisplatin. All enrolled cases received the same chemo regimens. All blood samples were collected prior to TACE treatment. This study was approved by the Institutional Review Board of Shandong Cancer Hospital, Shandong Academy of Medical Sciences, and the signed informed consent was obtained from each patient.
RNA isolation and genome-wide microRNA expression profiling
The whole blood was separated into plasma and cellular fractions within 24 h after sample collection. Plasma-derived total RNA was isolated using mirVana™ PARIS™ Kit (Ambion) according to the protocol supplied by the manufacturer for liquid specimens. Agilent 2100 Electrophoresis Bioanalyzer was used to examine quality of total RNA samples. Expression of miRNA was profiled utilizing Human miRNA OneArray® miRNA Profiling Chip (version HmiOA4.1, based on Sanger’s miRBase database release 19) (Phalanx Biotech). For miRNA profiling, we used two pooled serum samples; one was pooled from the five patients in the group with good response to TACE treatment and the other was from the five patients in the group with no response to TACE treatment. A total of 2.5 μg total RNA from these two mixed samples was used for profiling.
Quantitative reverse-transcription polymerase chain reaction
A miRNA quantification was performed by quantitative real-time polymerase chain reaction (qRT-PCR) using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) and FastStart Universal Probe Master (Roche) according to manufacturer’s protocol. Each TaqMan MicroRNA Assay primer set in this study was purchased from Applied Biosystems. For miRNAs with no ready-to-use TaqMan MicroRNA Assay primer sets, their expression levels were examined through using Bulge-Loop™ qRT-PCR Primer Set (RiBoBio), the ReverTra Ace qPCR RT Kit (TOYOBO), and Quantitect SYBR-Green Real-time PCR Master Mix (TOYOBO). All samples were run in triplicate. The expression levels of each circulating miRNA are presented as the threshold cycle (Ct) values, defined as the fractional cycle number at which the fluorescent signal surpasses the fixed threshold in qRT-PCR. During data analysis, we utilized the comparative Ct method (ΔCt), normalized by subtracting the Ct value of an endogenous reference (U6-small nuclear RNA) from each miRNA (i.e., ΔCt = CtmiRNA of interest − CtU6). Data were summarized as mean and standard deviation (SD) of ΔCt value for each miRNA by disease group; fold change (calculated as the relative value 2−(ΔCT (group1)−ΔCT (group2))) was also reported. Comparisons between groups were conducted using Student’s t-test.
Cell proliferation assay
In a 37 °C humidified incubator containing 5 % CO2, human HCC cells Hep3B and PLC/PRF/5 were cultured with RPMI 1640 supplemented with 10 % fetal bovine serum (FBS). Hep3B and PLC/PRF/5 cells were seeded in 96-well plates. After being cultured for 24 h, HCC cells were transfected with 30 nmol/L miR-1285-3p mimics or noncoding RNA (ncRNA). WST-1 cell proliferation assay (Roche) was utilized at various days after transfection.
Target prediction of miRNAs and luciferase activity assay
The miR-1285-3p targeting genes were predicted using the algorithm TargetScan Human 6.2 (http://www.targetscan.org). A 379-bp fragment of the human JUN 3′-UTR (spanning from nucleotides 2608–2986, NM_002228.3) was amplified by PCR using primers JUN-3F (5′-GCTTCATGCCTTTGTAAGT-3′) and JUN-3R (5′-CCCTCCTCCTCATATTGGAC-3′) and was cloned downstream of the firefly luciferase gene present in the pMIR-REPORT vector (Ambion) to develop the pMIR-JUN 3′-UTR-WT plasmid.
Hep3B and PLC/PRF/5 cells were seeded in 24-well plates and each transfected with 50 ng of either pMIR-JUN 3′-UTR-WT or pMIRREPORT together with 1 ng pRL-SV40 vector (Promega), which contains the Renilla luciferase gene, and 30 pmol miR-1285-3p mimics or ncRNA. All transfections were done using Lipofectamine® 2000 (Invitrogen). At 24 h after transfection, firefly and Renilla luciferase activities were examined using the Dual-Luciferase Reporter Assay (Promega). Each transfection was performed in triplicate.
