Abstract
Gallbladder cancer (GBC) is a violent neoplasm associated with late diagnosis, unsatisfactory treatment, and poor prognosis. The disease shows complex interplay between multiple genetic variants. We analyzed 15 polymorphisms in nine genes involved in various pathways to find out combinations of genetic variants contributing to GBC risk. The genes included in the study were matrix metalloproteinases (MMP-2, MMP-7, and MMP-9), tissue inhibitor of metalloproteinases (TIMP-2), cytochrome P450 (CYP)1A1, CYP1B1, phospholipase C epsilon 1 (PLCE1), liver X receptor (LXR)-alpha, and LXR-beta. Genotypes were determined by PCR-RFLP and TaqMan probes. Statistical analysis was done by SPSS version 16. Multilocus analysis was performed by Classification and Regression Tree (CART) analysis and multifactor dimensionality reduction (MDR) to gene–gene interactions in modifying GBC risk. In silico analysis was done using various bioinformatics tools (F-SNP, FAST-SNP). Single locus analysis showed association of MMP-2 (−735 C > T, −1306 C > T), MMP-7 − 181 A > G, MMP-9 (P574R, R668Q), TIMP-2 − 418 G > C, CYP1A1-MspI, CYP1A1-Ile462Val, PLCE1 (rs2274223 A > G, rs7922612 T > C) and LXR-beta T > C (rs3546355 G > A, rs2695121 T > C) polymorphisms with GBC risk (p < 0.05) whereas CYP1B1 and LXR-α variants were not associated with GBC risk. Multidimensional reduction analysis revealed LXR-β (rs3546355 G > A, rs2695121 T > C), MMP-2 (−1306 C > T), MMP-9 (R668Q), and PLCE1 rs2274223 A > G to be key players in GBC causation (p < 0.001, CVC = 7/10). The results were further supported by independent CART analysis (p < 0.001). In silico analysis of associated variants suggested change in splicing or transcriptional regulation. Interactome and STRING analysis showed network of associated genes. The study found PLCE1 and LXR-β network interactions as important contributory factors for genetic predisposition in gallbladder cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gallbladder cancer (GBC) is a violent neoplasm associated with late diagnosis, unsatisfactory treatment, and poor prognosis [1–3]. The worldwide variations in the GBC incidence, highest being in Native American and South American populations and people from Poland and Northern India [4]. GBC Prognosis at early stage is very poor and attributed to the lack of any specific symptoms. Gallbladder cancer (GBC) is a multifactorial disease with complex interplay between multiple genetic variants and environmental risk factors (dietary carcinogens exposure such as tobacco, alcohol, etc). Extensive epidemiological studies have demonstrated that genetic variants, mainly single nucleotide polymorphisms (SNPs), are likely to modulate the consequence of environmental risk factors through modifying functions of various biological pathways concerned in gallbladder carcinogenesis [5]. These variations along with familial and epidemiological data suggest the contribution of genetic components in its etiopathogenesis. Oncogenesis is a complex process involving interplay between multiple genetic variants along with the environmental and dietary factors as causing disease or acting as risk modifiers. Detection of these risk sets of genetic variants will facilitate in determining individuals at higher risk for developing GBC.
Previously, we have studied the role of individual genetic variants with GBC susceptibility in a North Indian population. These results suggested the important role of inflammatory and steroidal receptor pathways (MMPs, TIMPs, PLCE, LXRs, and CYPs) in GBC susceptibility [6–9]. Due to low impact of single polymorphisms in complex diseases such as cancer, the current focus is aimed on searching for gene–gene interactions as key contributory factors in the disease outcome. However, the analysis of such interactions in case–control studies is weighed down by one of the major problems, namely, the curse of dimensionality. Since, multifactor dimensionality reduction (MDR) approach and tree-based techniques, classification, and regression trees (CART) and random forest (RF) methodologies have ability to identify association in cases of small sample sizes and low penetrance of candidate single nucleotide polymorphisms (SNPs), these are widely used to detect interactions in association studies Therefore, we have extended our previous work by jointly investigating 15 polymorphisms in nine genes involved in inflammatory, xenobiotic, steroidal receptor pathways, and tumor suppressor genes to find out combinations of genetic variants contributing to GBC risk. The genes included in the study are matrix metallopeptidase (MMP)-2, MMP-7, MMP-9, tissue inhibitor of metalloproteinases (TIMP)-2, cytochrome P450 (CYP)1A1, CYP1B1, phospholipase C epsilon 1 (PLCE1), liver X receptor (LXR)-α, and LXR-beta.
