Abstract
Succinate dehydrogenases (SDH), including SDHA, SDHB, SDHC, and SDHD, form the respiratory complex II in the mitochondria and play an important role in cell growth and homeostasis. In order to evaluate the expression and functional significance of SDH in colorectal cancer, the expression of four SDH subunits was analyzed, and SDHB protein was found to be significantly lower in colorectal cancer tissues. In vitro experiments including cell growth assay, colony formation assay, cell cycle analysis, and nude mouse xenograft of SDHB-transfected colorectal cancer cell line SW620 were performed. Notably, reduced SDHB expression in tumor tissues was associated with tumor de-differentiation, and restoration of SDHB could inhibit the growth of cancer cells both in vitro and in vivo. Furthermore, cDNA microarray of SDHB-transfected cell line showed that most of the differentially expressed genes are related to cell cycle control and cell proliferation. Thus, we conclude that SDHB expression is significantly decreased in human colorectal cancer tissues, and reconstitution of SDHB in colorectal cancer may be a potential therapeutic approach to inhibit aggressiveness of colorectal cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most prevalent cancer in the world, accounting for approximately 1.2 million new cases and more than 600,000 cancer-related deaths each year worldwide [1]. This disease begins with adenomatous polyp, further advances to adenoma with high-grade dysplasia, and finally progresses to an invasive cancer. The behavior of CRC can be established as a progressively proliferating and invasive model of a group of uncontrolled cells which interacted at both genetic and environmental level [2]. At the molecular level, carcinogenesis is a multistep process featuring the accumulation of several genetic alterations, including the activation of oncogenes and the inactivation of tumor suppressor genes (TSG) [3]. The loss or inactivation of TSG plays an important role in the development and progression of CRC [4]. Previously, we sought to uncover the proteomic changes that took place during the formation and progression of CRC. Our proteomics data indicated a different group of proteins expressed dynamically through the course of cancer progression which implicate their oncogenic and/or tumor suppressor role in CRC tumorigenesis. Among these proteins, we found the four nuclear-encoded genes: succinate dehydrogenases (SDH) as a putative tumor suppressor candidate for further functional investigation [5].
SDH is a heterotetrameric protein which consists of A, B, C, and D subunits, localized on the inner mitochondrial membrane which catalyze the oxidative dehydrogenation of succinate and, as part of complex II of the electron transport chain, couple this to the reduction of ubiquinone (succinate–ubiquinone oxidoreductase) [6]. When succinate is oxidized to fumarate, two hydrogen atoms are removed from succinate and transferred to FAD, reducing it to FADH2 on the A subunit of SDH (SDHA). Electrons from FADH2 are sequentially transferred through three iron–sulfur centers in SDHB to the ubiquinone site associated with SDHC and SDHD, embedded in the mitochondrial inner membrane. Studies showed SDH plays a critical role in cellular energy metabolism through its dual function in the Krebs cycle and the respiratory chain [7–9], and many reports indicated that the genetic defects in SDH are associated with a diverse collection of diseases, including familial head and neck paragangliomas (PGL), pheochromocytoma, gastrointestinal stromal cell tumor (GIST), and renal cancer [10–14]. Although the mechanisms by which disruption of SDH leads to neoplasia are largely unknown, evidence has shown that activation of the hypoxia-response pathway is a common theme underlying SDH loss of function [15–18].
To date, little is known about the role of SDH in CRC tumorigenesis or how it regulates malignant behavior of tumor cells. Thus, in this study, we designed experiments to investigate the potential SDH tumor suppressor functions in CRC based on the unique background of this gene and our previous proteomics findings. We first assessed expression of these four subunit proteins of SDH in CRC tissue microarray (TMA) using immunohistochemistry and found that only SDHB protein was significantly lower in CRC tissues. After that, we stably expressed SDHB protein in human colon cancer cells to determine the effects of SDHB expression on tumor cell growth and tumorigenesis. Moreover, differential expression of genes induced by ectopic SDHB expression in cancer cells was screened by microarray analysis. These data could help us to understand the role of SDHB in CRC progression and future control of CRC in the clinic.