Furthermore, pMIR-JUN 3′-UTR-WT was mutated in the seed-match sequence for miR-1285-3p (original seed-match sequence: UGCCCAG; mutated seed-match sequence: GAAACGA). The mutated plasmid was named as pMIR-JUN 3′-UTR-M. pMIR-JUN 3′-UTR-M was cotransfected with miR-1285-3p mimics or ncRNA to Hep3B and PLC/PRF/5 cells as described above. At 24 h after transfection, luciferase activities were examined. Each transfection was also repeated in triplicate.
Real-time RT-PCR and Western blot analysis of JUN expression
Hep3B and PLC/PRF/5 cells either transfected with miR-1285-3p mimics or ncRNA were collected 24 h after drug treatment or transfection. JUN and GAPDH real-time PCR primer and probe sets (Assay ID Hs01103582_s1 and Hs02758991_g1, respectively) were inventoried products of Applied Biosystem. GAPDH mRNA was quantified as an endogenous control of internal RNA from each sample. Analyses were performed using a standard TaqMan PCR Kit protocol. For each data point, experiments were carried out in triplicate.
Hep3B and PLC/PRF/5 cells either transfected with miR-1285-3p mimics, JUN small interfering RNA (siRNA), or ncRNA were lysed for 15 min at 4 °C with the RIPA buffer. A total of 10 μg of extract was separated in 10 % SDS-PAGE and then blotted onto nitrocellulose membrane (Immobilon-NC, Millipore). JUN was detected with the monoclonal anti-JUN antibody (sc-1694; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4 °C overnight. For GAPDH detection, we used a rabbit polyclonal antibody together with an anti-rabbit HRP-conjugated secondary antibody (Santa Cruz). For the detection of blots, SuperSignal chemiluminescence kit (Pierce) was used according to the manufacturer’s instructions.
Statistical analyses
Results were expressed as mean ± standard deviation. t-tests were used to calculate differences between dual-luciferase reporter gene assays and qRT-PCR mRNA expression levels of JUN. Mann–Whitney U analyses of variance were used to evaluate statistical differences in plasma miRNA expression between unpaired samples. Bonferroni correction was used for multiple comparisons. The statistical analysis was performed in SPSS version 16.0.
Results
Characteristics of the study population
Demographic and clinical characteristics of the 97 HCC patients with TACE treatment are summarized in Table 1. For the total of 97 patients, the median age at the time of HCC diagnosis was 55 years (SD = 10.4 years). The majority of patients (81.4 %) were males and 83 (85.6 %) of all patients had hepatitis B virus infection. About 85.6 % (n = 83) of patients had liver cirrhosis and 82 patients (84.5 %) had a Child–Pugh score A. There were 20.6 % (n = 20) of patients with tumor size of <5 cm, 57.7 % (n = 56) with single tumor, and 47.4 % (n = 46) with significantly increased serum alpha-fetoprotein (AFP) (≥200 lg/L). According to the sixth edition of TNM Classification of International Union Against Cancer, the percentage (number) of patients with TNM stages I and II, III, and IV was 8.2 (n = 8), 36.1 (n = 35), and 55.7 % (n = 54), respectively. We divided the HCC cases as “good responders” and “no responders” as follows: good responders include ones achieved complete response (CR), partial response (PR), or stable sisease (SD); and no responders are cases with progressive disease (PD) after TACE.
Identification of TACE associated circulating miRNAs
We first examined differential expression of miRNAs between two pooled samples (five patients with good response to TACE treatment and the other five patients with no response to TACE treatment) using Human miRNA OneArray® miRNA Profiling Chip. After normalizing all data to the expression level of small RNA U6, we found 19 circulating miRNAs to be significantly differentially expressed between the TACE-response group and the TACE-nonresponse group (P < 0.0001 and fold change >2; still significant after Bonferroni correction) (Supplementary Table 1 and Supplementary Fig. 1).
