Abstract
Interleukin (IL) 17A is an inflammatory cytokine expressed by Th 17 cells and plays a role in tissue inflammation by inducing release of proinflammatory and neutrophil-mobilizing cytokines. We have investigated the association between colorectal cancer and polymorphisms of IL17A (rs2275913. G197A). The study was performed in 241 subjects (102 with colorectal cancer and 139 healthy controls). Genotypes were determined by fluorescent-based restriction fragment length polymorphism method. The association between the molecular features at the gene in relation to tumor and patient clinical characteristics was analyzed. There was a significant difference between the genotype frequencies of IL17A G197A of control subjects (GG 68.34 % and GA + AA 31.65 %) and patients with colorectal cancer (GG 47.05 % and GA + AA 52.94 %) (p = 0.001with odds ratio (OR) 2.45 (1.43–4.11)). IL17A G197A polymorphism is particularly associated with colon cancer. Indeed, the IL17A GG genotype could be considered as a protective factor against colon cancer (p = 0.00001) with OR 3.77 (2.04–6.99). We have noted a significant association of IL17A G197A polymorphism not only with tumor localization (p = 0.003) but also with tumor differentiation (p = 0.0005) in CRC patients. We have also showed a significant association of G197A variant with an increased risk of advanced stage (p = 0.005). Our result suggests that the A allele of IL17A gene is involved in susceptibility to colorectal cancer and is associated with clinical features as tumor location, tumor differentiation, and TNM stage. IL17A polymorphism may serve as biomarker of disease location and progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related death worldwide. Several factors can increase the risk of colorectal cancer [1]. The major causes of this cancer are environmental, genetic, epigenetic as well as inflammatory bowel disease (IBD).
The association between inflammation and cancer was illustrated by epidemiological and clinical studies. Indeed, there is a close link between inflammatory bowel diseases such as ulcerative colitis and Crohn’s disease and the subsequent development of colorectal cancer [2, 3]. Chronic inflammation is caused by persistent infection that could be parasitic, bacterial, or viral. However, the inadequate disposal of pathogens, prolonged inflammatory signaling as well as defects in the anti-inflammatory mechanisms may lead to chronic inflammation and promote tumor development [4–7]. Inflammation is a protective response to infection by the immune system that requires communication between different classes of immune cells to coordinate their actions. In chronic inflammation, the inflammatory foci are dominated by lymphocytes, plasma cells, natural killer cells, and macrophages. Macrophages and other inflammatory cells generate a great amount of growth factors, cytokines, chemokines, and reactive oxygen and nitrogen species that may cause DNA damage. If the macrophages are activated persistently, they may lead to continuous tissue damage [8].
In IBD, the chronic mucosal inflammation is caused by hyper-activation of effector immune cells, which produce high levels of pro-inflammatory cytokines like TNFα and IL6 resulting in colonic tissue damage. The nuclear transcription factor (NFκB) is considered as one of the key regulators in this immunological setting [9–11].
In addition to epidemiological data linking chronic infections to increased cancer risk [12], genetic links between cytokines and cancer also exist. Indeed, genes encoding cytokines that are genetically polymorphic have recently been considered as important determinants of disease susceptibility [13]. Indeed TNF-α, IL1β, IL10, IL8, and IL17 gene polymorphisms have been identified as risk factors for gastric cancer as well as with colorectal cancer except for IL17 polymorphism [14–21].
Interleukin-17 (IL17) is a cytokine secreted by T helper 17 (TH17), which are defined as a CD4 lymphocyte population. There are different types of T cell CD4 “helper” with different roles and which direct the immune response differently. TH17 are important for the immune response against bacteria and fungi and are involved in various autoimmune diseases. IL17 induces the recruitment of immune cells in peripheral tissues following activation of transcription factor NFκB, resulting in the expression of a series of genes encoding pro-inflammatory factors (TNF, IL6, IL1β) suggesting the important role of this cytokine in the localization and amplification of inflammation [22, 23]. The IL17 family of cytokines contains six members, IL17A, IL17B, IL17C, IL17D, IL17E, and IL17F [24, 25]. IL17A gene is located on chromosome 6p12. Genetic studies have shown that IL17A polymorphism is highly correlated with a high susceptibility to develop inflammatory bowel disease such as Crohn’s disease and ulcerative colitis [26, 27] and intestinal Behcet’s disease [28].