Materials and methods
Tissue microarray
A TMA containing 103 cases of CRC tissue specimens with a total of 336 spots was applied in this study [19]. In brief, the 336 tissue spots included 103 cases of paired CRC and mucosa specimens adjacent to the primary tumor (0.5 and 1.0 cm away from the primary tumor), 103 distant normal mucosa (more than 10 cm away from the primary tumor), and 27 cases of lymph node metastatic tumor tissues. Forty patients were older than 60 years and 63 younger than 60 years. Forty-two cases were well differentiated, 46 were moderately differentiated, and 15 were poorly differentiated. Of these patients, 3 were stage I, 45 were stage II, 30 were stage III, and 25 were stage IV, according to the clinical TNM stage revised by the International Union Against Cancer in 2003. Fifty-five patients had lymph node metastasis, and 48 had no lymph node metastasis, whereas 25 cases had distant metastasis. Commercially available TMA sections containing 32 cases of poorly differentiated colon cancer tissue specimens (Shanghai Outdo Biotech Co., Ltd, China) were also included in this study. Of the 32 cases, 2 were female and 30 were male patients, with 11 younger than 60 years old. Informed consent was obtained from each patient. This study was approved by the Institutional Review Board of the Third Xiangya Hospital, Central South University.
Immunohistochemistry
The TMA sections were first heated at 60 °C for 30 min, deparaffinized in xylene twice for 10 min each, and rehydrated with a graded series of ethanol. The sections were then subjected to antigen retrieval in citrate buffer (10 mM, pH 6.0) in a microwave oven for 20 min and incubated in 3 % H2O2 for 30 min to block potential activity of endogenous peroxidase. After washing in phosphate-buffered saline (PBS), the sections were incubated in normal goat serum (Zhongsha, Beijing, China) for 30 min to minimize nonspecific staining and incubated with a primary antibody overnight at 4 °C. The sections were incubated with a monoclonal anti-SDHA (1:20,000; Abcam, Hong Kong, China), an anti-SDHB (1:8,000; Abcam), an anti-SDHC (1:100; Santa Cruz, Heidelberg, Germany), or an anti-SDHD (1:2,000; Abcam) antibody. After washing with PBS, the sections were further incubated with a secondary antibody (Zhongsha, Beijing, China) for 30 min. The immunochemical reaction was visualized by diaminobenzidine (Zhongsha, Beijing, China) and hematoxylin counterstaining (Maixin, Fuzhou, China). The sections were dehydrated following a standard procedure and sealed with coverslips. Known positive sections served as the positive control. Sections incubated with PBS instead of the primary antibody were used as negative controls.
The stained TMA sections were reviewed under a microscope and scored by two pathologists independently according to the intensity of the immune-positivity and the number of cells stained. Immuno-staining intensity was scored as follows: 0, negative; 1, weak; 2, intermediate; and 3, strong. The number of cells stained was scored as follows: 0, <5 % of tumor cells stained positively; 1, 5–25 %; 2, 26–50 %; 3, 51–75 %; and 4, >75 %. After that, these two scores were added together to form a final score of each case: negative (0 score) and positive (1–7 scores) [0, negative (0 score); 1, weak positive (1–2 scores); 2, intermediate positive (3–5 scores); and 3, strong positive (6–7 scores)].
Cell lines and culture
Human colon cancer HT-29 and SW620 cell lines were provided by the Cancer Research Institute of the Central South University (Hunan, China) and cultured in RPMI 1640 medium containing 10 % fetal bovine serum at 37 °C in a humidified incubator with 5 % CO2.
Protein extraction and Western blot
Total cellular protein was extracted from cultured cells using a lysis buffer containing 50 mM Tris–HCl, 150 mM NaCl, 0.02 % NaN3, 0.1 % SDS, 0.5 % sodium deoxycholate, 0.1 % EDTA, 1 % NP-40, 5 % glycerol, and a complete protease inhibitor mixture (Roche, Rotkreuz, Switzerland). After centrifugation, the supernatant was collected and analyzed for protein concentration using the BCA Protein Assay Kit (Pierce, Rockford, USA). Equal protein samples (50 μg) were loaded onto SDS-PAGE gels and then transferred onto PVDF membranes, which were blocked with 5 % fat-free milk/0.1 % Tween-20 in PBS for 1 h at room temperature. Membranes were then incubated with primary antibodies at 4 °C overnight. In the next day, the membranes were washed three times with PBS containing 0.1 % Tween-20 and then incubated with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). The immunoreactive bands were then detected by enhanced chemiluminescence (GE healthcare, Buckinghamshire, UK). The primary antibodies were a mouse monoclonal anti-SDHB (1:1,000; Abcam), anti-cyclinD1 (1:200; Santa Cruz Biotechnology), or anti-α-tubulin antibody (1:4,000; Santa Cruz Biotechnology).
