Abstract
Growing number of long noncoding RNAs (lncRNAs) are emerging as new modulators in cancer origination and progression. A lncRNA, ADAM metallopeptidase with thrombospondin type 1 motif, 9 (ADAMTS9) antisense RNA 2 (ADAMTS9-AS2), with unknown function, is the antisense transcript of tumor suppressor ADAMTS9. In the present study, we investigated the expression pattern and functional role of ADAMTS9-AS2 in glioma by using real-time PCR and gain-/loss-of-function studies. The results showed that the ADAMTS9-AS2 expression was significantly downregulated in tumor tissues compared with normal tissues and reversely associated with tumor grade and prognosis. Multivariate analysis of the prognosis factors showed that low ADAMTS9-AS2 expression was a significant independent predictor of poor survival in glioma. Overexpression of ADAMTS9-AS2 resulted in significant inhibition of cell migration in glioma, whereas knockdown of ADAMTS9-AS2 showed the opposite effect. We also found that ADAMTS9-AS2 expression was negatively correlated with DNA methyltransferase-1 (DNMT1). In addition, DNMT1 knockdown led to remarkable enhancement of ADAMTS9-AS2 expression. By 5-aza-dC treatment, the ADAMTS9-AS2 expression was also reactivated. The results suggested that ADAMTS9-AS2 is a novel tumor suppressor modulated by DNMT1 in glioma. LncRNA ADAMTS9-AS2 may serve as a potential biomarker and therapeutic target for glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is the most common malignant tumor in the central nervous system [1]. The development of glioma is a complex process with multigene interaction and multimolecular modulation [2]. It has been proved that a large portion of human genome is transcribed into noncoding RNAs (ncRNAs). Long noncoding RNAs (lncRNAs) are those ncRNA transcripts larger than 200 bases in length [3]. Distinct from miRNAs and other smaller noncoding RNAs, lncRNAs can modulate the downstream target genes by multiple means via cis- and trans-regulatory effects, in both transcriptional and posttranscriptional levels [4, 5]. Although many lncRNAs have been demonstrated to exert crucial regulating activities in extensive cellular processes in human diseases, the function of most lncRNAs is still unknown. Recently, a growing number of lncRNAs have been found linked with various types of cancer [6-8]. Among them, a common type of cancer-related lncRNAs is an antisense partner of a protein coding gene. Examples include ANRIL (CDKN2B-AS1) [9, 10], CTBP1-AS [11], HNF1A-AS1 [12], and GAS6-AS1 [13].
By our previous bioinformatics analysis of the lncRNA-messenger RNA (mRNA) targeting relationships in lung, colon, gastric, and liver cancers (data not published), we noticed that a lncRNA ADAM metallopeptidase with thrombospondin type 1 motif, 9 (ADAMTS9) antisense RNA 2 (ADAMTS9-AS2; Ensembl, ENSG00000241684) may be closely related with some important cancer-related genes. LncRNA ADAMTS9-AS2 is an antisense transcript of protein coding gene ADAMTS9. In addition, transcript factors chip-seq data from encode/analysis (http://genome.ucsc.edu/) also showed that many cancer-related transcript factors interact with the ADAMTS9-AS2 locus, especially at the promoter region, which ADAMTS9-AS2 shares with its mRNA partner ADAMTS9. This lncRNA/mRNA gene pair is located at chromosome 3p14.1, a region known to be lost in hereditary renal cancers [14]. ADAMTS9 is an important anti-angiogenesis factor, which can inhibit tumor progression and metastasis by regulating fibroblast growth factor (FGF) and vascular endothelial growth factor (VEGF) [15]. ADAMTS9 has already been identified as a tumor suppressor in esophageal, gastric, breast, and nasopharyngeal cancers, the expression is mainly regulated by DNA hypermethylation [16-20], and in gliomas, inhibition of specific ADAMTS targets could slow glioma invasion [21]. However, whether the lncRNA ADAMTS9-AS2 is associated with cancer is still unknown, which prompted us to conduct the current study.
In this study, we first investigated the ADAMTS9-AS2 expression profile and its relationship with clinical pathological features in glioma patients. Moreover, we also evaluated its functional role in glioma cell migration and the possible modulating mechanism.