Furthermore, polymorphism of IL17A G197A has recently been identified to be associated with the susceptibility to rheumatoid arthritis, ulcerative colitis, breast cancer, bladder cancer, cervical cancer, and gastric cancer, respectively [29, 30, 21, 31–34]. However, it has not been investigated with respect to CRC risk.
Though, studies on the expression level of IL 17 were performed in colorectal cancer and in other tumors and expression excess of IL17A was found in various tumor tissues, including prostate cancer, breast cancer, gastric cancer, and colorectal cancer [35–38].
In the present study, we have examined the IL17A G197A polymorphism in relation to colorectal cancer risk and patient characteristics in Tunisian population.
Material and methods
Subjects
A group of patients/controls was collected from the Salah Azaiez Institute and Charles Nicolle Hospital of Tunis (Tunisia). The group of patients concerns 102 unrelated sporadic CRC cases (47 women, 55 men, with age range 58 ± 14) with no family cancer histories and classified on the bases of their histopathological profiles. The healthy group concerns 139 unrelated persons from the region of Tunis. Consent for the genetic study was obtained from volunteers. The characteristics of the study subjects are summarized in Table 1, where the colorectal cancer group and the control group are presented with respect to the age and sex distributions.
DNA extraction
Genomic DNA was extracted from peripheral blood leukocytes using conventional proteinase K digestion and phenol/chloroform extraction method. A NanoDrop (ND-1000) is used to quantify DNA.
Genotyping of polymorphisms
Polymorphisms have been genotyped by fluorescent-based restriction fragment length polymorphism method as reported previously [39]. DNA has been amplified using the following primer pairs: forward primer AACAAGTAAGAATGAAAAGAGGACATGGT, reverse primer CCCCCAATGAGGTCATAGAAGAATC around the IL17A G197A single nucleotide polymorphism (SNP). The forward primer is fluorescently labeled. The PCR amplification was performed in a total 10 μl mixture containing 50 ng genomic DNA, 0.4 μM of each primer, each deoxynucleotide triphosphate at 400 μM, 1.5 mM MgCl2 contained in 10× PCR buffer, 0.4 U AmpliTaq DNA polymerase (Applied Biosystems, Evry, France). Thermocycling conditions were 94 °C for 5 min followed by 30 cycles of 94 °C for 20 s, 58 °C for 30 s, and 72 °C for 20 s with a final extension step of 72 °C for 7 min. Fluorescently labeled PCR products were incubated overnight at 37 °C with EcoNI (New England BioLabs) to reveal IL17A G197A polymorphism. The digested PCR product was diluted 1:10 in ddH2O. A volume of 2 μl of the digested PCR product was mixed with 0.3 μl of internal size standard and 9.7 μl of deionised formamide, denatured and separated on ABI 3130 genetic analyzer (ABI). The two alleles were easily separated using this method: for IL17A G197A, the A Allele resulted in an undigested PCR product of 102 pb; the G allele, resulted in a digested PCR product with two fragments of 68 and 34 pb; as the 68 pb fragment contained the fluorescently labeled forward primer, it was visualized on the ABI 3130. Allele typing was performed with GeneMapper software (Applied Biosystems, Evry, France).
Statistical analysis
The data were analyzed using SPSS software (version 11.5). Significance of the association was determined by Pearson’s chi-squared test χ2 and Fisher’s exact test. A value of p ≤ 0.05 was considered significant.
Results
Genotypes and alleles frequencies of G197A variant of IL17A gene are shown in Table 2. Comparison between observed and theoretical distributions showed that genotypes distribution at SNP IL17A G197A is in agreement with Hardy-Weinberg equilibrium in controls group as well as in patients.