Construction of SDHB eukaryotic expression vector and gene transfection
The open reading frame of human SDHB gene (NM_003000.2) was amplified by a PCR with HT-29 cDNA as template. The primers used were 5′-TAAGATATCATGGCGGCGGTGGTCGCC-3′ and 5′-GCCGGATCCTTAAACTGAAGCTTTCTT-3′. PCR conditions were 95 °C for 5 min and 30 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 2 min. After that, the PCR fragment was purified and ligated into EcoR V and BamH I-digested pIRESneo3 vectors (Clontech, San Diego, CA, USA). After amplification and sequence confirmation, this vector was named pIRESneo3-SDHB for gene transfection. The Escherichia coli-competent cells were obtained from Takara (Dalian, China).
For gene transfection, CRC cells were grown and transfected with 3 μg pIRESneo3-SDHB using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Twenty-four hours after transfection, cells were passaged at 1:3 into new dishes with fresh growth medium. G418 (400 μg/ml, Invitrogen) was added to the growth medium the following day. After 6 weeks of selection, stably transfected cells were obtained and maintained in RPMI 1640 containing 400 μg/ml G418. Cells transfected with the empty pIRESneo3 vector served as the control.
Cell growth assay
Cell growth rate was determined by using direct cell counting. Specifically, the stably SDHB-transfected CRC cells were seeded in 24-well plates in triplicate at a density of 1 × 104 cells/well and grown overnight. Cells were harvested at intervals of 24 h for up to 7 days for cell counting. The number of cells in each well was determined by harvesting the cells with trypsin and counting cells in an aliquot using a Z1 Particle Counter (Beckman-Coulter, Fullerton, CA, USA).
Colony formation assay
The anchorage-dependent growth of tumor cells was investigated using the monolayer colony formation assay [20]. Briefly, SW620 cells were seeded at a density of 1,000 cells per well in a six-well plate and maintained at 37 °C in a sterile 5 % CO2 incubator. Colonies were counted under a microscope at 14 days after being fixed with methanol and being stained with 5 % Giemsa (Sigma-Aldrich). Only those colonies containing at least 50 cells were considered to be viable survivors. Each assay was performed in triplicate.
Cell cycle analysis
Cell cycle distribution was assessed using a Cell Cycle Analysis Kit from Beyotime Institute of Biotechnology (Haimen, China) following the manufacturer’s instructions. In brief, cells were collected and fixed in 70 % ice-cold ethanol overnight. In the next day, the cells were stained with propidium iodide working solution (25 μl propidium iodide stock solution and 10 μl RNase A into 0.5 ml buffer solution) for 30 min in the dark at 37 °C and then analyzed by flow cytometry (Beckman). The cell cycle distribution was calculated from the resultant DNA histogram using Multicycle AV software (Phoenix Flow System, San Diego, CA, USA).
Immunocytochemistry
Cells were trypsinized and plated onto coverslips and grown for 24 h. After that, cells were fixed in 95 % ethanol for 20 min, rinsed in PBS, and then incubated with 0.5 % Triton X-100 for 20 min and in 3 % hydrogen peroxide for 15 min. The cells were incubated with normal goat serum for 30 min in room temperature and then with an anti-PCNA antibody (Cell Signaling Technology, Danvers, MA, USA) in a humidified chamber at 4 °C overnight. In the next day, the cells were washed with PBS three times and incubated with a secondary antibody for 20 min at 37 °C. After washing in PBS, the color reaction was performed with a diaminobenzidine solution.
Nude mouse xenograft assay
Twenty 4- to 6-week-old male Balb/c athymic nude mice, bred in specific pathogen-free conditions, were purchased from the Laboratory Animal Unit at the Central South University (Changsha, China) and housed in our animal facility. The mice were then randomly divided into two groups. Stable transfected CRC cells with vector only or SDHB cDNA vector (2 × 106) in 0.2 ml volume of RMPI 1640 were injected subcutaneously into the left flank of the mice. Tumor formation and growth were monitored. Tumor volume was measured and calculated by the formula: tumor volume = 0.5 × length × width2. At the end of the experiment, the tumors were excised and the tumor weight was measured. The animal experiment protocol was approved by the ethics committee of Third Xiangya Hospital, Central South University.
cDNA microarray
Total RNA was extracted and purified from SDHB and control vector-transfected cells, and then subjected to Illumina Human HT-12 v4 Expression BeadChip (Illumina, San Diego, CA, USA) analysis. Briefly, RNA was converted into biotin-labeled cRNA, which was used as probes for hybridization in the cDNA microarray. The data were scanned by Beadarray Reader (Illumina). The array intensity data were analyzed using the Illumina GenomeStudio Gene Expression Module (v1.1.1) for visualization and normalization. The average normalization method was applied, and average background correction was performed using Beadstudio software. Analysis of differential gene expression was performed using GeneSpring GX 11 (Agilent Technologies, Santa Clara, CA, USA). Differentially expressed genes with a fold change >2 or <0.5 and a p < 0.01 were included in further analyses. Functional profiling of gene lists was performed using DAVID as described [21]. All procedures were carried out by Beijing Emei Tongde Technology Development Co. Ltd (Beijing, China).