Materials and methods
Patient samples
Patients with glioma (n = 70) who underwent initial surgery in Chinese PLA General Hospital from 2005 to 2007 were selected for this study. Clinical pathology information was available for all samples (Table 1). No patients had received therapy before resection. Each patient participated after providing informed consent, and the use of the tumor samples for research was approved by the ethical committee of Chinese PLA General Hospital. All tumors were classified on the basis of the WHO criteria for tumors of the central nervous system and quick frozen at the time of resection until analysis. The series consisted of 46 cases of low-grade astrocytoma (17 cases of WHO grade I and 29 cases of WHO grade /II) and 24 cases of high-grade disease (14 cases of anaplastia astrocytoma (WHO grade III) and 10 cases of glioblastoma multiforme (WHO grade IV)).
Cancer cell lines
The human glioma cell lines T98G, A172, SNB-19, U87, and U251 were purchased from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) medium (Gibco, Carlsbad, CA, USA) containing 10 % fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37 °C with 5 % CO2.
Real-time quantitative PCR
Total RNA was isolated from glioma tumor tissue, adjacent normal tissue, and glioma cell lines using the Trizol Total RNA Reagent (Invitrogen). Complementary DNA (cDNA) synthesis was performed with 2 μg total RNA using the RevertAidTM H Minus First Strand cDNA Synthesis Kit (Takara, Otsu, Japan). The primers were obtained from GenePharma (Shanghai, China), and the sequences were shown in Table 2. Quantitative PCR was performed using the SYBR PrimeScript RT-PCR kit (Takara) in an Applied Biosystems 7500 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA). The reaction mixtures were incubated at 95 °C for 30 s, followed by 40 amplification cycles of 95 °C for 5 s and 60 °C for 34 s. The quantification of gene expression was performed by using the ΔΔCT calculation with CT as the threshold cycle. The expression level of a target gene in a patient was calculated as the ratio: target in tumor tissue/target in nontumorous tissue (R (T/N)).
Western blot analysis
Cells were washed twice with ice-cold PBS and cell lysates were harvested by the addition of lysis buffer (40 nM Tris (pH 7.4), 150 mM NaCl, 10 mM ethylene diamine tetraccetic acid, 10 % glycerol, 1 % Triton X-100, 10 mM glycerophosphate, 1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride) supplemented with protease inhibitor (Roche). Thirty micrograms of protein lysates were separated on a NuPAGE 4–12 % Bis-Tris Gel (Invitrogen), and the separated proteins were transferred onto a polyvinylidene difluoride membrane (Invitrogen). After blocking for 60 min with 5 % nonfat dry milk, membranes were incubated with the primary antibody overnight at 4 °C followed by incubation with corresponding secondary antibody for 60 min at room temperature. The membranes were developed using enhance chemiluminescence solutions (Thermo Fisher Scientific).
Overexpression of ADAMTS9-AS2 in glioma cells
Plasmid cDNA-ADAMTS9-AS2 was constructed by introducing aBamHI-EcoRI fragment containing the ADAMTS9-AS2 cDNA into the same site in pcDNA3.1. The ADAMTS9-AS2 low-expressed T98G cells were transfected with pcDNA-ADAMTS9-AS2 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were collected after transfection for RNA isolation, scratch wound-healing assay, and Matrigel invasion assay.
Transfection of siRNAs in glioma cells
Small interfering RNA (siRNA) sequences for target genes and negative-control (NC) siRNA were obtained from GenePharma (Shanghai, China). The sequences were shown in Table 3 [22, 23]. For ADAMTS9-AS2 siRNA transfection, ADAMTS9-AS2 high-expressed U87 was selected, for DNA methyltransferases (DNMTs) siRNA transfection, both T98G and U87 cells were selected. In brief, approximately 5 % cells were plated to each well of 12-well plates at least 24 h before transfection to achieve 30–50 % confluency. SiRNA transfection was done with X-tremeGENE transfection sreagent (Roche) according to the manufacturer's instructions. Forty-eight hours after transfection, RNA isolation, scratch wound-healing assay, and Matrigel invasion assay were performed.