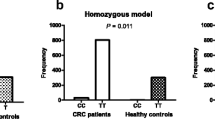
The G allele for this SNP in healthy subjects tented to be higher than in patients (82 vs 68 %), unlike the A allele that is more frequent in patients compared to controls (18 vs 32 %). Indeed, statistical analyses showed significant association between the A allele and risk of colorectal cancer in Tunisian population (p = 0.023; OR 2.14 (1.10–4.15)). Moreover, we have noted that frequencies of IL17A G197A genotypes in cases (GG 47.05 % and GA + AA 52.94 %) were significantly different from those in controls (GG 68.34 % and GA + AA 31.65 %) (p = 0.002; OR 2.45 (1.43–4.11)). This result shows that most of the healthy subjects carries the wild type genotype (GG 69 %) in contrast to patients (GG 48 %) suggesting that GG genotype seems to confer protection against colorectal cancer. While the IL17A/AA genotype could be considered as susceptibility factor for developing colorectal cancer (p = 0.002; OR 2.45 (1.43–4.11)) (Table 2).
We next examined the possible association of G197A SNP with gender and clinicopathologic features of patients (tumor location, tumor differentiation, TNM stage, lymph node, and metastasis) (Table 3). G197A variant is almost equally detected in male and female (p = 0.4). However, we showed that IL17A/AA genotype is more frequent in colon compared to rectum and in patients with tumor of higher differentiation than low or moderate differentiation. This suggests that IL17A G197A polymorphism is significantly associated with tumor location and tumor differentiation (p = 0.003 and p = 0.0005, respectively). In addition, IL17A/AA genotype was found to be positively associated with TNM III/IV stage colorectal cancer, showing a significant risk for the late stage (p = 0.005) (Table 3). On the other hand, we observed no evidence of association between this variant and tumor architecture and lymph node and metastasis.
To confirm that the IL17A polymorphism is associated with tumor location, we investigated this association with colon cancer and rectum cancer separately (Table 4). We showed that IL17A G197A genotypes frequencies in cases (GG 34.84 % and GA + AA 65.15 %) were significantly different from those in controls (GG 68.34 % and GA + AA 31.65 % ) (p = 0.00001; OR 3.77 (2.04-6.99)). In fact, IL17A AG + AA genotypes were more frequent in patients with colon cancer than in controls. In addition, the frequency of the IL17A GG genotype is rather higher in controls. However, there is no significant difference between the genotypes of rectal cancer patients and controls (p = 0.6).
This result suggests that IL17A polymorphism is rather associated with colon cancer and not with rectal cancer. Indeed, IL17A GG genotype could protect against colon cancer unlike AA genotype that could increase the susceptibility of colon cancer.
Discussion
It has recently been reported that the immune markers expression was different in adjacent mucosa and tumoral colorectal tissues suggesting that specific T helper subsets are recruited at the tumor colorectal site. Thus, immune coordination between T helper subsets was found within the tumors. Th1, cytotoxic, and TH17 clusters were associated with a particular clinical outcome [40]. IL17A secreted by TH17 cells, innate lymphoid cells exerts pro-inflammatory proprieties essential to the host protection against extracellular pathogens. On the other hand, IL17A has been implicated in the pathogenesis of several chronic inflammatory disorders. Moreover, IL17 is considered as an imported mediator in inflammation associated cancer. Indeed, Il17 secreting cells have been identified in several types of human cancers and in murine models as well. Furthermore, it has recently been reported that IL17 producing cells may facilitate development of colorectal carcinoma by fostering angiogenesis via promoting VEGF production from cancer cells. This proposes IL17 as a novel indicator of prognosis in the patients with colorectal carcinoma and could serve as a novel therapeutic target for colorectal carcinoma [41].
The present study has been performed taking into account the lack of report about the association between IL17A polymorphism and colorectal cancer.
In fact, we found a significant relationship between polymorphism of Il17A G197A and colorectal cancer in low-risk Tunisian population. The IL17A/AA genotype was positively correlated with increased risk of developing colorectal cancer (p = 0.002) (Table 2).
Previous results have found a significant relationship between polymorphism G197A of IL17A gene promoter and cancer. In fact, the IL17A/197A allele was positively correlated with increased risk of developing gastric cancer in Japanese people [21]. In addition, Tahara et al. reported the implication of IL17A 197G polymorphism in CpG island hyper-methylation (CIHM) in gastric cancer. In fact, CIHM is one of the major events in the gastric carcinogenesis [32]. In contrast, Wu et al. showed that IL17A G197A polymorphism was not associated with gastric cancer susceptibility in Chinese population [31]. Other studies have also shown that IL17A G197A/AA genotype was associated with susceptibility to develop bladder cancer, cervical cancer, and breast cancer [34, 33, 42].