Reverse transcription PCR and real-time PCR
Total RNA was isolated from cultured cells using a Trizol reagent (Invitrogen) and treated with DNase (Takara, Dalian, China) to eliminate contaminated DNA in the samples. One microgram of total RNA sample was then reversely transcribed into cDNA with oligo(dT) as primers using the cDNA synthesis kit (Promega, Beijing, China). Expression levels of genes were determined by real-time PCR with a Bio-Rad CFX Real-Time PCR Detection System using SYBR® Premix Ex Taq™ (Takara). The genes analyzed were ARMCX3 (5′-GGCGGAGGGAAAAAGGAAGAGGA-3′ and 5′-TCAAGACAAGCAGTTCGCCCCT-3′), ETS1 (5′-TGTTCGGGACTGGGTGATGTGG-3′ and 5′-AGCGGGATTCTGGATAGGCTGG-3′), CSF2 (5′-GAACCTGAGTAGAGACACTGCTGC-3′ and 5′-AGGTGATAATCTGGGTTGCACAGGA-3′), PTEN (5′-TGGGGAAGTAAGGACCAGAGA-3′ and 5′-ACTTGTCTTCCCGTCGTGTG-3′), CUL5 (5′-ATGCTGAGAGATTGGGAGAAG-3′ and 5′-TACGAGGGTGCTTGTGTTCT-3′), CAV1 (5′-CAAGGAGGGGCTGTAAAATGGAGG-3′ and 5′-ACTGCCTCCTCCCCCATCTTCTT-3′), C10orf125 (5′-ACTTGAACTTCCCGGCCTCCT-3′ and 5′-CAGCAAAAGCCTTCTTAGCCCGT-3′), and TPGS1 (5′-ATCGCCTTCCTGGCTCACTACTTC-3′ and 5′-GGCCACGCTCACGTTGTTGTT-3′). GAPDH was amplified as an endogenous control.
Statistical analysis
Statistical analysis was carried out using SPSS software (version 17.0; SPSS Inc, Chicago, IL, USA). Comparisons of categorical variables were made using the Chi-squared test. Differences between the mean of two groups were compared using Student’s t test. A p value <0.05 was regarded statistically significant.
Results
Protein expression of SDHA, B, C, and D in CRC
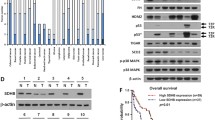
We first evaluated the expression of SDHA, SDHB, SDHC, and SDHD proteins in CRC. All SDH proteins were localized in the cytoplasm of colorectal cells (Fig. 1). Expression of SDHB protein was found to be significantly lower in CRC tissues (92/135 cases, 68.1 %) compared to the adjacent normal mucosa (93/103 cases, 90.3 %, p < 0.01) or the distant normal mucosa (97/103 cases, 94.2 %, p < 0.01), whereas no significant difference was found in that of SDHA, SDHC, or SDHD (Table 1). Therefore, SDHB was put into further investigation.
Expression of SDHA, SDHB, SDHC, and SDHD proteins in normal colorectal mucosa, adjacent normal mucosa, and colorectal cancer tissues. a Immunohistochemical detection of SDHA, SDHC, and SDHD proteins. b Immunohistochemical detection of SDHB protein in normal colorectal mucosa (a), adjacent normal mucosa (b), well differentiated colorectal cancer (c), moderately differentiated colorectal cancer (d), poorly differentiated colorectal cancer (e), and moderately differentiated colorectal cancer with infiltrating lymphocytes tissues (f) (magnification, ×400)
Correlation between clinicopathological features and SDHB expression in CRC
Correlation between clinicopathological parameters and SDHB expression was analyzed. Only differentiation state was associated with the expression level of SDHB (poorly vs. moderately differentiated, p < 0.01, or vs. well differentiated, p < 0.01). Significantly, the expression of SDHB was negatively correlated with the differentiation status of CRC tissues. The poorer the differentiation state of CRC cells, the lower the expression of SDHB. Expression of SDHB protein was not associated with age, gender, TNM stage, lymph node metastasis, distant metastasis, tumor size, or tumor location (Table 2, p > 0.05).