Treatment of T98G and U87 cells with 5-aza-dC
T98G and U87 cells were planted into plastic 6-well plates on day 0 and exposed to 5 μM 5-aza-dC (Sigma, St. Louis, MO, USA) for 3 days. The cells treated with 5-aza-dC were collected for RNA isolation and detection of ADAMTS9-AS2 expression.
Cell proliferation assay
After transfection, cell proliferation was assessed by MTS assay (Promega) according to the manufacturer’s protocol. T98G or U87 cells (2,000 cells per well) in each group were plated in 96-well plates. Twenty microliters of the MTS reagent was added to each well containing 100 μl culture medium. The plate was incubated for 2 h at 37 °C in a humidified, 5 % CO2 atmosphere. The plate was then read at 490 nm using a plate reader.
Scratch wound-healing assay
Uniform wounds were scraped in T98G and U87 cells grown on plastic 6-well plates using a pipette tip before transfection. The initial gap length (0 h) and the residual gap length 48 h after wounding were calculated from photomicrographs.
Matrigel invasion assays
A cell invasion assay was carried out using modified Boyden Chambers consisting of transwell-precoated Matrigel membrane filter inserts with 8-mm pores in 24-well tissue culture plates (BD Biosciences, Bedford, MA, USA). DMEM containing 10 % fetal bovine serum in the lower chamber served as the chemoattractant. Cells that had migrated through the filter were stained and counted. The average migration rate was calculated as the increasing radius of the entire cell population over time.
Statistical analysis
Differences between groups were analyzed using Student's t test or Chi-square test. Correlation between genes expression was studied by using Pearson's correlation. Survival analysis was performed using the Kaplan–Meier method, and the log-rank test was used to compare the differences between patient groups. Statistical analyses were performed using SPSS version 18.0 (SPSS, Chicago, IL). For all statistical analyses, P < 0.05 was considered statistically significant.
Results
ADAMTS9-AS2 was downregulated in glioma tissue samples and associated with prognosis
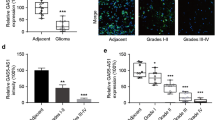
The ADAMTS9-AS2 expression levels were assessed in a panel-paired specimen obtained from 70 patients with glioma. ADAMTS9 expression in glioma tumor tissues was significantly downregulated compared with matched nontumorous tissues (Fig. 1a, b). The expression level of ADAMTS9-AS2 was significantly lower in tumors with higher WHO grade (III/IV) than that in tumors with lower WHO grade (I/II).
Relative ADAMTS9-AS2 expression in 70 glioma tissue samples was examined by real-time PCR and normalized to GAPDH expression. Data were presented as fold change in tumor tissues relative to normal tissues. a ADAMTS9-AS2 expression level was significantly lower in tumor tissues than that in normal tissues (*P < 0.05). b ADAMTS9-AS2 expression level was significantly lower in tumors of higher WHO grade (III/IV) than those in tumors of lower WHO grade (I/II) (*P < 0.05). c Kaplan–Meier overall survival curves according to ADAMTS9-AS2 expression level. The overall survival of the high-ADAMTS9-AS2 group (n = 35, ADAMTS9-AS2 expression ratio ≥ median ratio) was significantly higher than that of the low-ADAMTS9-AS2 group (n = 35, ADAMTS9-AS2 expression ratio ≤ median ratio; P = 0.0326, log-rank test)
Kaplan–Meier survival analysis and log-rank tests were performed to assess the association between ADAMTS9-AS2 expression and glioma patient prognosis. According to the mean ratio of relative ADAMTS9-AS2 expression (median ration of 0.315-fold) in glioma tissue specimens, the 53 glioma cancer patients were divided into two groups: relatively high-ADAMTS9-AS2 group (n = 35, ADAMTS9-AS2 expression ratio ≧ median ratio) and relatively low-ADAMTS9-AS2 group (n = 35, ADAMTS9-AS2 expression ratio ≦ median ratio). We found the overall survival of the high-ADAMTS9-AS2 group was significantly higher than that of the low-ADAMTS9-AS2 group (P = 0.0326). The results suggested that the downregulation of ADAMTS9-AS2 might have a critical role in the origination and progression of glioma. Univariate analysis identified five prognostic factors: WHO grade (I/II or III/IV), KPS (<80 or≧80), TNM stage (I/II or III/IV), tumor size (<5 or ≧5 cm), and ADAMTS9-AS2 expression. The other clinicopathological characteristics, such as age, gender, tumor size, extent of resection, whether or not accepted radiotherapy, and chemotherapy, were not statistically significant prognosis factors. Multivariate analysis of the prognosis factors confirmed that low-ADAMTS9-AS2 expression was a significant independent predictor of poor survival in glioma (P = 0.013), in addition to WHO grade (P = 0.021), KPS (P = 0.002), and tumor size (P = 0.045) (Table 4).
