Abstract
Cancer stem cells (CSCs) in most human tumors are commonly identified and enriched using similar strategies for identifying normal stem cells, including flow cytometry assays for side population, high aldehyde dehydrogenase (ALDH) activity, and CD133 positivity. Thus, development of a method for isolating a specific cancer using cancer-specific characteristic appears to be potentially important. Here, we reported extremely high Lgr5 levels in the specimen from skin squamous cell carcinoma (SCC) in patients. Using SCC cell line A431, we detected high Lgr5 and CD133 levels in ALDH-high or side population from these cancer cells. To figure out whether Lgr5 is a marker of CSCs in SCC, we transfected A431 cells with a Lgr5-creERT-2A-DTR/Cag-Loxp-GFP-STOP-Loxp-RFP plasmid and purified transfected cells (tA431) based on GFP by flow cytometry. 4-Hydroxytamoxifen (4-OHT) was given to label Lgr5-positive cells with RFP, for comparison to GFP-positive Lgr5-negative cells. Lgr5-positive cells grew significantly faster than Lgr5-negative cells, and the fold increase in growth of Lgr5-positive vs Lgr5-negative cells is significantly higher than SP vs non-SP, or ALDH-high vs ALDH-low, or CD133-positive vs CD133-negative cells. Moreover, in Lgr5-negative population, Lgr5-positive re-appeared in culture with time, suggesting that Lgr5-positive cells can be regenerated from Lgr5-negative cells. Furthermore, the growth of tA431 cells significantly decreased upon a single dose of diphtheria toxin (DT)/4-OHT to eliminate Lgr5-positive cell lineage, while multiple doses of DT/4-OHT nearly completely inhibited tA431 cell growth. Taken together, our data provide compelling data to demonstrate that Lgr5-positive cells are CSCs in skin SCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carcinoma is a cancer of epithelial tissue origin, which account for more than 80 % of all cancers worldwide. The incidence of the commonest carcinoma, squamous cell carcinoma (SCC), is continuing to rise, with skin SCC continuing to be a cause of mortality. Therefore, understanding the cancer molecular biology of skin SCC is extremely important for developing efficient therapy [1].
Cancer stem cells (CSCs) are cancer cells that possess characteristics associated with normal stem cells, specifically the ability to give rise to various cell types detected in a particular cancer [2, 3]. CSCs are thus tumorigenic, compared with other nontumorigenic cancer cells. CSCs may generate tumors through the stem cell processes of self-renewal and differentiation into multiple cell types. CSCs are proposed to persist in tumors as a distinct population and cause relapse and metastasis by giving rise to new tumors [2, 3]. Recently, CSCs have received substantial attention, and treatment targeting CSCs are believed to be potential to cure rapidly growing cancers, as well as highly metastatic cancers.
Dissection of the CSCs rely largely on the dissociation of solid cancers and sorting of tumor cells into subpopulations. To date, investigators have relied on cell surface proteins to isolate specific cell types by flow cytometry. It has been typically applied in proteins expressed by normal tissue stem cells in isolation of CSCs. Among those, the most population methods are prominin-1 (CD133), side population (SP), and increased activity of aldehyde dehydrogenase (ALDH). CD133 is expressed in hematopoietic stem cells, endothelial progenitor cells, and neuronal and glial stem cells. It has been recently shown to be expressed in the CSCs from various tumors [4–8]. However, subsequent studies have indicated the difficulty in isolating pure CSC populations by CD133 [9]. SP is a subpopulation of cells with distinguishing biological characteristics. SPs have been identified in cancer cells that efflux chemotherapy drugs, accounting for the resistance of cancer to chemotherapy. The experimentally used effluxing molecules is Hoechst (HO) [10]. Increased activity of ALDH, a detoxifying enzyme responsible for the oxidation of intracellular aldehydes [11, 12], has been detected in some stem/progenitor cells. For example, high ALDH activity has been found in murine and human hematopoietic and neural stem and progenitor cells [13–16] and in proliferating pancreatic beta cells [17, 18]. Recent evidence suggests that enhanced ALDH activity is also a hallmark of CSC measurable by the Aldefluor assay [19, 20]. However, all these methods are not perfect for isolation of CSCs, and moreover, are lack of cancer specificity.
