Abstract
Sema domain of semaphorin 4B (SEMA4B), which is an interacting protein of LNM35, plays an important role in lung cancer invasion. However, the regulation mechanism of this protein is completely unknown. Here, we report that hypoxia and hypoxia mimic reagent could downregulate the expression of SEMA4B in human non-small cell lung cancer (NSCLC) lines. We provide evidences that SEMA4B is a direct target of hypoxia-inducible factor 1 (HIF-1). Silencing the expression of HIF-1α in cancer cells by RNA interference abolished hypoxia-repressed SEMA4B expression. Using luciferase reporter assay, we showed that HIF-1α recognized a hypoxia-responsive element (HRE) of SEMA4B gene, which is required for HIF-1-repressed SEMA4B expression. Moreover, ectopic expression of SEMA4B abolished invasion of hypoxia-induced NSCLC cells. Taken together, these data would shed novel insights on the mechanisms for invasion of hypoxia-induced NSCLC cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of cancer death worldwide, with more than 1 million deaths annually. Non-small cell lung cancer (NSCLC), which includes squamous cell carcinoma, adenocarcinoma, and large cell carcinoma, accounts for approximately 85 % of all lung cancers [1]. Despite all efforts at management, prognosis of advanced lung cancer is extremely poor, with a 5-year survival rate of 17 %. About half of the radical resection of NSCLC relapses with metastases within 5 years, indicating that the tumor cell invasion, migration, and micrometastases may occur before surgical treatment, and the TNM classification may not be sufficient to accurately predict [2] the resected tumors that are more likely to relapse with metastases. Therefore, understanding the molecular biology of NSCLC is important for diagnosis, prevention, and treatment of NSCLC [3].
Accumulated evidence showed that hypoxia-inducible factor 1 (HIF-1) is involved in key aspects of cancer biology, including angiogenesis, proliferation, energy metabolism, and invasion [4–8]. The relevance of HIF-1 on prognosis in NSCLC was studied [9, 10]. HIF-1 is a heterodimeric transcription factor, consisting of two basic helix-loop-helix proteins of the PAS (Per/Arnt/Sim) family—an α-subunit (HIF-1α) which is regulated by hypoxia and a constitutive β-subunit (HIF-1β) [11]. Under normoxia, HIF-1-α was hydroxylated by PHD1 (prolyl hydroxylase domain-containing protein, also called EGLN2) for proteasomal destruction on two specific residues within the oxygen-dependent degradation domain [12], which leads to recognition by the VHL protein and recruitment of the E3 ubiquitin ligase complex, composed of elongin B, elongin C, Rbx1, and Cul2 [13]. Under hypoxic conditions, HIF-1 translocates to the nucleus and activates/represses the expression of downstream genes by binding to the core sequence (RCGTG) of the hypoxia-responsive element (HRE) [14].
It has been reported that tumor invasion and metastasis are controlled by the activation of HIF-1, enabling tumor cells to dissociate and migrate to distant organ sites [15]. HIF-1 is believed to control the invasive and metastatic potential of tumor cells partially through inducing the expression of several transcriptional repressors, including Snail, TWIST, and SIP1, and subsequent induction of an epithelial mesenchymal transition (EMT) [16–18]. In addition, HIF-1 also regulates the expression of LOX, the proinvasive extracellular matrix protein, to promote tumor invasion [19]. Sema domain of semaphorin 4B (SEMA4B), which is an interacting protein of LNM35, plays an important role in lung cancer invasion [20]. However, whether SEMA4B contributes to HIF-1-induced lung cancer cell invasion is completely unknown.
Here, we showed that SEMA4B is a direct target of HIF-1 in lung cancer cells. HIF-1α directly bound to a HRE of SEMA4B gene to repress SEMA4B gene expression. As a consequence, decreased SEMA4B expression played a role to hypoxia and HIF-1-induced lung cancer cell invasion.
Material and methods
Cell culture
The H157 and A549 NSCLC cell lines were obtained from ATCC. These lines were cultured in RPMI 1640 medium supplemented with 10 % fetal bovine serum (FBS) (Gibco, CA, USA) as well as 100 U/ml penicillin and 100 μg/ml streptomycin. For hypoxia treatment, cells were cultured in a hypoxia chamber flushed with 0.1 % O2 and 5 % CO2 with balance of 95 % nitrogen at 37 °C. Control cells were placed in a 5 % CO2 and 95 % air incubator (20 % O2) at 37 °C.