Expression of ADAMTS9-AS2 correlated with expression of ADAMTS9 and DNMT1
To identify the relationship between ADAMTS9-AS2 and ADAMTS9, we assessed the correlation of their expression levels using real-time PCR. The result indicated that the ADAMTS9-AS2 expression was positively correlated with ADAMTS9 expression (R = 0.49, P < 0.01; Table 5).
By reviewing the UCSC data, we noticed that extensive DNA methylation and histone modification can be found at the promoter region of ADAMTS9-AS2 and ADAMTS9 (http://genome.ucsc.edu/). To test the potential association between ADAMTS9-AS2 and DNA methylation-related genes in glioma, we assessed the expression correlation of ADAMTS9-AS2 with three DNMTs, DNMT1, DNMT3A, and DNMT3B. In Table 3, the results showed a positive correlations of ADAMTS9-AS2 expression with DNMT3A (R = 0.44, P = 0.004; Table 5) and a negative correlation with DNMT1 (R = −0.45, P = 0.004; Table 5).
ADAMTS9 expression was coregulated together with the ADAMTS9-AS2 overexpression or knockdown in glioma cell lines
By real-time PCR, we found that the expression of ADAMTS9-AS2 was the highest in U87 and the lowest in T98G among the 5 human glioma cell lines (Fig. 2a).
ADAMTS9 mRNA expression level was coregulated together with the ADAMTS9-AS2 lncRNA overexpression or knockdown in glioma cell lines. a The expression of ADAMTS9-AS2 was tested in five human glioma cell lines by real-time PCR. b, c ADAMTS9 mRNA and ADAMTS9-AS2 lncRNA level were measured by real-time PCR in T98G cells transfected with plasmid pcDNA-ADAMTS9-AS2. ADAMTS9-AS2 lncRNA expression was markedly elevated by pcDNA-ADAMTS9-AS2 (*P < 0.05), and ADAMTS9 mRNA expression level was also evidently upregulated concomitantly (*P < 0.05). Western blot showed that the ADAMTS9 protein expression was also markedly elevated. d, e ADAMTS9 mRNA and ADAMTS9-AS2 lncRNA level knockdown were measured by real-time PCR in U87 cells transfected with ADAMTS9-AS2 siRNA. Expression of ADAMTS9-AS2 and ADAMTS9 were significantly reduced in ADAMTS9-AS2 siRNA groups compared with negative control group (*P < 0.05)
In the overexpression experiment, ADAMTS9-AS2 cDNA plasmid was constructed and stably transfected into T98G cells. By real-time PCR, we found the lncRNA ADAMTS9-AS2 expression was markedly elevated, and the ADAMTS9 mRNA expression level was also evidently upregulated by ADAMTS9-AS2 cDNA plasmid (Fig. 2b, c). By Western blot, we found that the ADAMTS9 expression was also markedly elevated in protein level.
In the knockdown experiment, ADAMTS9-AS2 siRNAs was transfected into U87 cells. Quantification PCR and Western blot analysis showed that ADAMTS9-AS2 expression level was significantly knocked down in ADAMTS9-AS2 siRNA group and ADAMTS9 expression level was also downregulated by ADAMTS9-AS2 siRNA, both in mRNA level and protein level (Fig. 2d, e). The results indicated that the ADAMTS9 expression may be modulated by lncRNA ADAMTS9-AS2.