Materials and methods
Ethics statement
Ethics approval for the work was granted by local ethics committee of the institutes, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS) and Department of Surgical Oncology, KGMU Lucknow, India. All participants provided written informed consent for the study. The recruitment of subjects was carried out according to the norms of Helsinki Declaration.
Study population
The present study included 600 subjects comprising 400 consecutive newly diagnosed GBC patients (FNAC and histopathologically proven) and 200 controls. Patients were consecutively diagnosed between June 2008 and September 2012 from the Surgical Oncology, KGMU and Gastrosurgery, SGPGIMS, Lucknow. Staging of cancer was documented according to the AJCC/UICC staging. Inclusion criteria for controls were absence of prior history of cancer, precancerous lesions, and absence of gallstones proven by ultrasonography and were frequency-matched to cancer cases on age, gender, and ethnicity (Table 1). After obtaining informed consent, all the individuals were personally interviewed. Ethics approval for the work was granted by local ethics committee of the institutes. The recruitment of subjects was carried out according to the norms of Helsinki Declaration.
Selected SNPs
MMP-2 c.735 C > T, MMP-2 c.1306 C > T, MMP-9 (p.P574R, p. R668Q, p.R279Q), TIMP-2 c. 418 G > C, CYP1A1-MspI (rs4646903), CYP1A1-Ile462Val (rs1048943), CYP1B1-Val432Leu (rs1056836), and LXR-α T > C (rs7120118) and LXR-β (rs35463555 G > A and rs2695121 T > C) were selected (Table 2).
Genotyping
Genomic DNA was isolated from peripheral blood leukocytes. The polymorphisms were genotyped using the PCR restriction fragment length polymorphism and TaqMan® assays (Applied Biosystems 7500 Fast Real-Time PCR) method. The details of genotyping for studied polymorphisms are as reported in previous studies [6–8]. As a negative control, PCR mix without DNA sample was used to ensure contamination-free PCR product. Samples that failed to genotype were scored as missing. Genotyping was performed without knowledge of the case or control status. The 10 % of the samples were sequenced and showed 100 % concordance.
