Abstract
CUB and sushi multiple domain protein 1 (CSMD1) is a candidate tumor suppressor gene. The three members of CSMD family have very similar structures, each consisting of 14 CUB domains separated from one another by a sushi domain, an additional uninterrupted array of sushi domains, a single transmembrane domain, and a short cytoplasmic tail. In this work, we aimed to study the protein and mRNA levels of the CSMD1, CSMD2, and CSMD3 and evaluate their prognostic importance in colorectal cancer. Reduced expressions of these three proteins were detected in colorectal cancer tissues by comparing matched normal tissues. Low CSMD2 expression was significantly associated with differentiation, lymphatic invasion, and tumor size. CSMD3 was associated with differentiation and lymphatic invasion. CSMD1 and CSMD2 expressions were associated with overall survival. This study offers convincing evidence for the first time that the three genes of CSMD family were downregulated in the patients with colorectal cancer and may be used as predictors of colorectal cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer is the third most commonly diagnosed cancer in males and the second in females, with over 1.2 million new cancer cases and 608,700 deaths estimated to have occurred in 2008 [1].
The CUB and sushi multiple domains 1 (CSMD1) gene encodes a large, type I transmembrane protein located on the surfaces of neuronal and epithelial cells [2]. The human CSMD1 gene, consisting of 70 exons, spans two megabases in chromosome region 8p23.2 and encodes an 11.5-kb transcript [3]. CSMD1 expression is frequently lost in breast cancer [4], whereas CSMD1 loses allelic balance in head and neck squamous cell carcinomas (HNSCC) and lung cancers [5]. The full-length CSMD2 cDNA sequence is 12,486 bp long and is predicted to encode a protein with a molecular weight of 383 kDa [6]. Another CSMD family member is CSMD3. The expression of CSMD3 on the fetal and adult brain suggests that this gene is a good candidate for the pathogenesis of autistic spectrum disorders (ASDs) [7]. The role of CSMD2 and CSMD3 in carcinogenesis has not yet been studied so far. The great similarity between all three CSMD genes begs the question of whether CSMD2 and 3 are also likely to be tumor suppressors [6]. In a recent study of Tang et al. [8], they also found lower CSMD1 level in melanoma cells than in normal skin cells. Furthermore, they confirmed that CSMD1 exhibits antitumor activity through activation of the Smad pathway [8].
In this study, we observed significant correlations of CSMD1, CSMD2, and CSMD3 loss of function with clinical presentation of colorectal cancer patients. Therefore, our results support the idea that CSMD1, CSMD2, and CSMD3 may be used as new prognostic biomarkers for colorectal cancer.
Patients and methods
Subjects
A total of 52 patients with colorectal cancer were obtained from the Department of Intestine Surgery, Liaoning Cancer Hospital and Institute (Jan. 2008 to Dec. 2012). All patients underwent standard laboratory tests (cytology and histology). None of the patients underwent radiotherapy or chemotherapy before the operation. Informed consent was provided by all patients according to the Helsinki Declaration.
Extraction of total RNA
In this work, RNA was extracted using TRIzol solution (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the protocols recommended by the manufacturer, and RNAse-free DNase I was used to remove DNA contamination. Total RNA concentration and quantity were assessed by absorbency at 260 nm using a DNA/Protein Analyzer (DU 530, Beckman, Fullerton, CA, USA).
Real-time PCR
Real-time PCR was performed on a Rotor-Gene RG-6000A apparatus (Corbett Research, Cambridge, UK) for 40 cycles of 94 °C for 10 s, 60 °C for 10 s, and 72 °C for 15 s. Reactions (20 μl) included 2 μl of cDNA, target-specific primers, and the QuanTitect SYBR green PCR kit (QIAGEN, Valencia, CA, USA). The temperature range for analysis of melting curves was 55 to 99 °C over 30 s. The primer sequences were listed in Table 1. Relative quantitation was calculated by ΔΔCt method. Each reaction was repeated independently at three times in triplicate.