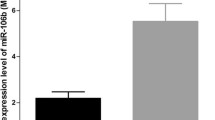
We then validated expression of the 19 circulating miRNAs identified by the expression profiling in the remaining 87 samples through TaqMan MicroRNA Assays (hsa-let-7f-5p, hsa-miR-1285-3p, hsa-miR-1306-3p, hsa-miR-1306-5p, hsa-miR-2681-3p, hsa-miR-3156-5p, hsa-miR-432-5p, hsa-miR-4419a, hsa-miR-4458, hsa-miR-4498, hsa-miR-4514, hsa-miR-4676-5p, hsa-miR-4690-5p, hsa-miR-4741, hsa-miR-5096, and hsa-miR-620) or SBRY Green miRNA assays (hsa-miR-4745-5p, hsa-miR-5196-3p, and hsa-miR-5585-3p). We observed that the expression levels of miR-1285-3p and miR-4741 are significantly downregulated in plasma samples of HCC case with bad response to TACE compared to those in the good response group (3.1-fold, P = 0.023 and 4.6-fold, P = 0.001) (Fig. 1).
miR-1285-3p inhibits proliferation of HCC cells
To reveal the potential role of miR-1285-3p in HCC, we firstly examined the inhibition effects of miR-1285-3p on Hep3B or PLC/PRF/5 cells. Cell proliferation rates were measured by WST-1 assay for 3 days after transfection of miR-1285-3p mimics into Hep3B or PLC/PRF/5 cells. It was found that miR-1285-3p mimics could inhibit Hep3B or PLC/PRF/5 cell proliferation significantly compared to NC RNA (Fig. 2), demonstrating that miR-1285-3p might act as a tumor-suppressor in HCC.
miR-1285-3p downregulates JUN expression by directly targeting its 3′-UTR
To reveal the potential role of miR-1285-3p in HCC, putative human protein-coding gene targets of miR-1285-3p were identified by using the TargetScan algorithm. It has been showed that the JUN gene 3′-UTR harbors a putative binding site for miR-1285-3p (position 616–622 of JUN 3′-UTR). To examine the potential interaction between miR-1285-3p and JUN 3′-UTR, the human JUN 3′-UTR was subcloned in pMIR-REPORT and cotransfected into Hep3B and PLC/PRF/5 cells with miR-1285-3p mimics. miR-1285-3p-transfected Hep3B or PLC/PRF/5 cells showed a 56 or 49 % decrease of relative luciferase activity compared with the ncRNA-transfected cells (both P < 0.05) (Fig. 3b). When HCC cells were transfected with both miR-1285-3p mimics and plasmids harboring mutated miR-1285-3p seed-match sequence (pMIR-JUN 3′-UTR-M), no significantly decreased relative luciferase activity was found compared with the ncRNA control group (both P > 0.05) (Fig. 3b), suggesting that miR-1285-3p may inhibit gene expression through both miR-285-3p-binding sequences at the 3′-UTR of JUN.
The dual-luciferase reporter gene assay. a The 3′-UTR of JUN was inserted in the pMIR-REPORT reporter plasmid after the luciferase coding sequence (pMIR-JUN 3′-UTR-WT). pMIR-JUN 3′-UTR-WT was cotransfected into Hep3B and PLC/PRF/5 cells with 30 pmol miR-1285-3p mimics or ncRNA. Luciferase activity was normalized relative to a simultaneously transfected Renilla expression plasmid. b When the seed-match sequence of pMIR-JUN 3′-UTR-WT was mutated, the plasmid was named as pMIR-JUN 3′-UTR-M. Inhibition effects of miR-1285-3p mimics on pMIR-JUN 3′-UTR-M are shown in b. Results of the mean of triplicate assays with standard deviation of the mean are presented. * P < 0.05
To further study whether JUN was a target gene of miR-1285-3p, we examined JUN mRNA and protein expression levels in Hep3B and PLC/PRF/5 cells with or without miR-1285-3p mimic or JUN siRNA expression. As shown in Fig. 4a, JUN siRNA significantly inhibits JUN mRNA expression in both Hep3B and PLC/PRF/5 cells (both P < 0.05). Overexpression of miR-1285-3p in HCC cells could also significantly reduce JUN mRNA expression compared with cells transfected with negative control small RNA (both P < 0.05). Western blot analysis showed that forced expression of miR-1285-3p or JUN siRNA in HCC cells resulted in a significant reduction of JUN expression (Hep3B: miR-1285-3p, 82 %, JUN siRNA, 85 %; PLC/PRF/5: miR-1285-3p, 90 %, JUN siRNA, 97 %) (Fig. 4b). These data elucidate that JUN might be a direct gene of miR-1285-3p in HCC.