Further, exploring the relationship between IL17A G197A genotypes with CRC stratified by tumor location, tumor differentiation, tumor architecture, TNM stage, lymph node, and metastasis, we observed that this polymorphism was significantly associated with tumor location (p = 0.003) and tumor differentiation (p = 0.0005). Moreover, IL17A/AA genotype was found to be positively associated with TNM III/IV colorectal cancer, showing a significant risk for the late stage (p = 0.005) unlike architecture tumor, lymph node, and metastasis (Table 3). Moreover, Zhang et al. showed that patients with the mutant A allele of IL17G197A tended to suffer mild active ulcerative colitis (UC) suggesting that the IL17A variant had a weak association with the severity of UC [27]. Furthermore, Zhou et al. demonstrated that IL17A/AA genotype was associated with tumor stage of bladder cancer taking into account gender and smoking status [34]. It has also been shown that IL17 gene polymorphism G197A was associated with the susceptibility of positive peritumor intravascular cancer emboli and with high clinical stage of cervical cancer in Chinese women as well [33]. Shibata et al. investigated the association between the severity of histologic gastritis and IL17AG197A. IL17A/A allele had an increased risk of the development of gastric mucosal atrophy, and a positive link between the inflammation score and IL17A/AA genotype was observed [21]. Finally, Wang et al. showed that IL17A G197A polymorphism is associated with occurrence and progression of breast cancer considering tumor suppressor protein 53 (P53) and progesterone receptor (PR) status [42].
In fact, we showed that this association between IL17A G197A polymorphism and colorectal cancer is rather specific to the colon cancer (p = 0.00001) and not to rectal cancer (p = 0.8). In colorectal cancer samples, clinical research showed increased IL17A expression [38, 43]. Chae et al. showed that IL17A deficiency caused significantly reduced intestinal tumorigenesis in Apc Min/+ mice [44]. In addition, Wu et al. noted that IL17A blocking antibody could inhibit enterotoxigenic bacteroides fragilis (ETBF)-induced colon carcinogenesis [45]. Therefore, IL17A may play a pathogenic role in colon cancer [46].
In conclusion, our results provided the first evidence that the IL17 polymorphism, whose function is still unclear, is significantly associated with colorectal cancer susceptibility especially with colon cancer risk. This leads to the conclusion that tumors of colon and rectum are different physiopathological entities. We also showed a significant association of IL17A with high tumor differentiation and with the late stage of CRC. IL17A cytokine could be serving as biomarker of disease location and progression? Further investigations in other cohorts are now needed and a comparative analysis of the expression level of wild type and mutated gene are essential in patients with different stage to clarify the involvement of IL17A polymorphism in colorectal carcinogenesis.
Abbreviations
- CRC:
-
Colorectal cancer
- IBD:
-
Inflammatory bowel disease
- UC:
-
Ulcerative colitis
- Th1/17 cells:
-
T helper 1/17 cells
- IL17A/B/C/D/E/F:
-
Interleukin A/B/C/D/E/F
- IL1β/6/8/10:
-
Interleukin IL1β/6/8/10
- SNP:
-
Single nucleotide polymorphism
- NF-κB:
-
Nuclear factor κB
- TNFα:
-
Tumor necrosis factor α
- TNM stage:
-
Tumor node metastasis stage
- ETBF:
-
Enterotoxigenic Bacteroides fragilis
- CIHM:
-
CpG island hyper-methylation
- PR:
-
Progesterone receptor
- VEFG:
-
Vascular endothelial growth factor
- p53:
-
Tumor suppressor protein53
References
Vasen HF, van der Meulen-de Jong AE, de Vos Tot Nederveen Cappel WH, Oliveira J. Familial colorectal cancer risk: ESMO clinical recommendations. Ann Oncol. 2009;20 Suppl 4:51–3.
Carter AB, Misyak SA, Hontecillas R, Bassaganya-Riera J. Dietary modulation of inflammation-induced colorectal cancer through PPARgamma. PPAR Res. 2009;2009:498352.
