Abstract
Reduced levels of specific microRNA in cancer are frequently reported and associated with attenuated cancer genes and associated pathways. We previously reported a loss of miR-124a in glioblastoma (GBM) patient specimens; however, the upstream causes of this loss are largely unknown. Loss of miR-124a has been attributed to hypermethylation while other studies have shown miR-124a to be regulated by the repressor-element-1-silencing transcription factor (REST, also known as neuron-restrictive silencing factor). This current study looked at both epigenetic and transcription factor regulation as potential mechanisms resulting in the loss of miR-124a expression in GBM patient specimens and cell lines. Hypermethylation of miR-124a was observed in 82 % of GBM patient specimens (n = 56). In vitro miR-124a expression levels also increased after treatment of several patient-derived cell lines with 5-aza-2′-deoxycytidine. Additionally, we also demonstrated a positive interaction between REST activity and miR-124a using a luciferase-binding assay and we correlated the reciprocal expression of REST and miR-124a in our clinical cohort. This result indicates that miR-124a expression may also be modulated through the upstream targeting of REST. Preclinical studies involving inhibitors of REST and treatment with demethylating agents with the intent to increase miR-124a levels could be interesting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The survival trends for glioblastoma (GBM) patients have remained largely static, reflecting a lack of improvements in the therapeutic options for patients. Less than 20 % of newly diagnosed GBM patients survive more than 5 years (CBTRUS) [1]. Although glioma cells rarely metastasize outside the brain, they have an extraordinary capacity to migrate long distances away from the primary tumour site and infiltrate into surrounding normal brain tissue. GBM is highly refractory to standard radiotherapy and chemotherapy making treatment very difficult. Novel therapeutic agents are needed as well as agents that can add to the efficacy of conventional therapies.
Aberrant microRNA (miRNA) expression has been associated with a range of different cancers, including GBM (reviewed in [2, 3]). MiRNAs post-transcriptionally regulate the expression of specific target mRNAs, and individual miRNAs may regulate entire gene networks [4] making them attractive therapeutic targets in cancer. Previously, we and others found GBM to be devoid of miR-124a expression despite enrichment of miR-124a expression in the normal brain [5–10]. As a result of transfecting cells with precursor miR-124a, a significant decline in tumour cell proliferation, migration and invasion was noted [7, 10, 11]. MiR-124a has been shown to negatively regulate SLC16A1, PIK3CA, IQGAP1, LAMC1, ITGB1, STAT3 and CDK4 [5, 10, 12–16]. High expression of ITGB1 and STAT3 have been associated with cancer stem cells (CSCs) leading to increased invasiveness [16].
The mechanism(s) by which miR-124a expression is significantly reduced in GBM compared to normal brain has not been fully elucidated. Although it has been shown that miR-124a is frequently hypermethylated and treatment with the demethylating agent 5-aza-2′-deoxycytidine (5-aza-dc) restores miR-124a levels in hepatocellular carcinoma [17], non-Hodgkin's lymphoma [18], and cervical cancer [19], this was not the case in two widely used, immortalized GBM cell lines, U87 and U251 [9].
Repressor element-1-silencing transcription factor (REST) is a well-characterised regulator of neuronal programs in non-neuronal tissues that bind to a highly conserved DNA sequence called neuron-restrictive silencer element (NRSE, also known as RE-1) [20, 21]. REST silences the transcription of neuronal genes through recruitment of specific co-repressor proteins and aberrant overexpression of REST has been associated with neurological disorders [22–24]. High REST expression has also been implicated in several human neural-specific cancers including neuroblastoma [25], medulloblastoma [26], and more recently, GBM [7, 27, 28]. Inhibition of REST in GBM isolated CSCs and subsequent implantation of these cells into mice resulted in tumors that were significantly less invasive, highly apoptotic and slower growing [29]. Studies of medulloblastoma have provided strong evidence that miR-124a is under direct regulation by REST [26, 27]. In a serial analysis of chromatin occupancy in a non-neuronal murine kidney cell line, miR-124a was expressed as a result of the dismissal of REST from its binding sites on the chromatin during neuronal differentiation [30]. Conti and colleagues inhibited REST in self-renewing tumorigenic-competent GBM cells [7] and its knockdown strongly reduced their self-renewal in vitro and tumour-initiating capacity in vivo. Inhibition of REST also affected the level of miR-124 [7].