Statistical analysis
Single locus analysis
Descriptive statistics were presented as mean and standard deviation (SD) for continuous measures while absolute value and percentages were used for categorical measures. The chi-square goodness of fit test was used for any deviation from Hardy–Weinberg equilibrium in controls. Differences in genotype and allele frequencies between study groups were estimated by chi-square test. Unconditional multivariate LR was used to estimate odds ratios (ORs) and their 95 % confidence intervals (CIs) adjusting for age and sex. The ORs were adjusted for confounding factors such as age and gender. A two-tailed p value of less than 0.05 was considered a statistical significant result. All statistical analyses were performed using SPSS software version 16.0 (SPSS, Chicago, IL, USA). The sample size was calculated considering the minor allele frequency (MAF) of the studied polymorphisms in Caucasian population. The sample size of 400 cases and 200 controls was adequate to give us a power of 90 % (inheritance mode = log-additive, genetic effect = 2, type-I error rate = 0.05). Unconditional univariate and multivariate logistic regression analysis was used to estimate odds ratio (OR) and 95 % confidence interval (CI) adjusted for age and gender to estimate the risk of gallbladder cancer with the polymorphisms. Risk estimates were also calculated for a codominant genetic model using the most common homozygous genotype as reference. Tests of linear trend using an ordinal variable for the number of copies of the variant allele (0, 1, or 2) genotype score were conducted to assess potential dose–response effects of genetic variants on gallbladder cancer risk
Multilocus analysis
Multifactor dimensionality reduction
Multifactor dimensionality reduction (MDR) method is non-parametric, genetic model-free method for overcoming some of the limitations of logistic regression (i.e. sample size limitations) for the detection and characterization of gene–gene interactions [10]. In MDR, multilocus genotypes are pooled into high-risk and low-risk groups, effectively reducing the genotype predictors from n-dimensions to one dimension (i.e., constructive induction). The new one-dimensional multilocus genotype variable is evaluated for its ability to classify and predict disease status through cross-validation and permutation testing. The MDR software (version 2.0 beta8) was applied to identify higher order gene–gene interactions associated with GBC risk. In this study, the best candidate interaction model was selected across all multilocus models that maximized testing accuracy and the cross-validation consistency (CVC). Furthermore, validation of models as effective predictors of disease status was derived empirically from 1,000 permutations, which accounted for multiple comparison testing as long as the entire model fitting procedure was repeated for each randomized dataset to provide an opportunity to identify false positives. The MDR permutation results were considered to be statistically significant at the level of 0.05. All the variables identified in the best model were combined and dichotomized according to the MDR software and their ORs and 95 % CIs in relation to GBC risk were calculated. Finally, joint effect of the variables in the best model by the number of risk genotypes was evaluated using logistic regression analysis.
Classification and Regression Tree Analysis
Classification and Regression Tree (CART) analysis was performed using the SPSS version 19 software to build a decision tree via recursive partitioning. For the analysis, decision tree was created by splitting a node into two child nodes repeatedly, beginning with the root node that contains the total sample. Before growing a tree, we choose measure for goodness of split using Gini criteria, by which splits were found that maximize the homogeneity of child nodes with respect to the value of the target variable. After the tree was grown to its full depth, a pruning procedure was performed to avoid over fitting the model. Finally, the risk of various genotypes was evaluated by using the logistic regression analysis. The ORs and 95 % CIs were adjusted for age and sex, with treating the least percentage of cases (case rate) as the reference.
In silico analysis and functional prediction of multilocus-associated SNPs through web-based software
The putative functional effects of were determined by using various online prediction tools viz. FASTSNP (http://fastsnp.ibms.sinica.edu.tw) and F-SNP (http://compbio.cs.queensu.ca/F-SNP/) [11, 12]. In addition, interaction network of all associated gene was determined through by GENEMANIA (http://www.genemania.org/) and String database http://string-db.org/.
Results
Among 400 GBC cases and 200 controls, the mean age was 52.19 ± 10.4 and 45.87 ± 11.5 years, respectively. Most of the GBC patients were in advanced stages of cancer (stage III and stage IV). In GBC cases, 24 (6 %) had stage II adenocarcinomas, 176 (44.0 %) stage III, and 200 (50 %) stage IV. Among GBC, 31 % of the cases were tobacco users and 37 % of the cases had early age of onset, i.e., <50 years. Gallstones were present in 49.2 % of GBC, 192 (48 %) were gallstones negative, and 2.8 % cases had unknown gallstone status. Characteristic of GBC patients and age–sex-matched controls are shown in Table 1.
Single locus analysis of all selected variants
Table 2 shows the GBC risk related to the studied polymorphisms. On comparing the genotype frequency distribution in GBC patients with that of controls, the heterozygous variant containing genotypes of MMP-2 (−735 C > T, −1306 C > T), MMP-7 − 181 A > G, MMP-9 R668Q, TIMP-2 − 418 G > C, CYP1A1 Msp1, CYP1A1-Ile462Val, PLCE1 rs2274223, PLCE1 rs7922612, LXR-beta (rs2695121, rs35463555) showed significant association with GBC risk (adjusted OR > 1; p < 0.05) whereas MMP-9 P574R, MMP-9 R279Q, CYP1B1-Val432Leu, LXR-alpha rs7120118 T > C variations were not associated with the risk of GBC.