SiRNA, plasmid, and transfection
Small interfering (si) RNA against HIF-1α, siHIF-1α-1 GGACACAGAUUUAGACUUG and siHIF-1α-2 GAUGGAAGCACUAGACAAA, was purchased from Dharmacon (USA). For control siRNA, a random siRNA sequence was selected. The cDNA of SEMA4B was amplified from A549 cells by PCR and cloned into the pcDNA3.1 vector. The cDNA was completely sequenced.
All transient transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
RNA isolation and real-time PCR
The total RNA from H157 to A549 NSCLC cells were isolated using the TRIzol reagent, and reverse transcriptions were done using the Takara RNA PCR kit following the manufacturer’s instructions. To quantify the transcription of genes of interest, real-time PCR was performed using SYBR Green Premix Ex Taq (Takara, Japan) on LightCycler 480 (Roche, Switzerland). The sequences of primers used are available upon request.
Western blot
Cells were harvested and lysed with an ice-cold lysis buffer (150 mM Tris–HCl, pH 7.5, 100 mM DTT, 2 % sodium dodecyl sulfate (SDS), 10 % glycerol). After centrifugation at 20,000g for 10 min at 4 °C, proteins in the supernatants were quantified and separated by 10 % SDS-PAGE and transferred to PVDF membrane. After blocking with 5 % nonfat milk, membranes were immunoblotted with antibodies as indicated, followed by horseradish peroxidase (HRP)-linked secondary antibodies. The signals were detected by Millipore SuperSignal® HRP Substrate kit according to the manufacturer’s instructions.
Chemicals and antibodies
Deferoxamine mesylate salt (DFO) was purchased from Sigma-Aldrich (St. Louis, MO, USA), dissolved in DMSO. SEMA4B antibody (AV49485) was purchased from Sigma-Aldrich (St. Louis, MO, USA); HIF-1α antibody (#3716) was purchased from Cell Signaling Technology. Actin (A2228) antibody was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Adenovirus infection
Adenoviruses encoding HIF-1α or vector control were propagated in HEK293T cells, which were collected upon detection of viral cytopathic effect. Multiplicity of infection (MOI) was estimated by OD260/280. For infection, 1 × 106 cells were plated per well in a six-well dish and overlaid with 0.5 mL serum-free media containing 10 mM MgCl2 and concentrated adenovirus at the indicated titer. Following incubation at 37 °C for 30 min, 2 mL of fresh full media was added, and the cells were grown until harvest.
Luciferase assays
The SEMA4B promoter was amplified from A549 cells genomic DNA template and inserted into pGL3 basic vector (Promega). Mutations were generated using a PCR mutagenesis kit (Toyobo) following the manufacturer’s instructions. For luciferase reporter assays, A549 cells were seeded in 24-well plates and transfected with the indicated plasmids. Cells were harvested 36 h after transfection. Luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, USA).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assay kits (Upstate, USA) were used to make chip test. Briefly, A549 cells were fixed with 1 % formaldehyde to cross-link proteins and DNA, after which the DNA was sheared to fragments of 200–800 bp by sonication in an ultrasonic bath on ice. The chromatin was then incubated and precipitated with IgG or HIF-1α antibody. The immunoprecipitated DNA fragments were detected by real-time PCR analysis. To calculate the fold enrichment of the precipitated SEMA4B HRE, sample was normalized with the corresponding input (sample before IP). The corresponding IgG Ct (IgG IP) values were subtracted from each sample (HIF-1α IP) before calculating the dCt and ddCt values.
Statistical analysis
Data were summarized as mean ± SEM. Statistical significance was analyzed by one-way ANOVA using SPSS 11.0 software (Chicago, IL, USA). A value of P < 0.05 was considered significant. Statistical significance is displayed as *P < 0.05, **P < 0.01, or ***P < 0.001.