Restoration of SDHB inhibits growth in CRC cells
Encouraged by the result of TMA analysis, we postulated that SDHB protein might exert a tumor suppressor role in the regulation of CRC cells. Therefore, we constructed SDHB expression vector and stably transfected SDHB into colon cancer cell line SW620, namely SW620-SDHB and SW620-Vector as negative control, to evaluate its possible tumor suppressor function of SDHB protein in vitro (Fig. 2a).
Restoration of SDHB expression inhibited colorectal cancer cell growth. a SDHB expression in colon cancer cells SW620 transfected with empty vector (pIRESneo3) and SW620 cells stably overexpressing SDHB was tested by Western blot. HT-29 cells were used as control, and each sample was tested in duplicate. b Cell growth curve of SW620 cells transfected with control vector and SDHB measured by cell count. c Cell cycle analysis of transfected cells by flow cytometry. SDHB induced colon cancer cell line SW620 G1 arrest
Cell growth assay showed that the growth rate of CRC cells SW620 was significantly reduced after the transfection of SDHB compared to the vector control cells (Fig. 2b). Flow cytometry results of the cell cycle distribution showed that SDHB induced cell cycle arrest at the G1 phase (Fig. 2c, p < 0.05). It is also noticed that the ectopic expression of SDHB substantially inhibited tumor cell colony formation (p < 0.01) (Fig. 3a) and downregulated the expression of PCNA and cyclinD1 (Fig. 3b, c). SW620-Vector cells showed strongly positive nuclear expression of PCNA (dark brown yellow), whereas the SW620-SDHB cells showed only weakly positive nuclear expression (light brown yellow, Fig. 3b). Western blot showed that SW620-SDHB cells expressed significantly less cyclinD1 than the vector control cells (Fig. 3c).
Effect of ectopic SDHB expression on the regulation of tumor cell colony formation and gene expression. a Control SW620 cells and SDHB-transfected SW620 cells were used for anchorage-dependent colony formation assay. Assays were performed in triplicate, and the average numbers of clones from one whole well are shown in the graph, with bars representing standard deviation. b Immunocytochemistry staining showed intense positive expression of PCNA in the nuclei in the empty vector-transfected SW620 cells and weak expression of PCNA in SDHB-transfected cells (magnification, ×200). c The overexpression of SDHB inhibited the cyclinD1 expression in cytoplasmic extract of SW620 cells measured by Western blot. **p < 0.01 compared with the vector-only-transfected cells
Effect of SDHB was also evaluated in vivo by SW620-SDHB and SW620-Vector cell xenograft model of nude mice. An average of 2 × 106 cells were injected subcutaneously into athymic nude mice and monitored for 7 weeks. Eight out of ten nude mice injected with SW620-Vector cells and six out of ten nude mice injected with SW620-SDHB cells developed grossly visible tumors. Result showed that average tumor weight of SW620-SDHB xenografts was significantly less than that of the vector control xenografts (Fig. 4, p < 0.01). Thus, both in vitro and in vivo experiments confirmed that the restoration of SDHB may contribute to growth inhibitory effect in CRC and the possible tumor suppressor role of SDHB protein.
Effect of SDHB expression in the suppression of nude mouse xenograft formation and growth. a SDHB-transfected SW620 cells or control vector-transfected SW620 cells were injected subcutaneously into nude mice, and the graph shows tumor volume as measured on the indicated days. b, c Overexpression of SDHB led to a decrease in in vivo tumor size compared with vector control. d The average weight of tumors of each group was shown as columns, with bars representing standard deviation. **p < 0.01 compared with the vector-only-transfected cells
Gene expression profile
Confirmed by the functional results of SDHB restoration, we further examine the gene expression profile of SW620-SDHB and SW620-Vector cell line to obtain more possible information that may be related to the downstream regulation of SDHB. cDNA microarray analysis showed a total of 1,653 differentially expressed genes with more than a twofold change (p < 0.01), of which 1,382 gene expressions were upregulated and 271 gene expressions were downregulated in SW620-SDHB cells compared with vector control cells. Gene ontology (GO) and pathway analysis were carried out to classify these differentially expressed genes into functional categories. As expected, most of the genes were grouped in GO with regard to “cell cycle” or “cellular process” (Fig.5a), which might be the downstream target of SDHB. Furthermore, microarray data involving those significant differentially expressed genes especially involving cell cycle and cellular process were validated by real-time RT-PCR. Upregulated genes including ARMCX3, CSF2, ETS1, CAV1, CUL5, and PTEN, whereas downregulated genes such as TPGS1, and C10orf125 were all in accordance with the microarray result (Fig. 5b). For related information, please refer to attachment file of the manuscript.