Multilocus analysis
Multifactor dimensionality reduction
For higher order gene–gene interaction, multifactor dimensionality reduction (MDR) was performed. The one-factor model for predicting GBC risk was PLCE1 rs2274223 SNP (testing accuracy = 0.548, CVC = 9/10, p < 0.001). The two-factor model of LXR-β rs35463555 and PLCE1 rs2274223 had the testing accuracy of 0.526 but with CVC = 4/10 (p < 0.001). The three-factor model including MMP-9 R668Q, LXR-β rs2695121, and PLCE1 rs2274223 SNPs, which yielded the testing accuracy of 0.512 and the CVC of 7/10 (p = <0.001). Furthermore, the four-factor interaction model consisted of MMP-9 R668Q, LXR-β rs2695121, LXR-β rs35463555, and PLCE1 rs2274223 polymorphisms with an improved testing accuracy of 0.542 and CVC = 7/10 with p < 0.001) (Table 3).
As presence of accompanying gallstones is major risk for GBC, the MDR was performed in case-only analysis based on the presence or absence of gallstones. After the analysis, one-factor model for predicting cholelithiasis-induced GBC risk was LXR rs2695121 SNP (testing accuracy = 0.48, CVC = 7/10, p = 0.03). The two-factor model consisting of LXR rs2695121 and PLCE1 rs2274223 had the testing accuracy of 0.47 but with CVC = 7/10 (p = 0.004). The three-factor model, including MMP-9 R668Q, LXR-β rs2695121, and PLCE1 rs2274223 SNPs, yielded the best interaction model with testing accuracy of 0.612 and the CVC of 10/10 (p < 0.0001). Furthermore, the four-factor interaction model consisted of MMP-9 R668Q, MMP-9 R279Q, LXR-β rs2695121, and PLCE1 rs2274223 polymorphisms with the testing accuracy of 0.563 but CVC = 10/10 with p < 0.0001) (Table 4).
CART results
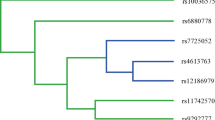
Figure 1 depicts the tree structure generated using the CART, which included all investigated genetic variants of the inflammatory, xenobiotic, steroidal receptor, and tumor suppressor genes. Table 5 shows the classification and regression tree analysis, which includes all investigated genetic variants of the selected pathways. The final tree structure contained nine terminal nodes as defined by single-nucleotide polymorphisms of the overall pathway genes. The initial split of the root node on the decision tree was PLCE1 polymorphism, suggesting that this SNP is the strongest risk factor for GBC among the polymorphisms examined. Individuals carrying LXR-β rs2695121 (W) and PLCE1 rs2274223 (W + M) genotypes had the lowest case rate of 52 %, considered as reference. Further inspection of the tree structure revealed distinct interaction patterns between individuals carrying the wild and variant genotypes of LXR-α rs7120118 (W + M), MMP-2(1306C > T) (W), LXR-β rs35463555 (W), and PLCE1 rs2274223 (H) gene polymorphisms. Table 5 summarizes the risk associated with all the terminal subgroups compared with the subgroup with the least case percentage (node 1). Using the terminal node with lowest case rate as reference, individuals carrying the combination of genotypes exhibited a significantly higher risk for GBC (adjusted OR = 1.9; p = 0.0007). It is apparent that all terminal risk nodes include variants of PLCE1 and LXR-β (Table 5)
In silico analysis
Multilocus analysis revealed that PLCE1 rs2274223 is the major contributing factor in GB carcinogenesis. Molecular phenotype by SNPEffect showed change in secondary structure of protein and solvent accessibility by PLCE1 rs2274223 variations and as well as prediction result to be deleterious [7]. The “PMUT” server predicted the mutation to be pathological, and the results of SNAP prediction by this variation is rs2274223 (H1927R) is non-neutral, and having a predicted accuracy of 70 % showing considerable change in structure [7]. Cyto-HUBBA topological analysis algorithm showed PLCE1 is crucial in protein–protein interaction network telling the PLCE1 as a major gene, and its deregulation may lead to disturbed protein–protein interaction network as shown in our previous studies [7]. Table 6 is showing in silico analysis of associated variants.