The Wnt target gene Lgr5 has been recently identified as a novel stem cell marker of the intestinal epithelium and the hair follicle. In the stem cell niche of the murine hair follicle, Lgr5 is expressed in actively cycling cells. Transplantation and lineage tracing experiments have demonstrated that these Lgr5-positive cells maintain all cell lineages of the hair follicle throughout long periods of time and can build entire new hair follicles [21, 22]. However, whether Lgr5 may be activated in skin SCC has not been reported before.
Here, we reported extremely high Lgr5 levels in the specimen from skin SCC patients. Using a SCC cell line A431, we detected high Lgr5 and CD133 levels in ALDH-high or side population from these cancer cells. To figure out whether Lgr5 is a marker of CSCs in SCC, we transfected A431 cells with a Lgr5-creERT-2A-DTR/Cag-Loxp-green fluorescence (GFP)-STOP-Loxp-red fluorescence (RFP) plasmid and purified transfected cells (tA431) based on GFP by flow cytometry. 4-Hydroxytamoxifen (4-OHT) was given to label Lgr5-positive cells with RFP, for comparison to GFP-positive Lgr5-negative cells. Lgr5-positive cells grew significantly faster than Lgr5-negative cells, and the fold increase in growth of Lgr5-positive vs Lgr5-negative cells is significantly higher than SP vs non-SP, ALDH-high vs ALDH-low, or CD133-positive vs CD133-negative cells. Moreover, in Lgr5-negative population, Lgr5-positive re-appeared in culture with time, suggesting that Lgr5-positive cells can be regenerated from Lgr5-negative cells. Furthermore, the growth of tA431 cells significantly decreased upon a single dose of diphtheria toxin (DT)/4-OHT to eliminate Lgr5-positive cell lineage, while multiple doses of DT/4-OHT nearly completely inhibited tA431 cell growth. Taken together, our data provide compelling data to demonstrate that Lgr5-positive cells are CSCs in skin SCC.
Materials and methods
Specimens from patients
A total of 15 resected SCC specimens (paired SCC and the adjacent normal skin tissue (NST)) was collected for this study. All specimens had been histologically and clinically diagnosed at the Department of Burn and Plastic from General Hospital of Jinan Military Region from 2010 to 2013. For the use of these clinical materials for research purposes, prior patient’s consents and approval from the Institutional Research Ethics Committee were obtained.
Cell line culture, plasmid transfection, cell labeling, and elimination by DTA
A431 cells are a human skin SCC cell line, purchased from ATCC, and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20 % fetal bovine serum (Invitrogen, Carlsbad, CA, USA).
Lgr5-creERT-2A-DTR/Cag-Loxp-GFP-STOP-Loxp-RFP plasmid was prepared and used for transfection of A431 cells with Lipofectamine 2000 reagent (Invitrogen), according to manufacturer’s instructions. The small 2A peptide sequences, when cloned between genes, allow for efficient, stoichiometric production of discrete protein products within a single vector through a novel “cleavage” event within the 2A peptide sequence. Stable transfected cells (tA431) expressing the plasmid were selected by flow cytometry based on GFP.
4-OHT (Sigma) solution (800 nmol/l) was freshly prepared and given to the cultured tA431 cells to label Lgr5-positive cells with RFP, to compare with GFP-positive Lgr5-negative cells (D-2), 2 days before FAC sorting (D0). 4-OHT was also given at D0/D2/D4 for multiple doses’ experiments. To avoid detection of cells expressing both GFP and RFP [23, 24], at least 2 days were spanned between cell labeling and analysis.
Diphtheria toxin (DT) was freshly prepared and given to the cultured tA431 cells at a concentration of 40 nmol/l to specifically eliminate cells expressing diphtheria toxin receptor (DTR). DT was also given at D0/D2/D4 for multiple doses’ experiments.
Analysis of CD133, SP, and Aldefluor by flow cytometry
For flow cytometry analysis and sorting, tA431 cells were either labeled with PEcy7-conjugated anti-human CD133 antibody, or incubated with HO red and HO blue, or labeled with the Aldefluor Kit (StemCell Technologies, China) according to the manufacturer’s instructions. Flow cytometry was performed using a FACSAria (Becton Dickinson) flow cytometer.
Cell proliferation assay
For assay of cell proliferation, pretreated cells were seed into 24-well plate at 10,000 cells per well and subjected to a Cell Proliferation Kit (MTT, Roche, USA), according to the instruction of the manufacturer.