Results
Hypoxia and hypoxia reagent decrease SEMA4B expression
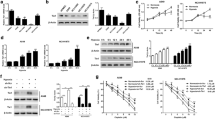
To test whether SEMA4B expression is regulated by hypoxia, A549 and H157 NSCLC cells were incubated under normoxia (21 % O2), hypoxia (1 % O2), or medium containing 100 μM DFO for 24 h [21]. Western blot data revealed a decreased SEMA4B protein level in the cells under hypoxic conditions or DFO treatment, accompanying increased levels of HIF-1α protein (Fig. 1a, b). Indeed, hypoxia decreased the expression on SEMA4B in A549 cells in a time-dependent manner (Fig. 1c). Taken together, these results demonstrated that SEMA4B level is downregulated by hypoxia.
Hypoxia and hypoxia reagent decrease SEMA4B expression. a Lysate from A549 cells under either normoxia or hypoxia condition was detected with indicated antibodies, and beta-actin was used as a loading control. b Lysate from H157 cells under either normoxia or hypoxia condition was detected with indicated antibodies. c Lysate from A549 cells treated with DFO was detected with indicated antibodies. d Lysate from A549 cells under hypoxia condition for the indicated time course was detected with indicated antibodies
Downregulation of SEMA4B during hypoxia is mediated by HIF-1
As our experiments demonstrated an inverse correlation between HIF-1α and SEMA4B levels, we then asked whether the decrease in SEMA4B levels is mediated by HIF-1. To this end, A549 and H157 cells were infected with adenovirus expressing HIF-1α or control. SEMA4B levels decreased in a HIF-1α titer-dependent fashion under normoxic conditions in A549 (Fig. 2a) and H157 cells (Fig. 2b), suggesting that HIF-1α overexpression is sufficient to induce SEMA4B suppression. To further investigate the role of HIF-1 in SEMA4B expression, endogenous HIF-1α in A549 cell was silenced by siRNAs targeting HIF-1α. HIF-1α siRNAs could achieve about 70–80 % silencing effect on HIF-1α as evaluated by real-time PCR and Western blot assays (Fig. 2c, d). As expected, SEMA4B was markedly increased when HIF-1α was silenced (Fig. 2d). Taken together, these experiments demonstrate that HIF-1α induction is sufficient for SEMA4B downregulation and suggest that hypoxic suppression of SEMA4B is significantly regulated by HIF-1.
Downregulation of SEMA4B during hypoxia is mediated by HIF-1. a Western blot analysis of SEMA4B and HIF-1α levels in A549 cells for 48 h after infection with control or HIF-1α adenoviruses at increasing MOIs as indicated. b Western blot analysis of SEMA4B and HIF-1α levels in H157 cells for 48 h after infection with control or HIF-1α adenoviruses at increasing MOIs as indicated. c A549 cells were transfected with con-siRNA or siRNAs against HIF-1α for 36 h. The mRNA level of HIF-1α was detected by real-time PCR. d A549 cells were transfected with con-siRNA or siRNAs against HIF-1α for 24 h and treated with or without DFO for 12 h. Cells were harvested, and the protein level of SEMA4B was detected by Western blot
Downregulation of SEMA4B under hypoxia is caused by reduced transcription of SEMA4B gene
To investigate the mechanism through which SEMA4B protein levels decrease following hypoxia, SEMA4B mRNA levels were quantified by quantitative RT-PCR. The abundance of SEMA4B mRNA in A549 cells treated with DFO was found to diminish in a time-dependent manner (Fig. 3a). Similarly, SEMA4B mRNA levels decreased in hypoxia-treated and adenoviral HIF-1α-infected cells compared with controls (Fig. 3b, c). Moreover, knockdown of HIF-1α levels significantly rescued SEMA4B mRNA expression, compared with cells transfected with control siRNA (Fig. 3d). Collectively, these data indicate that the decrease in SEMA4B expression under hypoxia results from reduced mRNA level.