cDNA microarray analysis of SDHB-transfected and control vector-transfected SW620 cells. a Biological process (BP) of GO showed most of the differentially expressed genes were enriched in GO with regard to “cell cycle” or “cellular process.” b Real-time PCR confirmation of differentially expressed genes. The upper panel shows the gene array data, while the lower panel shows the gene expression levels detected by qRT-PCR. *p < 0.05 compared with the vector-only-transfected cells
Discussion
The tumor suppressor functions of SDH proteins have been observed in various human malignancies, and SDH proteins were identified as the first nuclear-encoded mitochondrial tumor suppressor genes. A series of studies indicated that the mutations in SDH were associated with tumor formation including PGLs [11, 12, 22], GIST [10, 23], thyroid cancer [24], renal tumors, and neuroblastoma [14, 25, 26]. Studies on biochemical mechanisms of SDH proteins suggested that loss of SDH functions could cause accumulation of succinate and production of reactive oxygen species (ROS) [27, 28]. The excessive succinate leaves the mitochondria and can inhibit the activity of different cellular enzymes [such as HIF-α or prolyl hydroxylases] in the cytosol, leading to unhydroxylated and undegraded HIF-α, and in turn heterodimerizes with HIF-β to form an active complex to induce gene expressions for cell proliferation, survival, glycolysis and angiogenesis [29, 30]. Furthermore, accumulation of succinate could also induce production of the ROS, and the latter has a role in tumorigenesis and progression. Indeed, a number of studies showed that loss of SDH functions led cells to having oxidative stress, reduced lifespan in model organisms, genomic instability, and cancer development [31–36]. Thus, both succinate and ROS could independently or synergistically induce hypoxic response under normoxic condition (pseudo-hypoxia), a phenomenon that has been widely observed in a variety of malignancies. However, the completely defined mechanisms of SDH deficiency in human tumors remain to be determined, and there was no extensive functional assay performed in CRC. In our previous study on CRC, the proteomic profile of a different stage of CRC was conducted, and we found a group of proteins were dynamically (followed by TNM stage series) expressed, including SDH [5]. Specifically, SDH protein was significantly downregulated during CRC development and progression, thus leading us to hypothesize the possible tumor suppressor role of SDH in this high incidence gastroenterological disease.
In our initial study, we simultaneously screened the expression of all four different SDH proteins in CRC, adjacent mucosa, and distant normal colorectal mucosa. Our data showed that SDHA, SDHC, and SDHD proteins were ubiquitously expressed in both tumor and normal colorectal mucosa, whereas only SDHB expression was found to be significantly reduced in CRC compared to their normal counterpart. Notably, reduced SDHB protein expression was more frequently observed in young patients and in poorly differentiated CRC than in moderately and well differentiated CRC. These results imply that SDHB can serve as a potential colorectal-specific biomarker for aggressiveness phenotype of CRC, and its reduced expression might be a poor indicator of clinical outcome of CRC patients.
Focused on the cellular function of SDHB in CRC cells, we applied the SDHB-cDNA recombinated SW620 cell line to reconstitute the expression of SDHB. SW620 was derived from a highly metastatic CRC patient and was initiated by Leibovitz et al. [37] from a lymph node in the same manner as was the primary adenocarcinoma from which SW480 was derived the previous year. Due to the aggressiveness of this CRC cell line, we carried out all our in vitro and in vivo experiments in context of SW620-SDHB and SW620-Vector to validate our previous speculation on SDHB. Apparently, restoration of SDHB expression would suppress the tumor cell growth by a significant extent. Both cell growth and colony formation assay showed significant difference between SDHB-transfected and vector control cells. Flow cytometry data showed overexpressed SDHB would result in an increase number of cells in the G0/G1 phase of the cell cycle and reduced S phase population. This indicates the SDHB would cause G1/S phase arrest which might trigger tumor cell apoptosis or necrosis. When this cellular model was transferred to the in vivo state of the xenograft of nude mice, the SW620-SDHB tumor mass greatly decreased in volume, and microscopic examination showed most of the SW620-SDHB tumor cells were going through necrosis or apoptosis (data not shown) which was in accordance with our in vitro results.