Western blot
Protein was extracted from the SCC or the adjacent NST from the same patient with RIPA buffer. Primary antibodies were rabbit anti-Lgr5 (Santa Cruz, USA) and anti-α-tubulin (Cell Signaling, USA). Secondary antibody is HRP-conjugated anti-rabbit (Jackson Labs). α-Tubulin was used as protein loading controls. The protein levels were first normalized to α-tubulin, and then normalized to control NST.
RT-qPCR
RNA was extracted from cultured cells with RNeasy kit (Qiagen, Hilden, Germany). cDNA was then synthesized by reserve transcription (Qiagen). Quantitative real-time PCR (RT-qPCR) was performed in duplicates with QuantiTect SYBR Green PCR Kit (Qiagen). All primers were purchased from Qiagen. Values of genes were first normalized against α-tubulin, and then compared with controls.
Statistical analysis
All statistical analyses were carried out using the SPSS 18.0 statistical software package. All values are depicted as mean ± standard deviation from five individuals and are considered significant if p < 0.05. All data were statistically analyzed using one-way ANOVA with a Bonferroni correction.
Results
Extremely high Lgr5 levels were detected in skin SCC from patients
Lgr5 has been recently identified as a novel stem cell marker of the intestinal epithelium and the hair follicle, where Lgr5 is expressed in actively cycling cells. However, whether Lgr5 may be activated in skin SCC has not been reported before. We, thus, examined Lgr5 levels in 15 resected SCC specimens with paired adjacent NST from the patients. We found a significant increase in Lgr5 levels in SCC, compared with NST (Fig. 1; 6.8 ± 1.2-fold increase), which prompted us to analyze the role of Lgr5 in the SCC.
ALDH-high or SP SCC cells expressed high levels of CD133 and Lgr5
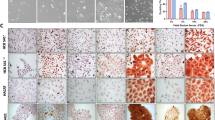
We used a human SCC cell line A431 for our study. Since Lgr5 is a stem cell marker, we aimed to figure out whether Lgr5 expression in SCC may be heterozygous, and moreover, whether Lgr5 may be a marker for CSCs in SCC. Thus, we used flow cytometry to isolate SP and non-SP cells (Fig. 2a), or Aldefluor (representing ALDH)-high and Aldefluor-low cells (Fig. 2b), in A431 cells. We found that ALDH-high or SP A431 cells expressed high levels of CD133 and Lgr5, compared with the control ALDH-low or non-SP A431 cells, respectively (Fig. 2c). These data highly suggest that Lgr5 are predominantly expressed by CSCs in SCC.
ALDH-high or SP SCC cells expressed high levels of CD133 and Lgr5. a–c A human SCC cell line A431 was used in flow cytometry to isolate SP (rectangled) and non-SP cells (a), or Aldefluor (representing ALDH)-high (rectangled) and Aldefluor-low cells (b). c RT-qPCR for CD133 and Lgr5 in SP/non-SP cells and in ALDH-high/ALDH-low cells. HO Hoechst, SSC sideward scatter. *p < 0.05
Preparation of SCC with inducible Lgr5-positive cell lineage tracing and elimination
To figure out whether Lgr5-positive SCC cells are really CSCs, we transfected A431 cells with a Lgr5-creERT-2A-DTR/Cag-Loxp-GFP-STOP-Loxp-RFP plasmid and purified the transfected cells (tA431) based on GFP by flow cytometry (Fig. 3a, b). tA431 cells expressed strong GFP in all cells. However, when 4-OHT (the active form of tamoxifen) was given, the expressed creERT was translocated into nuclei to remove the construct (GFP and a stop codon) between two Loxps, resulting in expression of RFP in Lgr5-positive cells (Fig. 3b). Tamoxifen is a prodrug with relatively little affinity for its target protein. It is metabolized in the liver by the cytochrome P450 isoform CYP2D6 and CYP3A4 into active metabolite 4-OHT which has 30–100 times more affinity with the receptor than tamoxifen. In the in vitro study, since no liver metabolism is involved, the direct active form of tamoxifen, 4-OHT, is needed. In Lgr5-negative cells, since the Lgr5 promoter is inactivated, the cells will remain GFP due to presence of GFP and the stop codon (Fig. 3a). In this system, all Lgr5-positive cells that received 4-OHT will not only change GFP to RFP but also start to express a DTR on the cell surface. These cells can be efficiently eliminated when exposed to DT treatment (Fig. 3a).