Western blot analysis
Tissues were lysed in lysis buffer (20 mM Tris–HCl, 150 mM NaCl, 2 mM EDTA, 1 % Triton-X100) containing a protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, MO, USA). Cell extract protein amounts were quantified using the BCA protein assay kit. Equivalent amounts of protein (30 μg) were separated using 12 % SDS-PAGE and transferred to a PVDF membrane (Millipore Corporation, Billerica, MA, USA). Western blot was performed using primary antibodies (Table 2). Each specific antibody binding was detected with horseradish peroxidase (HRP)-conjugated respective secondary antibodies (Amersham Biosciences, UK) and ECL solutions (Amersham Biosciences).
Statistics and survival analysis
Overall survival (OS) was determined using the Kaplan-Meier estimator. Kaplan-Meier survival plots were generated, and comparisons were made with log-rank statistics. Cox’s proportional hazards model was employed for multivariate analysis. For all analyses, only p < 0.05 was considered significant. All the statistical analyses and graphics were performed with GraphPad Prism 5.
Results
CSMD expression in human colorectal cancer specimens
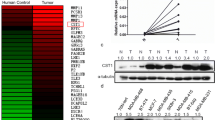
To determine whether CSMD transcription was reduced, real-time PCR analysis for CSMD1, CSMD2, and CSMD3 expression was performed in 52 gastric cancer specimens. Results show that the levels of CSMD1, CSMD2, and CSMD3 mRNA in cancer tissue were significantly lower than in normal tissue (Fig. 1, p < 0.05). Western blot analysis was performed in order to determine the protein expression level of CSMD1, CSMD2, and CSMD3 protein. CSMD1, CSMD2, and CSMD3 protein expression in cancer tissue was significantly lower than in normal tissue (Fig. 2, p < 0.05).
CSMD expression and the clinicopathological variables
We then analyzed the potential relationship between the expression of CSMD and the clinicopathological characteristics of these patients. Unfortunately, CSMD1 expression was not associated with the clinicopathological characteristics of the patients with colorectal cancer (Table 3, p > 0.05). However, CSMD2 was associated with differentiation, lymphatic invasion, and tumor size (Table 3, p < 0.05). CSMD3 was associated with differentiation and lymphatic invasion (Table 3, p < 0.05). Cox’s proportional hazard analysis indicated that sex, age, differentiation, lymphatic invasion, tumor size, and pN category were not independent prognostic factors for colorectal cancer with CSMD protein expression (Table 4, p > 0.05). To investigate the level of CSMD with the patient survival, the survival data from 52 patients with colorectal cancer were assessed. Comparison by the Kaplan-Meier method for low versus high CSMD1 or CSMD2 expression showed a significant difference in the 5-year survival rate of the patients with colorectal cancer (Fig. 3, p < 0.05). However, CSMD3 was not associated with the patient survival (Fig. 3, p > 0.05). Furthermore, we found that the survival rates of the triple-positive patients (CSMD1+, CSMD2+, and CSMD3+) were far higher than the single-positive (CSMD1+, CSMD2+, or CSMD3+) or double-positive (CSMD1+CSMD2+, CSMD2+CSMD3+, or CSMD1+ CSMD3+) ones (Fig. 3, p < 0.05).
Discussion
CSMD1 was cloned as a candidate suppressor of head and neck squamous cell carcinomas [3]. The great similarity between all three CSMD genes begs the question of whether CSMD2 and 3 are also likely to be tumor suppressors. In this study, we detected the first investigation of the role of all three CSMD genes in colorectal cancer. We confirmed that all three CSMD were lower in colorectal cancer tissues than in matched normal tissues. Our results agree with other studies that report reduced CSMD1 mRNA expression in prostate cancer, hepatocellular carcinoma, and non-small cell lung cancer [9–11]. Shull et al. [12] found that somatic mutations, allele loss, and DNA methylation were the main reasons for low CSMD1 expression in colorectal cancer. We will detect the cause of low CSMD1 expression in our future study. Liu et al. [13] found that CSMD3 is the second most frequently mutated gene in lung cancer. Kamal et al. [4] confirmed that loss of CSMD1 expression is associated with high tumor grade and poor survival in invasive ductal breast carcinoma. In this study, we did not find any associations of CSMD1 with the clinicopathological features of the patients with colorectal cancer. However, CSMD1 is associated with the survival rates of these patients.