Downregulation of SEMA4B under hypoxia is caused by reduced transcription of SEMA4B gene. a A549 cells were treated DFO for the indicated time, and the mRNA level of SEMA4B was detected by real-time PCR. b Real-time PCR was used to detect the mRNA level of SEMA4B in A549 cells under either normoxia or hypoxia condition. c A549 cells were infected with vector or HIF-1α plasmid for 48 h. The mRNA level of SEMA4B was detected by real-time PCR. d A549 cells were transfected with con-siRNA or siRNAs against HIF-1α for 24 h and incubated under hypoxia condition for additional 24 h. The mRNA level of SEMA4B was detected by real-time PCR
Identification and validation of a functional HRE in SEMA4B promoter region
We next determined whether SEMA4B could be a transcriptional target of HIF-1. There are two potential HREs between 0.5- and 1-kb region upstream to the transcription start site of SEMA4B (HRE1, -542-gcgtg-538, and HRE2, -820-gcgtg-816) (Fig. 4a). We generated a series of truncated human SEMA4B promoter-driven luciferase reporter genes and found that SEMA4B promoter region around the predicted HRE core sequence was required for hypoxia response. Moreover, we confirmed the HRE sites located in SEMA4B promoter by using a mutation approach. As shown in Fig. 4b, mutation of any one of the two HRE sites of SEMA4B promoter significantly affected its response to hypoxia, and the two-site mutant lost all hypoxia-repressed SEMA4B transcriptions. To further demonstrate that HIF-1 binds to the HRE in SEMA4B promoter, we performed ChIP assay using antibodies against HIF-1α (IgG as a negative control) in normoxia- and hypoxia-cultured A549 cells. As depicted in Fig. 4c, HIF-1α but not IgG specifically bound to the HRE sequence in SEMA4B promoter. In summary, these results showed that SEMA4B was directly repressed by HIF-1 under hypoxia.
Identification and validation of a functional HRE in SEMA4B promoter region. a Schematic diagrams of the regulating sequences with the two putative HREs (black ovals) of SEMA4B. b The wild-type and potential HRE site mutant of the SEMA4B promoter-driven luciferase activity was measured at 12 h after hypoxia treatment or under normoxia. c Anti-IgG and anti-HIF-1α antibodies were used in the ChIP assays using A549 cells incubated under either normoxia or hypoxia condition
SEMA4B repression contributed to hypoxia-induced NSCLC invasion
Hypoxia could trigger metastasis and invasion of cancer cells. SEMA4B has been shown to be a potent inhibitor of tumor angiogenesis and metastasis. To investigate whether SEMA4B has a role in hypoxia-induced lung cancer invasion, SEMA4B was overexpressed in A549 cells. By using an invasion model, we show that hypoxia could significantly promote invasion of A549 cells. However, ectopic expression of SEMA4B significantly reversed this phenomenon (Fig. 5a–c). Taken together, these data suggested that SEMA4B played an important role in hypoxia-induced NSCLC invasion.
SEMA4B repression contributed to hypoxia-induced NSCLC invasion. a WB analysis for SEMA4B in A549 cells transfected with control or SEMA4B plasmid under normoxic or hypoxic condition. b Invasion assay in A549 cells transfected with control or SEMA4B plasmid under normoxic or hypoxic condition. c Quantification of the invasion assay in A549 cells transfected with control or SEMA4B plasmid under normoxic or hypoxic condition. Triple asterisks indicate hypoxic vs normoxic and double asterisk SEMA4B hypoxic vs hypoxic
Discussion
In the present study, we showed that SEMA4B was downregulated by hypoxia in NSCLC. We proved that SEMA4B was a direct target gene of HIF-1. Hypoxia and ectopically expressed HIF-1α protein significantly decreased SEMA4B expression at both mRNA and protein levels in NSCLC cell lines. Silencing the expression of HIF-1α by specific siRNA antagonized hypoxia-repressed SEMA4B expression. Using luciferase reporter and ChIP assay, we found that SEMA4B was a direct target gene of HIF-1.
The semaphorin family characterized by a conserved domain of about 500 amino acids that forms a seven-bladed β-propeller structure has been implicated in axon guidance, cell migration, angiogenesis, and the immune response [22–24]. There are 20 genes in semaphorin family which can be subdivided into seven distinct classes based on the presence of class-specific carboxy-terminal sequences and contains both secreted and membrane-bound proteins [25]. However, the function of the large number of membrane-bound semaphorins is not fully understood. In this study, we found that in the overexpression of SEMA4B, hypoxia-induced NSCLC cell invasion was dramatically decreased, indicating that SEMA4B repression may be necessary for hypoxia-induced NSCLC cell invasion.
In summary, our results identify SEMA4B, as a target of transcriptional repression by hypoxia and HIF-1. We conclude that hypoxia regulates invasion of NSCLC cells through transcriptional repression of SEMA4B. We elucidate a novel hypoxia–HIF-1–SEMA4B axis and a previous unknown role of SEMA4B in the invasion of hypoxia-induced NSCLC cells.