The majority of in vitro studies examining miR-124a expression have been conducted on commercially obtained cell lines such as U251. Studies of miR-124a using patient-derived tumor cell lines (primary GBM cells) and patient specimens have been limited. In the current study, we confirmed very low expression of miR-124a in primary cell lines and post-transfection with precursor miR-124a, supported the inhibitory effects of miR-124a on proliferation, migration and invasion previously observed with commercial, immortalised cells. To determine if miR-124a loss was a consequence of methylation, we measured the degree of miR-124a methylation in both patient specimens and cell lines using methylation-specific PCR (MSP) and evaluated the effect of the demethylating agent, 5-aza-dc, on miR-124a expression in primary cells. Finally, we investigated whether miR-124a and REST reciprocally regulate each other in primary cell lines and patient specimens.
Materials and methods
Tumour collection from patient cohort
GBM tumor samples were collected from patients undergoing surgical resection at The Prince of Wales Private Hospital (Randwick, Australia). The Human Research Ethics Committee South Eastern Sydney Local Health District–Northern Sector approved the collection and use of freshly frozen human GBM tissue for this project. A board-certified pathologist reviewed all specimens and only primary GBM samples were included in the study. Three human normal brain RNAs were purchased from Ambion for use as control samples in all RT-qPCR experiments; namely: human parietal cortex superior brain total RNA, human orbital frontal cortex brain total RNA, and human parietal cortex posterior total RNA (Ambion, Austin, TX, USA).
Glioma cell lines and culture
The commercial GBM cell lines, U251, U87 and T98, were acquired from the American Type Culture Collection and were cultured under sterile condition at 37 °C, 5 % CO2 in minimum essential medium (Life Technologies, USA) supplemented with 10 % foetal bovine serum (Life Technologies, USA) and 2 mM l-glutamine (Life Technologies, USA). Low-passage primary cell lines derived from human biopsies (annotated BAH1, RN1, HW1 and PB1) [31] were provided from the Queensland Institute of Medical Research and cultured in RHB-A stem cell culture medium (Stemcells Inc., Newark, CA, USA) supplemented with β-FGF and EGF (20 ng/ml respectively, Sigma-Aldrich, St Louis, MO, USA) on 1 % matrigel-coated flasks or plates. BAH1 cells were cultured from a GBM patient, aged 76 and survived only 94 days. BAH1 was MGMT methylated and PTEN mutant. RN1 cells were cultured from a GBM patient, aged 57 years and survived 243 days after diagnosis (MGMT unmethylated). The clinical details for the patients from which PB1 and HW1 cells were generated were unknown. Both cell lines were MGMT methylated.
Transient transfection
For studies investigating miR-124a function in GBM cell lines in vitro, transient transfections were conducted in BAH1, U87 and U251 GBM cell lines according to previously published protocols [10]. For miR-124a–REST interaction studies, cells (6 × 103 cells) were plated into 96-well plates and transfected on the same day. Firefly luciferase promoter reporter constructs containing the REST binding (RE1) sites of miR-124a or, as a positive control for REST (RE1) binding, glutamate decarboxylase 1 (GAD1), cloned upstream of the thymidine kinase (TK) promoter (TK-miR-124a RE1 and TK-GAD1 RE1) and vectors in which the RE1 sites were deleted (TK-miR-124a ∆RE1 and TK-GAD1 ∆RE1) were kindly provided by Dr. Gail Mandel, Vollum Institute, Oregon Health & Science University, Portland, OR, USA. These vectors were co-transfected with the pRL-TK Renilla luciferase reporter (0.3 μg each plasmid per well of a 96-well plate; Promega) into cells using Lipofectamine 2000 (Invitrogen). An empty pGL2-basic vector and pGL2-control vector containing the SV40 promoter were used as negative and positive controls, respectively. Twenty-four hours post-transfection, firefly and Renilla luciferase activities were measured sequentially using the Dual-Glo Luciferase assay kit (Promega) with a 96 microplate luminometer (GLOMAX, Promega). Firefly luciferase activity was normalised to Renilla luciferase activity and the results represent as the mean of three independent experiments, each performed in triplicate.