To understand how SDHB exerts its tumor suppressive function, we performed microarray analysis to search the potential targets of SDHB. Array data of SW620-SDHB showed that restoration of SDHB in CRC would lead to significant gene expression change by which 1,653 differentially expressed genes with over twofold change. Bioinformatics analysis revealed that restoration of SDHB in CRC would cause upregulation of genes mainly involved in cellular process including cell cycle and cell proliferation. For example, we have confirmed some of them by qRT-PCR, such as CUL5, CAV1, and PTEN. CUL5 is involved in a variety of biological functions, including regulation of cell growth [38], and lost CUL5 expression promoted growth and invasion of human cervical cancer HeLa cell line [39]. CAV1 expression was also shown to be reduced in human colon cancers and ectopic expression of CAV1 reduced tumorigenicity of colon carcinoma cells [40]. Moreover, PTEN is a tumor suppressor gene to suppress the antiapoptotic and proliferative AKT (protein kinase B) [41] and mitogen-activated kinase (MAPK) pathways [42]. PTEN dysfunction was associated with activation of these gene pathways. Another study showed that Cowden syndrome/Cowden syndrome-like-related SDH mutations/variants were able to increase phosphorylation of AKT and/or MAPK, even in the absence of PTEN dysfunction [43], indicating the role of SDH in regulation of PTEN expression or activity. Taken altogether, our current data further indicated that SDHB plays an important role in suppression of CRC development and progression.
In conclusion, the data from our current study demonstrated that the SDHB is a putative CRC-specific tumor suppressor that mainly exerts the inhibitory effect to CRC growth. Reduced expression of SDHB in CRC were associated with poor differentiation of CRC cells and poor prognosis. Further investigation concerning the regulation of SDHB and larger clinical data pointing to this prominent tumor suppressor gene will be considered in our future investigation.
References
Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907.
Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67.
Macleod K. Tumor suppressor genes. Curr Opin Genet Dev. 2000;10:81–93.
Peng Y, Li X, Wu M, Yang J, Liu M, Zhang W, et al. New prognosis biomarkers identified by dynamic proteomic analysis of colorectal cancer. Mol Biosyst. 2012;8:3077–88.
Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta. 1807;2011:1432–43.
Rustin P, Munnich A, Rötig A. Succinate dehydrogenase and human diseases: new insights into a well-known enzyme. Eur J Hum Genet. 2002;10:289–91.
Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–93.
Scheffler IE. Molecular genetics of succinate:quinone oxidoreductase in eukaryotes. Prog Nucleic Acid Res Mol Biol. 1998;60:267–315.
Bolland M, Benn D, Croxson M, McCall J, Shaw JF, Baillie T, et al. Gastrointestinal stromal tumour in succinate dehydrogenase subunit B mutation-associated familial phaeochromocytoma/paraganglioma. ANZ J Surg. 2006;76:763–4.
Kantorovich V, King KS, Pacak K. SDH-related pheochromocytoma and paraganglioma. Best Pract Res Clin Endocrinol Metab. 2010;24:415–24.
Pasini B, Stratakis CA. SDH mutations in tumorigenesis and inherited endocrine tumours: lesson from the phaeochromocytoma-paraganglioma syndromes. J Intern Med. 2009;266:19–42.
Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase—assembly, regulation and role in human disease. Mitochondrion. 2010;10:393–401.
Vanharanta S, Buchta M, McWhinney SR, Virta SK, Peczkowska M, Morrison CD, et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am J Hum Genet. 2004;74:153–9.
Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, et al. A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1:72–80.
Gimenez-Roqueplo AP, Favier J, Rustin P, Mourad JJ, Plouin PF, Corvol P, et al. The R22X mutation of the SDHD gene in hereditary paraganglioma abolishes the enzymatic activity of complex II in the mitochondrial respiratory chain and activates the hypoxia pathway. Am J Hum Genet. 2001;69:1186–97.
Gimenez-Roqueplo AP, Favier J, Rustin P, Rieubland C, Kerlan V, Plouin PF, et al. Functional consequences of a SDHB gene mutation in an apparently sporadic pheochromocytoma. J Clin Endocrinol Metab. 2002;87:4771–4.
Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–9.
Peng Y, Li H, Wu M, Wang X, Fan S, Liu F, et al. NGX6 inhibits AP-1 and Ets-1 expression and down-regulates cyclin D1 in human colorectal cancer. Acta Biochim Biophys Sin (Shanghai). 2009;41:504–14.