Reference
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917.
Baltayiannis N, Chandrinos M, Anagnostopoulos D, Zarogoulidis P, Tsakiridis K, Mpakas A, et al. Lung cancer surgery: an up to date. J Thorac Dis. 2013;5:S425–39.
Cooper WA, Lam DC, O’Toole SA, Minna JD. Molecular biology of lung cancer. J Thorac Dis. 2013;5:S479–90.
Maxwell P, Salnikow K. HIF-1: an oxygen and metal responsive transcription factor. Cancer Biol Ther. 2004;3:29–35.
Yang Y, Sun M, Wang L, Jiao B. HIFs, angiogenesis, and cancer. J Cell Biochem. 2013;114:967–74.
Mamlouk S, Wielockx B. Hypoxia-inducible factors as key regulators of tumor inflammation. Int J Cancer. 2013;132:2721–9.
Goda N, Kanai M. Hypoxia-inducible factors and their roles in energy metabolism. Int J Hematol. 2012;95:457–63.
Calzada MJ, del Peso L. Hypoxia-inducible factors and cancer. Clin Transl Oncol. 2007;9:278–89.
Jackson AL, Zhou B, Kim WY. HIF, hypoxia and the role of angiogenesis in non-small cell lung cancer. Expert Opin Ther Targets. 2010;14:1047–57.
Graves EE, Maity A, Le QT. The tumor microenvironment in non-small-cell lung cancer. Semin Radiat Oncol. 2010;20:156–63.
Scheuermann TH, Yang J, Zhang L, Gardner KH, Bruick RK. Hypoxia-inducible factors Per/ARNT/Sim domains: structure and function. Methods Enzymol. 2007;435:3–24.
Metzen E, Ratcliffe PJ. HIF hydroxylation and cellular oxygen sensing. Biol Chem. 2004;385:223–30.
Arjumand W, Sultana S. Role of VHL gene mutation in human renal cell carcinoma. Tumour Biol. 2012;33:9–16.
Caro J. Hypoxia regulation of gene transcription. High Alt Med Biol. 2001;2:145–54.
Tsai YP, Wu KJ. Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci. 2012;19:102.
Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of twist by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305.
Evans AJ, Russell RC, Roche O, Burry TN, Fish JE, Chow VW, et al. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol Cell Biol. 2007;27:157–69.
Imai T, Horiuchi A, Wang C, Oka K, Ohira S, Nikaido T, et al. Hypoxia attenuates the expression of E-cadherin via up-regulation of snail in ovarian carcinoma cells. Am J Pathol. 2003;163:1437–47.
Schietke R, Warnecke C, Wacker I, Schodel J, Mole DR, Campean V, et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: insights into cellular transformation processes mediated by HIF-1. J Biol Chem. 2010;285:6658–69.
Nagai H, Sugito N, Matsubara H, Tatematsu Y, Hida T, Sekido Y, et al. CLCP1 interacts with semaphorin 4B and regulates motility of lung cancer cells. Oncogene. 2007;26:4025–31.
Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72.
Vadasz Z, Toubi E. Semaphorins: their dual role in regulating immune-mediated diseases. Clin Rev Allergy Immunol 2013. doi:10.1007/s12016-013-8360-4.
Yoshida Y. Semaphorin signaling in vertebrate neural circuit assembly. Front Mol Neurosci. 2012;5:71.
Neufeld G, Sabag AD, Rabinovicz N, Kessler O. Semaphorins in angiogenesis and tumor progression. Cold Spring Harb Perspect Med. 2012;2:a006718.
Gu C, Giraudo E. The role of semaphorins and their receptors in vascular development and cancer. Exp Cell Res. 2013;319:1306–16.
Acknowledgements
This work was supported by WU JIE PING Medical foundation. We thanks Dr. Liu Ping from Shanghai Institute of Materia Medica, Chinese Academy of Sciences for her kind assistant about the invasion assay.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jian, H., Liu, B. & Zhang, J. Hypoxia and hypoxia-inducible factor 1 repress SEMA4B expression to promote non-small cell lung cancer invasion. Tumor Biol. 35, 4949–4955 (2014). https://doi.org/10.1007/s13277-014-1651-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-1651-4