SCC with inducible Lgr5-positive cell lineage tracing and elimination. a A431 cells were transfected with a Lgr5-creERT-2A-DTR/Cag-Loxp-GFP-STOP-Loxp-RFP plasmid, resulting in tA431 cells expressing GFP in all cells. However, when 4-hydroxytamoxifen (4-OHT) was given, the expressed creERT was translocated into nuclei to remove the construct (GFP and a stop codon) between two Loxps, resulting in expression of RFP in Lgr5-positive cells. In Lgr5-negative cells, since the Lgr5 promoter is inactivated, the cells will remain GFP due to presence of GFP and the stop codon. In this system, all Lgr5-positive cells that received 4-OHT will not only change their green to red fluorescence but also start to express DTR on the cell surface. These cells can be efficiently eliminated when exposed to DT treatment. b Transfected cells (rectangled) were purified for based on GFP by flow cytometry. 4-OHT was given to tA431 cells to label Lgr5-positive cells with RFP, to allow separation of RFP-positive Lgr5-positive cells (rectangled) and GFP-positive Lgr5-negative SCC
Lgr5-positive cells grew significantly faster than Lgr5-negative cells
4-OHT was then given to tA431 cells to label Lgr5-positive cells with RFP and to allow separation of RFP-positive Lgr5-positive cells and GFP-positive Lgr5-negative SCC (Fig. 3b). We also isolated CD133-positive vs CD133-negative cells, SP vs non-SP cells, and ALDH-high vs ALDH-low cells by flow cytometry. Same number of these sorted cells was subjected to an MTT assay (Fig. 4a–d). We found that Lgr5-positve cells grew significantly faster than Lgr5-negative cells (Fig. 4a), CD133-positve cells grew significantly faster than CD133-negative cells (Fig. 4b), SP cells grew significantly faster than non-SP cells (Fig. 4c), and ALDH-high cells grew significantly faster than ALDH-low cells (Fig. 4d). Of note, the fold increase in growth of Lgr5-positive vs Lgr5-negative cells at D4 is significantly higher than SP vs non-SP cells, ALDH-high vs ALDH-low, or CD133-positive vs CD133-negative cells (Fig. 4e). These data suggest that Lgr5 may be a better CSC marker in SCC, than CD133, SP, or ALDH.
Lgr5-positive cells grew significantly faster than Lgr5-negative cells. (a-d) We isolated Lgr5-positive vs Lgr5-negative cells, CD133-positive vs CD133-negative cells, SP vs non-SP cells, and ALDH-high vs ALDH-low cells by flow cytometry. Same number of these sorted cells were subjected to an MTT assay. a Lgr5-positve cells grew significantly faster than Lgr5-negative cells. b CD133-positve cells grew significantly faster than CD133-negative cells. c SP cells grew significantly faster than non-SP cells. d ALDH-high cells grew significantly faster than ALDH-low cells. e The fold increase in growth of Lgr5-positive vs Lgr5-negative cells at D4 is significantly higher than SP vs non-SP cells, ALDH-high vs ALDH-low, CD133-positive vs CD133-negative cells. *p < 0.05. NS nonsignificant
Lgr5 expression in SCC can be dynamic
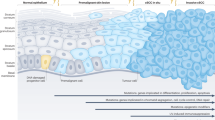
Interestingly, we detected increase in cell growth of Lgr5-negative tA431 cells with the time (Fig. 4a). This led to the hypothesis that Lgr5 expression in SCC may be dynamic. In order to figure out whether Lgr5-positive cells may re-appear in these Lgr5-negative populations, we examined the Lgr5 gene expression at different time points after seeding the sorted cells into culture. We found a continuous increase in Lgr5 levels with time (Fig. 5a). Moreover, when we gave more 4-OHT at D0/D2/D4, and analyzed these cells at D7 by flow cytometry, some Lgr5-positive cells were detected (Fig. 5b). These data thus confirmed our hypothesis, suggesting that the Lgr5 expression in SCC may be dynamic. These data also explained why the growth of Lgr5-negative cells seemed to increase after some time in culture in that new Lgr5-positive cells seemed to be regenerated from Lgr5-negative cells (Fig. 6).