CSMD2 and CSMD3 are expressed at low levels in many tissues, and that expression is highest in the central nervous system [14]. Previous studies mainly focused on the roles of CSMD2 or CSMD3 in psychosis. In the Alzheimer’s Disease Neuroimaging Initiative (ADNI) sample, genome-wide association study (GWAS) analysis of voxels of the entire brain in healthy subjects, mildly cognitively impaired patients, and Alzheimer’s patients identified the CSMD2 marker rs476463 among the most significantly associated SNPs to brain volume [15]. The expression of CSMD3 in fetal and adult brain suggests that this gene may be involved in the pathogenesis of autistic spectrum disorders (ASDs) [7]. This study offers convincing evidence for the first time that the three genes of CSMD family was downregulated in the patients with colorectal cancer and may be used as predictors of colorectal cancer.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, et al. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J Immunol. 2006;176:4419–30.
Sun PC, Uppaluri R, Schmidt AP, Pashia ME, Quant EC, Sunwoo JB, et al. Transcript map of the 8p23 putative tumor suppressor region. Genomics. 2001;75:17–25.
Kamal M, Shaaban AM, Zhang L, Walker C, Gray S, Thakker N, et al. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res Treat. 2010;121:555–63.
Ma C, Quesnelle KM, Sparano A, Rao S, Park MS, Cohen MA, et al. Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer Biol Ther. 2009;8:907–16.
Lau WL, Scholnick SB. Identification of two new members of the CSMD gene family small star, filled. Genomics. 2003;82:412–5.
Floris C, Rassu S, Boccone L, Gasperini D, Cao A, Crisponi L. Two patients with balanced translocations and autistic disorder: CSMD3 as a candidate gene for autism found in their common 8q23 breakpoint area. Eur J Hum Genet. 2008;16:696–704.
Tang MR, Wang YM, Guo S, Han SY, Wang D. CSMD1 exhibits antitumor activity in A375 melanoma cells through activation of the Smad pathway. Apoptosis. 2012;17:927–37.
Midorikawa Y, Yamamoto S, Tsuji S, Kamimura N, Ishikawa S, Igarashi H, et al. Allelic imbalances and homozygous deletion on 8p23.2 for stepwise progression of hepatocarcinogenesis. Hepatology. 2009;49:513–22.
Henshall SM, Afar DE, Hiller J, Horvath LG, Quinn DI, Rasiah KK, et al. Survival analysis of genome-wide gene expression profiles of prostate cancers identifies new prognostic targets of disease relapse. Cancer Res. 2003;63:4196–203.
Paris PL, Andaya A, Fridlyand J, Jain AN, Weinberg V, Kowbel D, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004;13:1303–13.
Shull AY, Clendenning ML, Ghoshal-Gupta S, Farrell CL, Vangapandu HV, Dudas L, et al. Somatic mutations, allele loss, and DNA methylation of the Cub and sushi multiple domains 1 (CSMD1) gene reveals association with early age of diagnosis in colorectal cancer patients. PLoS One. 2013;8:e58731.
Liu P, Morrison C, Wang L, Xiong D, Vedell P, Cui P, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis. 2012;33:1270–6.
Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, et al. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci U S A. 2002;99:11287–92.
Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, et al. Voxelwise genome-wide association study (vGWAS). Neuroimage. 2010;53:1160–74.
Acknowledgments
We appreciate the thoughtful advice of Ya-Nan Xing (China Medical University).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, R., Song, C. Loss of CSMD1 or 2 may contribute to the poor prognosis of colorectal cancer patients. Tumor Biol. 35, 4419–4423 (2014). https://doi.org/10.1007/s13277-013-1581-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-1581-6