In silico analysis of other multilocus associated SNPs is summarized in Table 6. In addition interaction network of all important associated genes is shown in Fig. 2. The interactome is showing interaction of MMP-9, MMP-2, NR1H2 (LXRβ), and NR1H3 (LXR-α). The PLCE1 network is showing most of the PI3K family of genes (Fig. 3).
Discussion
GBC is a complex multifactorial condition involving large number of risk alleles and their interactions acting in combination rather than individually. To date, several genetic variants are known to be associated, but these explain only a minority of the etiology of the GBC. In our previous single locus analysis, out of 15 SNPs, 11 were found to be significantly associated with increased risk of GBC [6–8]. Therefore, for more comprehensive assessment of GBC risk considering several genetic variants simultaneously, and to remove insignificant associations, we carried out multidimensional reduction (MDR) and corelation and regression (CART) analysis with the aim of identifying high-risk sets of genetic variants. The main finding of the study indicates that PLCE1 independently, and together with MMP-9 and LXR-β genetic variations, may be major risk factors for GBC susceptibility.
Both the MDR and CART are non-parametric methods; therefore, no hypothesis concerning the value of any statistical parameter is made. MDR detects multilocus genotype combinations which predict disease risk for common complex and multifactorial diseases. In this present MDR analysis, PLCE1 rs2274223 independently predicted best model with highest testing accuracy and cross-validation consistency. In addition, we observed the best four-factor interaction model consisting of MMP-9 R668Q, LXR-β rs2695121, LXR-β rs35463555, and PLCE1 rs2274223 polymorphisms with testing accuracy of 0.542 and CVC = 7/10 with p < 0.001. In subgroup analysis, LXR-β rs2695121 SNP (testing accuracy = 0.48, CVC = 7/10, p = 0.03) along with MMP-9 R668Q, LXR-β rs2695121, and PLCE1 rs2274223 SNPs still conferred a higher risk in GBC patients with stones as compared to cases without stones In CART analysis, the study subjects were partitioned according to different risk levels. The result from CART analyses again reiterates that the LXR-β rs2695121 and PLCE1 rs2274223 polymorphisms are the most important susceptibility factors for GBC progression. These results suggest that the interaction of above associated SNPs may have significant role in developing risk for GBC.
Both multianalytical approaches revealed that PLCE1 rs2274223 is the major contributing factor in GB carcinogenesis. We had previously reported an association between PLCE1 due to rs2274223 polymorphism in a single locus case–control study for GBC [7]. Three GWAS studies have previously identified significant association of genetic variants of phospholipase C epsilon 1 (PLCE1) with esophageal cancer risk (ESCC), [13–15]. Multiple polymorphisms within the PLCE1 are associated with esophageal cancer via promoting the messenger RNA and protein expression of PLCE1 [16], and its overexpression is associated with cancer metastasis and aggressiveness in esophageal squamous cell carcinoma in a Kazakh population [17]. Moreover, two recent meta-analysis studies had shown that PLCE1 variants are associated with upper gastrointestinal cancers [18] as well as other cancers [19]. PLCE1 gene encodes a phospholipase involved in intracellular signaling. It has been proposed that downregulation of PLCE1 rs2274223 variations may affect the PI3K signaling which has vital role in tumor cell proliferation, motility, metabolism, and survival, and hence could be an attractive therapeutic target in cancer [20].