Tanaka T. Development of an inflammation-associated colorectal cancer model and its application for research on carcinogenesis and chemoprevention. Int J Inflamm. 2012;2012:658786.
Bernstein CN, Blanchard JF, Kliewer E, Wajda A. Cancer risk in patients with inflammatory bowel disease: a population-based study. Cancer. 2001;91(4):854–62.
Lakatos PL, Lakatos L. Risk for colorectal cancer in ulcerative colitis: changes, causes and management strategies. World J Gastroenterol. 2008;14(25):3937–47.
Wilkening S, Tavelin B, Canzian F, Enquist K, Palmqvist R, Altieri A, et al. Interleukin promoter polymorphisms and prognosis in colorectal cancer. Carcinogenesis. 2008;29(6):1202–6.
Erdman SE, Poutahidis T. Roles for inflammation and regulatory T cells in colon cancer. Toxicol Pathol. 2010;38(1):76–87.
Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–81.
Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118(6):671–4.
Sakamoto K, Maeda S, Hikiba Y, Nakagawa H, Hayakawa Y, Shibata W, et al. Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin Cancer Res. 2009;15(7):2248–58.
Wang S, Liu Z, Wang L, Zhang X. NF-kappaB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol. 2009;6(5):327–34.
Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117(5):1175–83.
Macarthur M, Hold GL, El-Omar EM. Inflammation and cancer II. Role of chronic inflammation and cytokine gene polymorphisms in the pathogenesis of gastrointestinal malignancy. Am J Physiol. 2004;286(4):G515–20.
Li M, You Q, Wang X. Association between polymorphism of the tumor necrosis factor alpha-308 gene promoter and colon cancer in the Chinese population. Genet Test Mol Biomark. 2011;15(11):743–7.
Cen G, Wu W. Association between tumor necrosis factor-alpha 857C/T polymorphism and gastric cancer: a meta-analysis. Tumour Biol. 2013;47(9):1119–33.
Ito H, Kaneko K, Makino R, Konishi K, Kurahashi T, Yamamoto T, et al. Interleukin-1beta gene in esophageal, gastric and colorectal carcinomas. Oncol Rep. 2007;18(2):473–81.
El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404(6776):398–402.
Cacev T, Radosevic S, Krizanac S, Kapitanovic S. Influence of interleukin-8 and interleukin-10 on sporadic colon cancer development and progression. Carcinogenesis. 2008;29(8):1572–80.
Lu W, Pan K, Zhang L, Lin D, Miao X, You W. Genetic polymorphisms of interleukin (IL)-1B, IL-1RN, IL-8, IL-10 and tumor necrosis factor alpha and risk of gastric cancer in a Chinese population. Carcinogenesis. 2005;26(3):631–6.
Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, de Oca J, et al. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63(13):3560–6.
Shibata T, Tahara T, Hirata I, Arisawa T. Genetic polymorphism of interleukin-17A and -17 F genes in gastric carcinogenesis. Hum Immunol. 2009;70(7):547–51.
McGovern DP, Rotter JI, Mei L, Haritunians T, Landers C, Derkowski C, et al. Genetic epistasis of IL23/IL17 pathway genes in Crohn’s disease. Inflamm Bowel Dis. 2009;15(6):883–9.
Yamada H. Current perspectives on the role of IL-17 in autoimmune disease. J Inflamm Res. 2010;3:33–44.
Kawaguchi M, Adachi M, Oda N, Kokubu F, Huang SK. IL-17 cytokine family. J Allergy Clin Immunol. 2004;114(6):1265–73. quiz 74.
Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21(4):467–76.
Kim SW, Kim ES, Moon CM, Park JJ, Kim TI, Kim WH, et al. Genetic polymorphisms of IL-23R and IL-17A and novel insights into their associations with inflammatory bowel disease. Gut. 2011;60(11):1527–36.
Zhang X, Yu P, Wang Y, Jiang W, Shen F, Wang Y, et al. Genetic polymorphisms of interleukin 17A and interleukin 17 F and their association with inflammatory bowel disease in a Chinese Han population. Inflamm Res. 2013;62(8):743–50.