RNA extraction and reverse transcriptase-quantitative PCR
Total RNA (including miRNAs) was extracted from transfected cells using the miRNeasy Mini kit to collect both mRNA and miRNA (Qiagen, Germany). cDNA was synthesized using 10 ng of total RNA with a TaqMan MicroRNA Reverse Transcription kit and quantitative PCR analysis was performed using the Viia 7 Real-time PCR System (Life Technologies). The human RNU48 TaqMan probe was used as an endogenous control.
Cell proliferation and migration assays
Cell proliferation and migration were examined using the xCELLigence system (Roche, Basel, Switzerland) as per manufacturer's instructions and previously published [10].
miR-124a DNA methylation analysis
Primary and commercial GBM cells were seeded at 5 × 105 cells per well of a six-well plate, incubated for 24 h, supplemented with fresh media containing 2.5 μM of 5-aza-dc (Sigma-Aldrich) for 72 h and media was changed every 24 h. Methylation Specific PCR (MSP) was conducted from DNA extracted from fresh frozen GBM samples and cell lines. Extraction of DNA was carried out using the QIAmp DNA FFPE Tissue Kit (Qiagen, Hamburg, Germany) according to the manufacturer's recommendations. Bisulfite modification followed by CpG pyrosequencing was performed to assess the degree of miR-124a methylation for each tumor specimen. Chemically methylated or unmethylated universal human genomic DNA (Millipore, Billerica, MA, USA) controls were included with each batch. In brief, tumor DNA (500 ng) was bisulfite modified using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA) according to the manufacturer's recommendations. DNA methylation status was determined by bisulfite sequencing of both strands of the corresponding CpG islands using primers for miR-124a [32]. First, methylation status was analysed by bisulfite genomic sequencing of both strands of the corresponding CpG islands. miR-124a(1) sense: AAGGATGGGGGAGAATAAAGAGTTT and miR-124a(1) antisense: CTCAACCAACCCCATTCTTAACATT (annealing temperature, 55 °C). The second analysis used MSP using primers specific for either the methylated or modified unmethylated DNA: MiR-124a methylated sense: AAAGAGTTTTTGGAAGACGTC; miR-124a methylated antisense: AATAAAAAACGACGCGTATA and miR-124a unmethylated sense: AATAAAGAGTTTTTGGAAGATGTT; miR-124a unmethylated antisense: AAAAAAATAAAAAACAACACATATAC (annealing temperature, 52 °C). DNA from normal lymphocyte donors was used as a negative control, while enzymatically methylated control DNA (Universal Methylated Human DNA, Zymo Research) was used as positive control in all the experiments. MSP was performed with KapaTag ReadyMix DNA Polymerase (Kapabiosystems) as per manufacturer's instructions. Ten microliters of PCR product were electrophoresed on 2 % agarose gels pre-cast with GelRed and visualised under ultraviolet light with a Gel Doc instrument (Bio-Rad).
Statistical analysis
Data analysis was performed using Prism 5 (GraphPad Software). Results are represented as mean ± standard error mean and obtained from at least three independent experiments. Statistical analysis was performed by two-tailed unpaired t test and p values <0.05 were regarded as statistically significant. Pearson correlation coefficient was used to assess the relationship between miR-124a and REST expression in patient samples. Other experimental data was analysed by two-way analysis of variance or Student's t test.
Results
Significantly lower levels of miR-124a are found in GBM
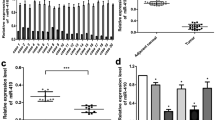
MiR-124a expression was measured in a small clinical cohort of GBM patients (n = 31) and three commercially obtained normal brain RNA (Fig. 1a). A significant reduction in miR-124a level in the GBM patient samples compared to normal brain was observed (Student's t test, p ≤ 0.001), a finding consistent with other reports. Previously, we showed that transfection of the immortalised GBM cell line A172 with synthetic precursor miR-124a resulted in a reduction of cell proliferation, invasion and migration [10]. Similar to these previous findings, endogenous miR-124a expression in the primary cell line BAH1 was also significantly lower compared to normal brain (Fig. 1b; untreated column). We restored miR-124a in BAH1 cells by transient transfection with precursor (pre-) miR-124a. Quantitative RT-PCR was used to confirm ectopic expression of miR-124a, 48 h post-transfection of BAH1 cells (Fig. 1b). Changes in cell proliferation were assessed 24 h post-transfection using xCELLigence live cell analysis (Fig. 1c). BAH1 cells transfected with miR-124a (pre-miR-124a; 60 nM) showed significantly lower cell proliferation rates when compared to cells transfected with a negative control miRNA (NC-1 miRNA, 60 nM; Fig. 1c). Tumor cell migration in pre-miR-124a transfected BAH1 cells was significantly slower when compared to cells transfected with negative control miRNA (p ≤ 0.001), untreated or cells treated with lipofectamine 2000 alone (LF2000 only; Fig. 1d).