Li W, Li X, Wang W, Tan Y, Yi M, Yang J, et al. NOR1 is an HSF1- and NRF1-regulated putative tumor suppressor inactivated by promoter hypermethylation in nasopharyngeal carcinoma. Carcinogenesis. 2011;32:1305–14.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–20.
Celestino R, Lima J, Faustino A, Máximo V, Gouveia A, Vinagre J, et al. A novel germline SDHB mutation in a gastrointestinal stromal tumor patient without bona fide features of the Carney-Stratakis dyad. Fam Cancer. 2012;11:189–94.
Zantour B, Guilhaume B, Tissier F, Louvel A, Jeunemaitre X, Gimenez-Roqueplo AP, et al. A thyroid nodule revealing a paraganglioma in a patient with a new germline mutation in the succinate dehydrogenase B gene. Eur J Endocrinol. 2004;151:433–8.
Armstrong R, Greenhalgh KL, Rattenberry E, Judd B, Shukla R, Losty PD, et al. Succinate dehydrogenase subunit B (SDHB) gene deletion associated with a composite paraganglioma/neuroblastoma. J Med Genet. 2009;46:215–6.
Housley SL, Lindsay RS, Young B, McConachie M, Mechan D, Baty D, et al. Renal carcinoma with giant mitochondria associated with germ-line mutation and somatic loss of the succinate dehydrogenase B gene. Histopathology. 2010;56:405–8.
Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85.
Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–4.
Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5:857–66.
Kaelin Jr WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402.
Adachi H, Fujiwara Y, Ishii N. Effects of oxygen on protein carbonyl and aging in Caenorhabditis elegans mutants with long (age-1) and short (mev-1) life spans. J Gerontol A Biol Sci Med Sci. 1998;53:B240–4.
Ishii T, Yasuda K, Akatsuka A, Hino O, Hartman PS, Ishii N. A mutation in the SDHC gene of complex II increases oxidative stress, resulting in apoptosis and tumorigenesis. Cancer Res. 2005;65:203–9.
Slane BG, Aykin-Burns N, Smith BJ, Kalen AL, Goswami PC, Domann FE, et al. Mutation of succinate dehydrogenase subunit C results in increased O2.-, oxidative stress, and genomic instability. Cancer Res. 2006;66:7615–20.
Smith EH, Janknecht R, Maher 3rd LJ. Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum Mol Genet. 2007;16:3136–48.
Goffrini P, Ercolino T, Panizza E, Giachè V, Cavone L, Chiarugi A, et al. Functional study in a yeast model of a novel succinate dehydrogenase subunit B gene germline missense mutation (C191Y) diagnosed in a patient affected by a glomus tumor. Hum Mol Genet. 2009;18:1860–8.
Szeto SS, Reinke SN, Sykes BD, Lemire BD. Ubiquinone-binding site mutations in the Saccharomyces cerevisiae succinate dehydrogenase generate superoxide and lead to the accumulation of succinate. J Biol Chem. 2007;282:27518–26.
Leibovitz A, Stinson JC, McCombs 3rd WB, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–9.
Lewis SP, Willis AN, Johnson AE, Resau J, Burnatowska-Hledin MA. Mutational analysis of VACM-1/cul5 exons in cancer cell lines. APMIS. 2011;119:421–30.
Xu XM, Wang XB, Chen MM, Liu T, Li YX, Jia WH, et al. Microrna-19a and -19b regulate cervical carcinoma cell proliferation and invasion by targeting CUL5. Cancer Lett. 2012;322:148–58.
Bender FC, Reymond MA, Bron C, Quest AF. Caveolin-1 levels are down-regulated in human colon tumors, and ectopic expression of caveolin-1 in colon carcinoma cell lines reduces cell tumorigenicity. Cancer Res. 2000;60:5870–8.
Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39.
Weng LP, Brown JL, Baker KM, Ostrowski MC, Eng C. PTEN blocks insulin-mediated ETS-2 phosphorylation through MAP kinase, independently of the phosphoinositide 3-kinase pathway. Hum Mol Genet. 2002;11:1687–96.
Ni Y, Zbuk KM, Sadler T, Patocs A, Lobo G, Edelman E, et al. Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am J Hum Genet. 2008;83:261–8.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (no. 81172300, 81000883). We are grateful for the support of Medjaden Bioscience Limited.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 1185 kb)
Rights and permissions
About this article
Cite this article
Zhang, D., Wang, W., Xiang, B. et al. Reduced succinate dehydrogenase B expression is associated with growth and de-differentiation of colorectal cancer cells. Tumor Biol. 34, 2337–2347 (2013). https://doi.org/10.1007/s13277-013-0781-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-0781-4