ADAMTS9-AS2 expression was restored by 5-aza-dC treatment or DNMT1 siRNA in glioma cell lines
UCSC data suggests the hypermethylation of the ADAMTS9-AS2 promoter region (http://genome.ucsc.edu/). To evaluated the role of DNA methylation in ADAMTS9-AS2 lncRNA inactivation in glioma. We assessed the impact of DNA demethylating agent (5-aza-dC) on ADAMTS9-AS2 expression. In both U87 and T98G cells, we found that ADAMTS9-AS2 expression levels were evidently increased in 5-aza-dC groups compared with control groups (Fig. 3b).
ADAMTS9-AS2 expression in glioma cells with 5-aza-dC treatment or DNMTs siRNA. a Real-time PCR analysis of ADAMTS9-AS2 expression following treatment with 5 μM of 5-aza-dC in U87 and T98G cells (*P < 0.05). b Real-time PCR analysis of ADAMTS9-AS2 expression following transfection of siRNA of three target DNA methyltransferases genes (DNMT1, DNMT3A, and DNMT3B) in U87 and T98G cells (*P < 0.05)
Subsequently, we investigated whether DNMTs might contribute to ADAMTS9-AS2 downregulation. By siRNA transfection of three target DNA methyltransferases genes (DNMT1, DNMT3A, and DNMT3B), we found that the DNMT1 siRNA let to remarkably elevation of ADAMTS9-AS2.
It suggested that the silencing of ADAMTS9-AS2 in glioma might partly resulted by hypermethylation of the ADAMTS9-AS2 promoter region. In addition, a DNA methyltransferases DNMT1 may play an important role in ADAMTS9-AS2 regulation.
T98G cell migration and invasion were inhibited by ADAMTS9-AS2 overexpression
To further identify the role of ADAMTS9-AS2 in T98G cells, functional assay was performed by transfecting plasmid cDNA ADAMTS9-AS2 and negative control plasmid. The results showed that the cells growth in pcDNA-ADAMTS9-AS2 group was not significantly altered compared with the cells in the pcDNA control group (Fig. 4a). The wound-healing assay showed remarkable cell migration retardation in pcDNA-ADAMTS9-AS2 group compared with in the pcDNA control group (Fig. 4b). The Matrigel invasion assay also showed significant cell invasion inhibition in the pcDNA-ADAMTS9-AS2 group compared with the pcDNA control group (Fig. 4c).
Cell migration and invasion were inhibited by pcDNA-ADAMTS9-AS2 in T98G cells. a Cell proliferation fold increase was tested using MTS with 1-day intervals. PcDNA-ADAMTS9-AS2 group did not show significant different growth rate compared with pcDNA control group in T98G cells (*P < 0.05). b, c Overexpression of ADAMTS9-AS2 inhibited the cell migration and invasion. Scratch wound healing assay and Matrigel invasion assay showed that the cell migration and invasion capacities in pcDNA-ADAMTS9-AS2 group were remarkably inhibited (*P < 0.05)
U87 cell migration and invasion were enhanced by ADAMTS9-AS2 knockdown
The results showed that the growth rates of U87 cells in ADAMTS9-AS2 siRNA-transfected groups were not significantly different from the negative controls group (Fig. 5a). The scratch wound-healing assay and Matrigel invasion assay were performed also using U87 cells. The wound-healing assay showed remarkable cell migration elevation in ADAMTS9-AS2 siRNA group compared with in the NC group (Fig. 5b). The Matrigel invasion assay also showed significant cell invasion elevation in the ADAMTS9-AS2 siRNA groups compared with the NC group (Fig. 5c).
Cell migration and invasion were elevated by ADAMTS9-AS2 siRNA in U87 cells. a Cell proliferation fold increase was tested using MTS with 1-day intervals. ADAMTS9-AS2 siRNA groups showed elevated growth rates compared with negative control groups in U87 cells (*P < 0.05). b, c Knockdown of ADAMTS9-AS2 enhanced the cell migration and invasion. Scratch wound healing assay and Matrigel invasion assay showed that the cell migration and invasion capacities in ADAMTS9-AS2 siRNA group were remarkably elevated (*P < 0.05)
Discussion
Recently, more and more studies have proved that the lncRNAs can play pivotal role in cancer development. LncRNAs has been regarded as the new frontier of translational research in cancer diagnosis and treatment [6-8]. There are a few prior microarray data on the association between glioma and lncRNAs [24-27]. In addition, it has been demonstrated that some well-known cancer-related lncRNAs, such as HOTAIR, H19, and MEG3, are also involved in the tumor progression of glioma [28-30]. The emerging role of lncRNAs in glioma prompted us to conduct the current study.