miR-124a expression in GBM patient specimens and the functional effects of miR-124a restoration in GBM patient-derived cell lines (a) miR-124a levels in GBM specimens (n = 30) and normal brain (n = 3). Bar graphs represent log10 of the relative quantification (RQ) values of miR-124a levels, normalised by RNU48. b Ectopic expression of miR-124a in BAH1 cells. BAH1 cells were transfected with precursor miR-124a (pre-miR-124a) (60 nM), pre-MiR negative control 1 (negative control 1) (60 nM), Lipofectamine 2000 control (transfection reagent) or non-transfected control cells (control). Column charts indicate log10 of the relative quantification (RQ) values of miR-124a levels, normalised by RNU48. Tumour proliferation (c) and migration (d) were measured at 24 h post-transfection using the xCelligence cell index (CI) impedance measurements in BAH1 cells (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
miR-124a is epigenetically silenced by DNA methylation in GBM cell lines and patient tumors
To determine if the loss of miR-124a in GBM is a result of epigenetic modulation, we treated BAH1, U251 and U87MG cells with the demethylating agent, 5-aza-dC. MiRNA-124a expression significantly increased in BAH1 and U87 cells after 5′aza-dC treatment (***p < 0.001); however, U251 cells did not show a significant increase in miR-124a following treatment with 5-aza-dC, even after 72 h of treatment (Fig. 2a). DNA from cell lines and patient specimens were bisulfite converted and subjected to MSP. All seven GBM cell lines (four primary GBM cell lines and three commercial GBM cell lines) showed miR-124a methylation. A high frequency of miR-124a hypermethylation was also observed in GBM clinical specimens (82 %; 46 out of 56 patients; Fig. 2c). We confirmed relative expression of miR-124a correlates with hypermethylation status (R 2 = 0.492; two-tailed p value, 0.004).
MiR-124a expression is regulated by hypermethylation. a GBM cells (BAH1; U251 and U87) were treated with 2.5 μM of 5-aza-DC for 72 h and miR-124a levels were measured. Vehicle is media only. Scatter plots represent relative quantification (RQ) values of miR-124a levels, normalised by RNU48. Methylation-specific PCR analyses of miR-124a methylation in GBM cell lines (b) and glioma patient specimens (c) where (U) denotes unmethylated or (M) represents methylated sequences. Normal lymphocytes (NL) and in vitro methylated DNA (IVD) are shown as negative and positive controls for unmethylated and methylated sequences, respectively (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
miR-124a expression is suppressed by the transcription factor, REST, in GBM
Utilising existing transcript data from the REMBRANDT (REpository for Molecular BRAin Neoplasia DaTa, National Cancer Institute) dataset, GBM and other gliomas showed increased REST transcript compared to normal brain [7]. We also found that the median intensity of the REST transcript was higher in GBM patients (n = 31; median, 9.80 (range, 6.34–12.67) compared to normal brain (n = 3; median, 1.17; range, 1.07–1.47).
Endogenous REST levels were high in BAH1 cells (13.75). Ectopic expression of miR-124a in these cells, significantly reduced REST transcript levels by threefold when measured at two time points after transfection (2 and 7 days; Fig. 3a). To determine if miR-124a expression was regulated by the transcription factor REST in GBM, a luciferase reporter assay was performed in the BAH1 cell line. Luciferase reporter constructs were cloned with flanking sequences containing the REST-binding site (RE1) from the miR-124a gene. As a positive control, the RE1 site from the GAD1 gene; each of which was cloned upstream of the TK promoter and designated the annotation TK-miR-124a RE1 and TK-GAD1 RE1 was used. Constructs bearing the REST-binding site of the miR-124a or, as a positive control GAD1 gene, showed significantly lower luciferase activity in BAH1 cells (Fig. 3a), compared with constructs in which the RE1 site had been deleted (TK-miR-124a ∆RE1 and TK-GAD1 ∆RE1) suggesting that the presence of the miR-124a-REST binding site enabled the reduced luciferase activity in these experiments. Therefore, to confirm that it was in fact the binding of the transcription factor REST to the RE1 sites of miR-124a that was responsible for the lower activity, BAH1 cells were co-transfected with either siRNA-targeting REST or a corresponding negative control siRNA. Specific knockdown of REST, as determined relative to a negative control siRNA, resulted in a doubling of luciferase activity for both constructs containing the miR-124a RE1. Again, GAD1-RE1 constructs were used as a positive control in these experiments. Constructs lacking RE1 sites were not affected by REST knockdown (Fig. 3b). This data suggests that REST does indeed regulate miR-124a expression in GBM through binding to the RE1 site.