Inverse correlation between miR-1285-3p and JUN mRNA and protein levels in Hep3B and PLC/PRF/5 cell lines. a Real-time RT-PCR analysis of JUN expression in Hep3B and PLC/PRF/5 cell lines either transfected with miR-1285-3p mimics, JUN siRNA, or ncRNA. GAPDH mRNA from each sample was quantified as an endogenous control of internal RNA. Each determination was done in triplicate. Mean ± standard deviation. * P < 0.05. b Western blot assay of JUN protein expression in Hep3B and PLC/PRF/5 cell lines normalized based on GAPDH expression. Hep3B and PLC/PRF/5 cell lines were transfected with miR-1285-3p mimics, JUN siRNA or ncRNA
Discussion
In the current study, we found that the HCC patients with good response to TACE or had response to TACE have different circulating miRNA signatures. The expression levels of differentially expressed miR-1285-3p and miR-4741 showed a consistent downregulation in HCC cases with bad response to TACE compared to ones with good response to TACE. We presume that the decrease of miR-1285-3p and miR-4741 expression in the good response group might be related to TACE, either a result of shrunken HCC tumors or other human body biological changes of the HCC patients. In addition, we found that miR-1285-3p might directly target JUN oncogene in HCC cells.
Tian et al. reported that miR-1285 may act as an oncogene in HCC HepG2 cells [22]. The well-known tumor suppressor p53 plays an essential part in the regulation of multiple cellular processes. However, we found that miR-1285-3p might play its role as tumor suppressor via repress JUN oncogene expression in HCC cells. Therefore, we speculate that the increased levels of miR-1285 in HCC patients with good response to TACE might be due to diminished controls of JUN expression by miR-1285 and enhanced JUN pathway in HCC lesions of these patients. This is critical because increased expression of circulating miRNAs could be indicative of miRNAs secreted from a tumor, raising the overall diagnostic specificity of the biomarker. For miR-4741, no reports on its role in HCC have been found up to now.
The activating protein 1 (AP-1) family proteins are diametric transcription factors, which consist of JUN and c-Fos [23, 24]. After binding to DNA [24, 25], the AP-1 family regulates multiple downstream genes which are important regulators of cell proliferation, differentiation, invasion, and metastasis and, thus, plays a crucial part in carcinogenesis [25]. In AP-1, although JUN is able to both homo- and heterodimerize, JUN–Fos heterodimers exist more stably and have stronger DNA-binding activity than JUN–JUN homodimers [26]. JUN is the cellular homologue of v-Jun, one of the transforming oncogenes from the avian sarcoma virus 17 [27, 28]. Accumulated evidences indicated that JUN is important in tumorigenesis and progression of HCC [29, 30]. Upregulation of JUN was identified in human HCC tissues [30]. Moreover, data of mouse models with inactivated JUN showed that the size of hepatic tumors induced by diethylnitrosamine significantly shrank, and the number of hepatic tumors reduced [29]. In HCC, JUN is involved in both ERK and JNK signaling pathways and impacts cancer cell proliferation as well as migration [30–32]. All aforementioned results suggest that JUN may have an important role in HCC. In the current study, we identified a JUN-regulatory miRNA (miR-1285-3p), which adds another layer of aberrant JUN expression regulation in HCC.
There are several limitations in the current study. First, there might be inherent selection bias since this was a hospital-based study including cases from one hospital. Therefore, these findings warrant validation in a population-based prospective study. Second, the statistical power might be limited because of the sample size. Third, future studies with the follow-up data will need to address impacts of circulating miR-1285-3p and miR-4741 on patient survival in the future.