Elimination of Lgr5-positive cells inhibited SCC growth. Multiple doses of 4-OHT and DT were given to cultured tA431 or Lgr5-negative tA431 cells (control), to continuously remove Lgr5-positive cells and the Lgr5-positive cells that were regenerated from Lgr5-negative cells. When multiple doses of DT and 4-OHT were given (D-2/D0/D2/D4), tA431 cells nearly stopped growing (p < 0.05 compared with single dose to tA431, or purified Lgr5-negative cells). *p < 0.05
Elimination of Lgr5-positive cells inhibited SCC growth
To further confirm the importance of Lgr5-positive cells to the SCC growth, and the dynamic expression of Lgr5 in SCC, we gave multiple doses of 4-OHT and DT, to continuously remove Lgr5-positive cells and the Lgr5-positive cells that were regenerated from Lgr5-negative cells. The growth of tA431 cells significantly decreased when a single dose of DT was given with 4-OHT to eliminate Lgr5-positive cell lineage, similar to that in Lgr5-negative cells. When multiple doses of DT and 4-OHT were given, tA431 cells nearly stopped growing (p < 0.05 compared with single dose to tA431 or purified Lgr5-negative cells). Taken together, our data provide compelling data to demonstrate that Lgr5-positive cells are CSCs in skin SCC.
Discussion
Understanding the cancer molecular biology of skin SCC is extremely important for developing efficient therapy [1]. Recently, the importance of CSCs to cancer growth and metastasis has been generally realized [2, 3]. Efficiently manipulating CSCs rely largely on the dissociation of solid cancers and sorting of tumor cells into subpopulations. To date, isolation of CSCs is mainly dependent on cell surface markers, including CD133, SP, and ALDH. Although these methods have been shown to substantially enrich the CSCs from various cancers, more and more evidence has revealed their limitation, and even improper application in some situations. For example, CD133-positive cells are not all CSCs [9] and increased ALDH activity can be also detected in nonstem/progenitor and noncancer cells [17, 18]. These findings demonstrate that all these methods are not perfect for isolation of CSCs, and moreover, are lack of cancer specificity. Lgr5 has been recently identified as a novel stem cell marker of the intestinal epithelium and the hair follicle, in which Lgr5 is expressed in actively cycling cells. However, whether Lgr5 may be activated in skin SCC has not been reported before.
In the current study, Lgr5 levels in SCC specimens from the patients were examined, showing a significant increase in Lgr5 levels in SCC, compared with NST. Since Lgr5 is a stem cell marker, we aimed to figure out whether Lgr5 expression in SCC may be heterozygous, and moreover, whether Lgr5 may be a marker for CSCs in SCC. We used flow cytometry to isolate SP and non-SP cells, or ALDH-high and ALDH-low cells in A431 cells. We found that ALDH-high or SP SCC cells expressed high levels of CD133 and Lgr5, compared with control ALDH-low or non-SP SCC cells, respectively. These data suggest that Lgr5 are predominantly expressed by CSCs in SCC. We were thus prompted to examine using Lgr5 as a CSC marker for SCC, and tried to compare it with other methods.
For such purposes, we prepared a SCC line with inducible Lgr5-positive cell lineage tracing and elimination-tA431. In this system, all Lgr5-positive cells that received 4-OHT will not only change their green to RFP but also start to express DTR on the cell surface, which allows elimination of the lineage-tagged cells by DT treatment. Since the half-life of GFP protein is pretty long according to previous publications [23, 24], we thus analyzed the cells by flow cytometry at least 2 days after tomaxifen administration. This approach allows the complete degradation of GFP protein after Cre recombinase-mediated recombination has occurred.
When we isolated RFP-positive Lgr5-positive cells and GFP-positive Lgr5-negative SCC, which were subsequently used in an MTT assay, we found that Lgr5-positve cells grew significantly faster than Lgr5-negative. Of note, the fold increase in growth of Lgr5-positive vs Lgr5-negative cells at D4 is significantly higher than SP vs non-SP cells, ALDH-high vs ALDH-low, or CD133-positive vs CD133-negative cells, suggesting that Lgr5 may be a better CSC marker in SCC, than CD133, SP, or ALDH.
Since we also detected increase in cell growth of Lgr5-negative tA431 cells with the time, we hypothesized that the Lgr5 expression in SCC may be dynamic. This hypothesis was proved by several pieces of evidence. First, Lgr5 gene expression in sorted Lgr5-negative cells increased with time in culture. Second, Lgr5-positive cells were detected in the Lgr5-negative cell culture after 7 days. Finally, continuous elimination of Lgr5-positive cells by DT in tA431 cells nearly completely inhibited their growth. Of note, we have checked other SCC lines and essentially got the same results. These findings thus appear to be extremely important, suggesting that future Lgr5-directed therapy for SCC should be performed in a sufficiently frequent manner, to guarantee elimination of recurring Lgr5-positive CSCs during treatment.