Liver X receptors (LXR) act as “sensor” proteins that regulate cholesterol uptake, storage and efflux. In our previous studies, we also found significant association of LXR-β variations with gallstone associated GBC [21]. Studies have shown that liver X receptor (LXRs) are expressed in gallbladder cholangiocytes [21]. In animal study knockout of LXR-β (LXRβ −/− ) leads to development of gallbladder cancer in older female mice suggesting estrogen dependent gallbladder carcinogenesis [21]. Activation of liver X receptor-beta (LXR-β) induces transcription of genes associated with reduction of cellular cholesterol concentrations [22]. LXR-αβ −/−double knockout mice model shows elevation of circulating cholesterol and aberrant cholesterol ester accumulation [23]. Functional studied on LXR-β promoter variants had shown altered messenger RNA (mRNA) levels and reduced reporter gene activity, which suggests that variant is associated with lower mRNA levels [24]. The reduced expression of LXR-β results in increased cholesterol accumulation [22]. The LXR-β genetic variants may be responsible for supersaturation of cholesterol in gallbladder by inducing transporters like ABCG-8. The administration of LXR synthetic agonist GW4064 prevented gallstone formation in mice [25]. LXR agonists treatments (TO901317 at 20 μM and 22(R)-HC at 2 μg/ml) have been shown to inhibit the proliferation and apoptosis in MCF-7 cells in breast cancer [26]. Thus LXRs may also be considered as therapeutic candidate for GBC.
The MMPs also play role in cancer progressions which are generally expressed in lower levels under normal physiological conditions, but overexpression has been shown in various cancers [27–29]. MMPs are a family of proteolytic enzymes that are involved in many phases of cancer progression, including angiogenesis, invasiveness, and metastasis. MMPs have elevated level of intracellular expression in gallbladder tumor and gallbladder tumor cell lines [30, 31]. SNPs in the promoter regions of MMP-2 c.735 C > T, c.1306 C > T have allele-specific effects on regulation of MMP gene transcription [32–34]. In silico approaches also predicted significant change in structure of MMP-9 due to transition of R668Q [6]. MMP-9 R668Q variants located in the C-terminal hemopexin-like domain, affecting both substrate and inhibitor binding [35] and conversion of the positively charged amino acid arginine (R) to uncharged amino acid glutamine (G) which might affect the binding of tissue inhibitor of metalloproteinases (TIMPs) with MMP-9 leading to increased extracellular matrix degradation and hence increased inflammation. This can lead to more degradation of extracellular matrix (ECM) and disrupted maintenance and integrity of ECM which is an important event in carcinogenesis. Changes in the structure of the ECM are accompanied by physiological processes such as angiogenesis, apoptosis, and rebuilding of connective tissue [36]. We found significant association of the MMP-2 − 1306 C > T, MMP-9 R668Q with GBC susceptibility previously [6] and in present multianalytical approaches study also. Recently, MMPs are being evaluated as potential target molecules for development of anticancer drugs [37].
The in silco analysis of all multilocus-associated SNPs showed variable change in transcriptional regulation, splicing regulation, and protein coding (Table 6). Moreover, interactome analysis of all associated genes showed indirect connections involving LXRα, LXRβ, MMP-9, and MMP-2 while PLCE1 was out of network. The PLCE1 is a tumor suppressor gene and plays role through PI3K-mediated signaling (Fig. 3). Other genes are part of a network which may have important role in GB carcinogenesis. The observable fact that grouping of polymorphisms within pathway genes may elevate GBC risk can be explained by two hypotheses. One possibility is that some correlation among these genes or proteins exists. Another hypothesis, more expected, is that the genes influencing GBC risk may encompass a set of alterations situated within unrelated genes also. Such an adverse genetic profile could finally lead to appearance of the disease, though particular genes do not share any common functions and separately evoke a slight or unnoticeable effect. Furthermore, there may be multiple sufficient risk sets for GBC. Hence, it is worthwhile to look at many genes together rather than analyzing them individually that may improve identification of risk alleles. In the present study, both MDR and CART categorized the GBC patients into high- and low-risk groups on the basis of selected analyzed polymorphisms. In future, it would be worthwhile to explore other genes in the interacting pathways to further delineate sets of risk genes in GBC predisposition.