REST regulation of miR-124a. a Relative expression of REST in BAH1 cells after transfection with miR-124a. BAH1 cells were transfected with miR-124a (60 nM) and relative mRNA expression for REST were measured 2 days (2D) and 7 days (7D) post-transfection with quantitative real time reverse transcription polymerase chain reaction (qRT-PCR) and normalised by 18S. b Luciferase reporter analysis showed that RE1-containing constructs (TK-miR-124a RE1 and TK-GAD1 RE1), transfected into BAH1 cells, have lower luciferase activity compared with constructs lacking the RE1 site (TK-miR-124a ∆RE1 and TK-GAD1 ∆RE1). Relative luciferase activity was measured after 24 h (n = 3 ± SEM, **p ≤ 0.01). c Analysis using a luciferase reporter assay shows de-repression of reporters bearing the RE1s of miR-124a and GAD1 upon transfection of a siRNA targeting REST in BAH1 cells. Fold changes in luciferase activity are shown as the ratios of the geometric means of reporter activity with control siRNA and RESTsiRNA (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
Discussion
MiR-124a is one of the best-characterised miRNAs in the central nervous system [5, 9, 11, 12, 33, 34] with its loss shown to be significantly associated with tumorigenesis. Ectopic expression of miR-124a in GBM leads to significantly reduced tumor proliferation, migration and invasion [10].
We provide evidence that miR-124a expression is reduced in GBM via at least two mechanisms: (1) transcriptional inactivation by CpG island hypermethylation and (2) regulation by the transcription factor REST. Treatment of cells with the demethylating agent 5-aza-dC forced hypomethylation of cellular DNA and resulted in increased levels of miR-124a. We present similar findings to Silber et al. where no change was found when treating U251 with the demethylating agent. However, in contrast, we did find significant increases in miR-124a expression levels when U87 and the patient derived cell line, BAH1 were treated. This discrepancy could reside in the different 5-aza-dC scheduling; we used a lower dose of 2.5 μM for 72 h compared to 5 μM for 24 h. Endogenous methylation of miR-124a was confirmed in over 80 % of GBM patient specimens using MSP, providing strong evidence that miR-124a is epigenetically regulated.
That the expression levels of miR-124a could also be negatively regulated by the transcription factor, REST, was also confirmed with our luciferase reporter assay and the reciprocal expression of REST and miR-124a were further confirmed in patient specimens. REST dysfunction has been implicated in diverse diseases ranging from Down syndrome to cardiomyopathy and cancer. An oncogenic role for REST has been established in medulloblastoma [7, 26, 30, 35]. High REST levels in this childhood cancer, coupled with Myc overexpression promoted cells toward proliferation and tumorigenesis. In GBM, high expression of REST was enriched in tumor cells in the perivascular component of the tumor and knock down of REST strongly reduced tumor-initiating capacity in vivo [7, 30]. Targeting REST may prove to be an interesting avenue for future therapeutic research, particularly given the broad therapeutic index and toxicity of demethylating agents in vivo. The HDAC inhibitor, SAHA (suberoylanilide) promoted REST proteomic degradation and decreased medulloblastoma growth following treatment [26]. This approach might also be promising for GBM patients.
Moreover, significantly low miR-124a expression patterns observed in GBM can be attributed to both epigenetics and the transcription factor, REST. Treatment of GBM with a HDAC inhibitor such as SAHA might be a promising approach to modulate the expression of miR-124a.
References
CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2004–2008. 2012.
Tivnan A, McDonald KL. Current progress for the use of miRNAs in glioblastoma treatment. Mol Neurobiol. 2013. doi:10.1007/s12035-013-8464-0
Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386:1–5.
Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–55.
Silber J, Hashizume R, Felix T, Hariono S, Yu M, Berger MS, et al. Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro Oncol. 2013;15:83–90.
Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–71.
Conti L, Crisafulli L, Caldera V, Tortoreto M, Brilli E, Conforti P, et al. REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS One. 2012;7:e38486.
Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6:e24248.
Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14.
Fowler A, Thomson D, Giles K, Maleki S, Mreich E, Wheeler H, et al. miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur J Cancer. 2011;47:953–63.
Hua D, Mo F, Ding D, Li L, Han X, Zhao N, et al. A catalogue of glioblastoma and brain MicroRNAs identified by deep sequencing. OMICS: J Integr Biol. 2012;16:690–9.
Deng X, Ma L, Wu M, Zhang G, Jin C, Guo Y, et al. miR-124 radiosensitizes human glioma cells by targeting CDK4. J Neuro-Oncol. 2013;114(3):263–74.
Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun. 2012;426:247–52.
Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, et al. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73(13):3913–26.
Li D, Lu Z, Jia J, Zheng Z, Lin S. Changes in microRNAs associated with podocytic adhesion damage under mechanical stress. J Renin-Angiotensin-Aldosterone Syst JRAAS. 2013;14:97–102.
Lathia JD, Heddleston JM, Venere M, Rich JN. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell. 2011;8:482–5.
Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–76.
Wong KY, So CC, Loong F, Chung LP, Lam WW, Liang R, et al. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS One. 2011;6:e19027.
Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167.
Wang X, Ren J, Wang Z, Yao J, Fei J. NRSF/REST is required for gastrulation and neurogenesis during zebrafish development. Acta Biochim Biophys Sin. 2012;44:385–93.
Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci Off J Soc Neurosci. 2011;31:9772–86.
Soldati C, Bithell A, Johnston C, Wong KY, Stanton LW, Buckley NJ. Dysregulation of REST-regulated coding and non-coding RNAs in a cellular model of Huntington's disease. J Neurochem. 2013;124:418–30.
Yu M, Suo H, Liu M, Cai L, Liu J, Huang Y, et al. NRSF/REST neuronal deficient mice are more vulnerable to the neurotoxin MPTP. Neurobiol Aging. 2013;34:916–27.
Liu M, Sheng Z, Cai L, Zhao K, Tian Y, Fei J. Neuronal conditional knockout of NRSF decreases vulnerability to seizures induced by pentylenetetrazol in mice. Acta Biochim Biophys Sin. 2012;44:476–82.
Yu M, Cai L, Liang M, Huang Y, Gao H, Lu S, et al. Alteration of NRSF expression exacerbating 1-methyl-4-phenyl-pyridinium ion-induced cell death of SH-SY5Y cells. Neurosci Res. 2009;65:236–44.
Taylor P, Fangusaro J, Rajaram V, Goldman S, Helenowski IB, MacDonald T, et al. REST is a novel prognostic factor and therapeutic target for medulloblastoma. Mol Cancer Ther. 2012;11:1713–23.
Fuller GN, Su X, Price RE, Cohen ZR, Lang FF, Sawaya R, et al. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Mol Cancer Ther. 2005;4:343–9.
Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–78.
Kamal MM, Sathyan P, Singh SK, Zinn PO, Marisetty AL, Liang S, et al. REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells. 2012;30:405–14.
Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–7.
Day BW, Stringer BW, Al-Ejeh F, Ting MJ, Wilson J, Ensbey KS, et al. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell. 2013;23:238–48.
Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9.
Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol. 2008;90:1–7.
Fowler A, Cook R, Biggs M, Little N, Assaad N, McDonald K. Survival of patients following neurosurgical treatment of colorectal adenocarcinoma metastasis in the Northern Sydney-Central Coast area. J Clin Neurosci Off J Neurosurg Soc Australas. 2008;15:998–1004.
Das CM, Taylor P, Gireud M, Singh A, Lee D, Fuller G, et al. The deubiquitylase USP37 links REST to the control of p27 stability and cell proliferation. Oncogene. 2013;32:1691–701.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tivnan, A., Zhao, J., Johns, T.G. et al. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumor Biol. 35, 1459–1465 (2014). https://doi.org/10.1007/s13277-013-1200-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-1200-6