LncRNA ADAMTS9-AS2 is the antisense partner of protein coding gene ADAMTS9. ADAMTS9 has been recognized as an important tumor suppressor in various human malignancies, the expression is mainly controlled by promoter hypermethylation [10-21].
In the present study, we demonstrated the deregulation of lncRNA ADAMTS9-AS2 was linked with the tumor occurrence in glioma, and lower expression of ADAMTS9-AS2 was associated with higher WHO grades. Multivariate analyses showed that ADAMTS9-AS2 expression was an independent predictor for overall survival of the patients. Moreover, the results also showed that overexpression of ADAMTS9-AS2 inhibited migration and invasion abilities of the T98G cells allowing increased tumor progression. While knockdown of ADAMTS9-AS2 in U87 cells showed the opposite effect. To our knowledge, this maybe the first study about the expression pattern and functional role of lncRNA ADAMTS9-AS2 in cancer.
In other recent studies, some lncRNAs, like HOTAIR and H19, have also been linked with glioma cell invasion [28, 29], and HOTAIR has also been identified as a strong prognostic predictor in mesenchymal glioma [29]. In addition, our data may provide the first evidence that lncRNA ADAMTS9-AS2 might reversely correlate with the tumor progression and inhibit the cell invasion in glioma.
It has been proved that the antisense lncRNAs may often modulate their sense mRNA partners, either in discordant or concordant manner [31-33]. Thus far, multiple antisense lncRNA have been found to be associated with various malignant tumors [9-12]. Among them, some modulate their sense partners mainly by epigenetic regulation at the promoter regions, for example , ANRIL (CDKN2B-AS1) [9, 10], some exert their functions by regulating the sense transcripts by other methods, such as CTBP1-AS [11] and GAS6-AS1 [13], whereas some others exert their function without regulating their sense partner like HNF1A-AS1 [12].
Our results showed that the expression of ADAMTS9-AS2 was positively correlated with its sense partner, ADAMTS9. Then, to further assess the regulating role of lncRNA ADAMTS9-AS2 to the protein coding gene ADAMTS9, overexpression and knockdown experiments was performed. The results showed that the ADAMTS9 expression was altered concomitantly with the ADAMTS9-AS2 up- and downregulation in glioma cells. Based on these results, we speculated that lncRNA ADAMTS9-AS2 may fulfill its tumor suppressing function partly by modulating the ADAMTS9 expression.
In previous studies, many cancer-related lncRNAs have been revealed to be involved in epigenetic regulating network [6, 7]. According to the UCSC data, extensive DNA methylation and histone modification markers can be found at the promoter of ADAMTS9-AS2 and ADAMTS9 (http://genome.ucsc.edu/). Here, we showed that the DNA methylation may contribute to the ADAMTS9-AS2 lncRNA inactivation in glioma. By using DNA demethylating agent (5-aza-dC), we found the ADAMTS9-AS2 expression was evidently reactivated. In addition, by knocking down three DNA methyltransferases genes (DNMT1, DNMT3A, and DNMT3B), we found that the DNMT1 knockdown significantly restored the ADAMTS9-AS2 expression.
Taken together, this study provides the first evidence that the decrease of lncRNA ADAMTS9-AS2 expression is associated with the diagnosis, clinicopathological characteristics, and prognosis of glioma, and the function of ADAMTS9-AS2 is involved in suppressing the cell migration ability, partly by regulating the protein coding gene ADAMTS9. Our data also indicates that the expression of ADAMTS9-AS2 may be modulated by DNA methylation through the methyltransferases DNMT1. Tumor suppressor lncRNA ADAMTS9-AS2 may serve as a novel biomarker and therapeutic target for glioma in future cancer clinic.
References
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classifcation of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109.
Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN. Malignant glioma: genetics and biology of a grave matter. Gene Dev. 2001;15:1311–33.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7.
Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9.
Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–81.
Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:152–61.
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–87.