In conclusion, the present study suggests that interactions between PLCE1 and LXRβ networks are important risk factors for gallbladder cancer. These findings may have important implications in the understanding of pathobiology of gallbladder cancer.
References
Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167–76.
Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer J Int Cancer. 2006;118:1591–602.
Eslick GD. Epidemiology of gallbladder cancer. Gastroenterol Clin North Am. 2010;39:307–30. ix.
Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II, et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349–64.
Srivastava K, Srivastava A, Sharma KL, Mittal B. Candidate gene studies in gallbladder cancer: a systematic review and meta-analysis. Mutat Res. 2011;728:67–79.
Sharma KL, Misra S, Kumar A, Mittal B. Higher risk of matrix metalloproteinase (MMP-2, 7, 9) and tissue inhibitor of metalloproteinase (TIMP-2) genetic variants to gallbladder cancer. Liver Int. 2012;32:1278–86.
Sharma KL, Umar M, Pandey M, Misra S, Kumar A, Kumar V, Mittal B. Association of potentially functional genetic variants of plce1 with gallbladder cancer susceptibility in north indian population. J Gastrointest Cancer. 2013.
Sharma KL, Misra S, Kumar A, Mittal B. Association of liver x receptors (LXRs) genetic variants to gallbladder cancer susceptibility. Tumour Biol. 2013.
Sharma KL, Agarwal A, Misra S, Kumar A, Kumar V, Mittal B. Association of genetic variants of xenobiotic and estrogen metabolism pathway (CYP1A1 and CYP1B1) with gallbladder cancer susceptibility. Tumour Biol.
Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene–gene and gene–environment interactions. Bioinformatics. 2003;19:376–82.
Lee PH, Shatkay H. F-SNP: Computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–4.
Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–41.
Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759–63.
Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, et al. A shared susceptibility locus in plce1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764–7.
Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y, et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet. 2011;43:679–84.
Cui XB, Chen YZ, Pang XL, Liu W, Hu JM, Li SG, et al. Multiple polymorphisms within the PLCE1 are associated with esophageal cancer via promoting the gene expression in a Chinese Kazakh population. Gene. 2013;530:315–22.
Chen YZ, Cui XB, Hu JM, Zhang WJ, Li SG, Yang L, et al. Overexpression of PLCE1 in Kazakh esophageal squamous cell carcinoma: implications in cancer metastasis and aggressiveness. APMIS. 2013;121:908–18.
Hao NB, He YF, Zhang D, Luo G, Chen BJ, Zhang Y, et al. PLCE1 polymorphism and upper gastrointestinal cancer risk: a meta-analysis. PLoS One. 2013;8:e67229.
Umar M, Upadhyay R, Mittal B. PLCE1 rs2274223 A>G polymorphism and cancer risk: A meta-analysis. Tumour Biol. 2013.
Zhang J, Roberts TM, Shivdasani RA. Targeting PI3K signaling as a therapeutic approach for colorectal cancer. Gastroenterology. 2011;141:50–61.
Gabbi C, Kim HJ, Barros R, Korach-Andre M, Warner M, Gustafsson JA. Estrogen-dependent gallbladder carcinogenesis in LXRβ−/−female mice. Proc Natl Acad Sci U S A. 2010;107:14763–8.
Infante J, Rodriguez-Rodriguez E, Mateo I, Llorca J, Vazquez-Higuera JL, Berciano J, et al. Gene–gene interaction between heme oxygenase-1 and liver X receptor-β and Alzheimer's disease risk. Neurobiol Aging. 2010;31:710–4.