Kim ES, Kim SW, Moon CM, Park JJ, Kim TI, Kim WH, et al. Interactions between IL17A, IL23R, and STAT4 polymorphisms confer susceptibility to intestinal Behcet’s disease in Korean population. Life Sci. 2012;90(19–20):740–6.
Nordang GB, Viken MK, Hollis-Moffatt JE, Merriman TR, Forre OT, Helgetveit K, et al. Association analysis of the interleukin 17A gene in Caucasian rheumatoid arthritis patients from Norway and New Zealand. Rheumatology (Oxford). 2009;48(4):367–70.
Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17 F genes on the susceptibility to ulcerative colitis. J Clin Immunol. 2008;28(1):44–9.
Wu X, Zeng Z, Chen B, Yu J, Xue L, Hao Y, et al. Association between polymorphisms in interleukin-17A and interleukin-17 F genes and risks of gastric cancer. Int J Cancer. 2010;127(1):86–92.
Tahara T, Shibata T, Nakamura M, Yamashita H, Yoshioka D, Okubo M, et al. Effect of polymorphisms of IL-17A, -17 F and MIF genes on CpG island hyper-methylation (CIHM) in the human gastric mucosa. Int J Mol Med. 2009;24(4):563–9.
Quan Y, Zhou B, Wang Y, Duan R, Wang K, Gao Q, et al. Association between IL17 polymorphisms and risk of cervical cancer in Chinese women. Clin Dev Immunol. 2012;2012:258293.
Zhou B, Zhang P, Wang Y, Shi S, Zhang K, Liao H, et al. Interleukin-17 gene polymorphisms are associated with bladder cancer in a Chinese Han population. Mol Carcinog. 2012;52(11):871–8.
Steiner GE, Newman ME, Paikl D, Stix U, Memaran-Dagda N, Lee C, et al. Expression and function of pro-inflammatory interleukin IL-17 and IL-17 receptor in normal, benign hyperplastic, and malignant prostate. Prostate. 2003;56(3):171–82.
Zhu X, Mulcahy LA, Mohammed RA, Lee AH, Franks HA, Kilpatrick L, et al. IL-17 expression by breast-cancer-associated macrophages: IL-17 promotes invasiveness of breast cancer cell lines. Breast Cancer Res. 2008;10(6):R95.
Zhang B, Rong G, Wei H, Zhang M, Bi J, Ma L, et al. The prevalence of Th17 cells in patients with gastric cancer. Biochem Biophys Res Commun. 2008;374(3):533–7.
Le Gouvello S, Bastuji-Garin S, Aloulou N, Mansour H, Chaumette MT, Berrehar F, et al. High prevalence of Foxp3 and IL17 in MMR-proficient colorectal carcinomas. Gut. 2008;57(6):772–9.
Huber AK, Jacobson EM, Jazdzewski K, Concepcion ES, Tomer Y. Interleukin (IL)-23 receptor is a major susceptibility gene for Graves' ophthalmopathy: the IL-23/T-helper 17 axis extends to thyroid autoimmunity. J Clin Endocrinol Metab. 2008;93(3):1077–81.
Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–71.
Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13(5):715–25.
Wang L, Jiang Y, Zhang Y, Wang Y, Huang S, Wang Z, et al. Association analysis of IL-17A and IL-17 F polymorphisms in Chinese Han women with breast cancer. PLoS ONE. 2012;7(3):e34400.
Suzuki H, Ogawa H, Miura K, Haneda S, Watanabe K, Ohnuma S, et al. IL-23 directly enhances the proliferative and invasive activities of colorectal carcinoma. Oncol Lett. 2012;4(2):199–204.
Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A. 2010;107(12):5540–4.
Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–22.
Tong Z, Yang XO, Yan H, Liu W, Niu X, Shi Y, et al. A protective role by interleukin-17 F in colon tumorigenesis. PLoS ONE. 2012;7(4):e34959.
Acknowledgments
We would like to thank all the members of ‘Laboratoire Diagnostic Génétique et Moléculaire, Centre Jean Perrin, Clermont Ferrand, France for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Omrane, I., Marrakchi, R., Baroudi, O. et al. Significant association between interleukin-17A polymorphism and colorectal cancer. Tumor Biol. 35, 6627–6632 (2014). https://doi.org/10.1007/s13277-014-1890-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-1890-4