Tsai MC, Spitale RC, Chang HY. Long. Intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7.
Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–9.
Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6.
Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–80.
Yang X, Song JH, Cheng YL, Wu WJ, Bhagat T, and Yu YT. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut .2013;0:1–10.
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De W, et al. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol. 2013;30:694.
Clark ME, Kelner GS, Turbeville LA, Boyer A, Arden KC, Maki RA. ADAMTS9, a novel member of the ADAM-TS/ metallospondin gene family. Genomics. 2000;67(3):343–50.
DEMIRCAN K, GUNDUZ E, GUNDUZ M, BEDER LB, HIROHATA S, NAGATSUKA H, et al. Increased mRNA expression of ADAMTS metalloproteinases in metastatic foci of head and neck cancer. Head Neck. 2009;31:793–801.
Lo PH, Leung AC, Kwok CY, Cheung WS, Ko JM, Yang LC, et al. Identification of a tumor suppressive critical region mapping to 3p14.2 in esophageal squamous cell carcinoma and studies of a candidate tumor suppressor gene, ADAMTS9. Oncogene. 2007;26(1):148–57.
Lung HL, Lo PH, Xie D, Apte SS, Cheung AK, Cheng Y, et al. Stanbridge and M. L. Lung: Characterization of a novel epigenetically-silenced, growth-suppressive gene, ADAMTS9, and its association with lymph node metastases in nasopharyngeal carcinoma. Int J Cancer. 2008;123(2):401–8.
Sheu JJ, Lee CH, Ko JY, Tsao GS, Wu CC, Fang CY, et al. Chromosome 3p12.3-p14.2 and 3q26.2–q26.32 are genomic markers for prognosis of advanced nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2709–16.
OCAK Z, ACAR M, GUNDUZ E, GUNDUZ M, DEMIRCAN K, UYETURK U, et al. Effect of hypericin on the ADAMTS-9 and ADAMTS-8 gene expression in MCF7 breast cancer cells. Eur Rev Med Pharmacol Sci. 2013;17:1185–90.
Du W, Wang S, Zhou Q, Li X, Chu J, Chang Z, et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32:3319–28.
Viapiano MS, Hockfield S, Matthews RT. BEHAB/brevican requires ADAMTS-mediated proteolytic leavage to promote glioma invasion. J Neurooncol. 2008;88:261–72.
Zhao ZJ, Wu QX, Cheng J, Qiu XM, Zhang JQ, Fan H. Depletion of DNMT3A suppressed cell proliferation and restored PTEN in hepatocellular carcinoma cell. J Biomed Biotechnol. 2010;2010:737535.
Lee J, Jang SJ, Benoit N, Hoque MO, Califano JA, Barry T, et al. Presence of 5-methylcytosine in CpNpG trinucleotides in the human genome. Genomics. 2010;96:67–72.
Han L, Zhang KL, Shi ZD, Zhang JX, Zhu JL, Zhu SJ, et al. LncRNA profile of glioblastoma reveals the potential role of LncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40(6):2004–12.
Li R, Qian J, Wang YY, Zhang JX, YP Y, et al. Long noncoding RNA profiles reveal three molecular subtypes in glioma. CNS Neurosci Ther. 2014;2014:1–5.
Zhang XQ, Sun S, JK Pu ACT, Lee D, Man VY, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8.
Zhang XQ, Sun S, Lam KF, Kiang KY, Pu JK, Ho AS, et al. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis. 2013;58:123–31.
JX Zhang LH, Bao ZS, Wang YY, Chen LY, Yan W, et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-Oncology. 2013;15(12):1595–603.
Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, et al. Long non-coding RNA H19 promotes glioma cell Invasion by deriving miR-675. PLoS ONE. 2014;9:e86295.
Wang PJ, Ren ZQ, Sun PY. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113(6):1868–74.
Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–8.
Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;1:637–43.
Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–86.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Additional information
Jie Yao, Baosheng Zhou, Jian Zhang, and Peiliang Geng are co-first author
Rights and permissions
About this article
Cite this article
Yao, J., Zhou, B., Zhang, J. et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumor Biol. 35, 7935–7944 (2014). https://doi.org/10.1007/s13277-014-1949-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-1949-2