Pommier AJ, Dufour J, Alves G, Viennois E, De Boussac H, Trousson A, et al. Liver X receptors protect from development of prostatic intra-epithelial neoplasia in mice. PLoS Genet. 2013;9:e1003483.
Dahlman I, Nilsson M, Gu HF, Lecoeur C, Efendic S, Ostenson CG, et al. Functional and genetic analysis in type 2 diabetes of liver X receptor alleles—a cohort study. BMC Med Genet. 2009;10:27.
Wang J, Einarsson C, Murphy C, Parini P, Bjorkhem I, Gafvels M, et al. Studies on LXR- and FXR-mediated effects on cholesterol homeostasis in normal and cholic acid-depleted mice. J Lipid Res. 2006;47:421–30.
El Roz A, Bard JM, Huvelin JM, Nazih H. LXR agonists and ABCG1-dependent cholesterol efflux in MCF-7 breast cancer cells: Relation to proliferation and apoptosis. Anticancer Res. 2012;32:3007–13.
Stenvold H, Donnem T, Andersen S, Al-Saad S, Al-Shibli K, Busund LT, Bremnes RM. Overexpression of matrix metalloproteinase-7 and -9 in NSCLC tumor and stromal cells: correlation with a favorable clinical outcome. Lung Cancer. 75:235-241.
Yang JM, Xu Z, Wu H, Zhu H, Wu X, Hait WN. Overexpression of extracellular matrix metalloproteinase inducer in multidrug resistant cancer cells. Mol Cancer Res. 2003;1:420–7.
Perigny M, Bairati I, Harvey I, Beauchemin M, Harel F, Plante M, et al. Role of immunohistochemical overexpression of matrix metalloproteinases MMP-2 and MMP-11 in the prognosis of death by ovarian cancer. Am J Clin Pathol. 2008;129:226–31.
Wang JH, Li LF, Yu Y, Li B, Jin HJ, Shen DH, et al. Establishment and characterization of a cell line, EH-GB2, derived from hepatic metastasis of gallbladder cancer. Oncol Rep. 2012;27:775–82.
Koyama S. Intracellular localization of matrix metalloproteinases and their inhibitors in cultured tumor cell lines: flow cytometric analysis. Oncol Rep. 2006;15:735–8.
Decock J, Paridaens R, Ye S. Genetic polymorphisms of matrix metalloproteinases in lung, breast and colorectal cancer. Clin Genet. 2008;73:197–211.
Zhou G, Zhai Y, Cui Y, Qiu W, Yang H, Zhang X, et al. Functional polymorphisms and haplotypes in the promoter of the MMP2 gene are associated with risk of nasopharyngeal carcinoma. Hum Mutat. 2007;28:1091–7.
Qiu W, Zhou G, Zhai Y, Zhang X, Xie W, Zhang H, et al. No association of MMP-7, MMP-8, and MMP-21 polymorphisms with the risk of hepatocellular carcinoma in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2008;17:2514–8.
Murphy G, Knauper V. Relating matrix metalloproteinase structure to function: why the "Hemopexin" domain? Matrix Biol. 1997;15:511–8.
Trojanek J. matrix metalloproteinases and their tissue inhibitors. Postepy Biochem. 2012;58:353–62.
Hidalgo M, Eckhardt SG. Development of matrix metalloproteinase inhibitors in cancer therapy. J Natl Cancer Inst. 2001;93:178–93.
Acknowledgments
The funding for the study was provided by DST and DBT Government of India. Research fellowship to KLS by ICMR, New Delhi is gratefully acknowledged.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, K.L., Rai, R., Srivastava, A. et al. A multigenic approach to evaluate genetic variants of PLCE1, LXRs, MMPs, TIMP, and CYP genes in gallbladder cancer predisposition. Tumor Biol. 35, 8597–8606 (2014). https://doi.org/10.1007/s13277-014-2094-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2094-7