Taken together, our data provide compelling data to demonstrate that Lgr5-positive cells are CSCs in skin SCC and highlight Lgr5 as a promising therapeutic target of SCC.
References
Rahimi S. Squamous cell carcinoma of skin: a brief review. Clin Ter. 2013;164:143–7.
Petersson M, Niemann C. Stem cell dynamics and heterogeneity: implications for epidermal regeneration and skin cancer. Curr Med Chem. 2012;19:5984–92.
Perez-Losada J, Balmain A. Stem-cell hierarchy in skin cancer. Nat Rev Cancer. 2003;3:434–43.
Nagata T, Sakakura C, Komiyama S, Miyashita A, Nishio M, Murayama Y, et al. Expression of cancer stem cell markers CD133 and CD44 in locoregional recurrence of rectal cancer. Anticancer Res. 2011;31:495–500.
Fang DD, Kim YJ, Lee CN, Aggarwal S, McKinnon K, Mesmer D, et al. Expansion of CD133(+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–75.
Shi C, Tian R, Wang M, Wang X, Jiang J, Zhang Z, et al. CD44+ CD133+ population exhibits cancer stem cell-like characteristics in human gallbladder carcinoma. Cancer Biol Ther. 2010;10:1182–90.
Ottaiano A. Finding markers for cancer stem cells in renal cell carcinoma: looking beyond CD133. Cell Cycle. 2010;9:4431.
Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6:1146–53.
Wang J, Sakariassen PO, Tsinkalovsky O, Immervoll H, Boe SO, Svendsen A, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122:761–8.
Takubo K, Ohmura M, Azuma M, Nagamatsu G, Yamada W, Arai F, et al. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell. 2008;2:170–82.
Duester G. Families of retinoid dehydrogenases regulating vitamin a function: production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–24.
Magni M, Shammah S, Schiro R, Mellado W, Dalla-Favera R, Gianni AM. Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood. 1996;87:1097–103.
Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–51.
Hess DA, Craft TP, Wirthlin L, Hohm S, Zhou P, Eades WC, et al. Widespread nonhematopoietic tissue distribution by transplanted human progenitor cells with high aldehyde dehydrogenase activity. Stem Cells. 2008;26:611–20.
Hess DA, Meyerrose TE, Wirthlin L, Craft TP, Herrbrich PE, Creer MH, et al. Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood. 2004;104:1648–55.
Hess DA, Wirthlin L, Craft TP, Herrbrich PE, Hohm SA, Lahey R, et al. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–9.
Liu Y, Jiang X, Zeng Y, Zhou H, Yang J, Cao R: Proliferating pancreatic beta-cells upregulate ALDH. Histochem Cell Biol 2014
Zhang L, Wang L, Liu X, Zheng D, Liu S, Liu C. ALDH expression characterizes G1-phase proliferating beta cells during pregnancy. PLoS ONE. 2014;9:e96204.
Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001.
Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306.
Kinzel B, Pikiolek M, Orsini V, Sprunger J, Isken A, Zietzling S, et al. Functional roles of Lgr4 and Lgr5 in embryonic gut, kidney and skin development in mice. Dev Biol. 2014;390:181–90.
Haegebarth A, Clevers H. Wnt signaling, Lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–21.
Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605.
Xiao X, Chen Z, Shiota C, Prasadan K, Guo P, El-Gohary Y, et al. No evidence for beta cell neogenesis in murine adult pancreas. J Clin Invest. 2013;123:2207–17.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shunli Liu and Zhenyu Gong contributed equally to this work.
The Publisher and Editor retract this article in accordance with the recommendations of the Committee on Publication Ethics (COPE). After a thorough investigation we have strong reason to believe that the peer review process was compromised.
An erratum to this article can be found online at http://dx.doi.org/10.1007/s13277-017-5487-6.
About this article
Cite this article
Liu, S., Gong, Z., Chen, M. et al. RETRACTED ARTICLE: Lgr5-positive cells are cancer stem cells in skin squamous cell carcinoma. Tumor Biol. 35, 11605–11612 (2014). https://doi.org/10.1007/s13277-014-2488-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2488-6