In all, our observations strongly support the potential application of circulating miR-1285-3p and miR-4741 were predictive of response to TACE therapy among HCC patients in a minimally invasive way. miR-1285-3p might be a tumor suppressor in HCC cells.
References
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108.
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76.
Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye QH, et al. A decade’s studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130(4):187–96.
Vogl TJ, Naguib NN, Nour-Eldin NE, Rao P, Emami AH, Zangos S, et al. Review on transarterial chemoembolization in hepatocellular carcinoma: palliative, combined, neoadjuvant, bridging, and symptomatic indications. Eur J Radiol. 2009;72(3):505–16.
Gottesman S. Small RNAs shed some light. Cell. 2004;118(1):1–2.
Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat Med. 2005;11(7):712–4.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8.
Liu J, Tang X, Li M, Lu C, Shi J, Zhou L, et al. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res Treat. 2013;140(1):151–7.
Zhou L, Zhang X, Li Z, Zhou C, Li M, Tang X, et al. Association of a genetic variation in a miR-191 binding site in MDM4 with risk of esophageal squamous cell carcinoma. PLoS One. 2013;8(5):e64331.
Zhang X, Wei J, Zhou L, Zhou C, Shi J, Yuan Q, et al. A functional BRCA1 coding sequence genetic variant contributes to risk of esophageal squamous cell carcinoma. Carcinogenesis. 2013;34(10):2309–13.
Yao J, Liu L, Yang M. Interleukin-23 receptor genetic variants contribute to susceptibility of multiple cancers. Gene. 2014;533(1):21–5.
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58.
Kota J, Chivukula RR, O′Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–17.
Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12(4):390–9.
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–43.
Zhou C, Liu J, Li Y, Liu L, Zhang X, Ma CY, et al. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011;585(12):1828–34.
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006.
Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106(11):4402–7.
Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55(11):1977–83.
Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54(5):1767–76.
Tian S, Huang S, Wu S, Guo W, Li J, He X. MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3′ untranslated region. Biochem Biophys Res Commun. 2010;396(2):435–9.
Shaulian E. AP-1—the Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22(6):894–9.
Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–6.
Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334(6183):629–31.
Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41(16):2449–61.
Bohmann D, Bos TJ, Admon A, Nishimura T, Vogt PK, Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987;238(4832):1386–92.
Maki Y, Bos TJ, Davis C, Starbuck M, Vogt PK. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987;84(9):2848–52.
Eferl R, Ricci R, Kenner L, Zenz R, David JP, Rath M, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112(2):181–92.
Machida K, Tsukamoto H, Liu JC, Han YP, Govindarajan S, Lai MM, et al. c-Jun mediates hepatitis C virus hepatocarcinogenesis through signal transducer and activator of transcription 3 and nitric oxide-dependent impairment of oxidative DNA repair. Hepatology. 2010;52(2):480–92.
Watanabe T, Hiasa Y, Tokumoto Y, Hirooka M, Abe M, Ikeda Y, et al. Protein kinase R modulates c-Fos and c-Jun signaling to promote proliferation of hepatocellular carcinoma with hepatitis C virus infection. PLoS One. 2013;8(7):e67750.
Yang Z, Zhang Y, Wang L. A feedback inhibition between miRNA-127 and TGFβ/c-Jun cascade in HCC cell migration via MMP13. PLoS One. 2013;8(6):e65256.
Funding supports
This study is supported by the development projects of Shandong Province Science and Technology (2012GSF11837).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Jibing Liu, Jingchen Yan, and Changchun Zhou contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOC 13 kb)
Supplementary Fig. 1
(JPEG 115 kb)
Rights and permissions
About this article
Cite this article
Liu, J., Yan, J., Zhou, C. et al. miR-1285-3p acts as a potential tumor suppressor miRNA via downregulating JUN expression in hepatocellular carcinoma. Tumor Biol. 36, 219–225 (2015). https://doi.org/10.1007/s13277-014-2622-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2622-5