Abstract
There is a lack of understanding of the casual mechanisms behind the observation that some breast adenocarcinomas have identical morphology and comparatively different cellular growth behavior. This is exemplified by a differential response to radiation, chemotherapy, and other biological intervention therapies. Elevated concentrations of the free radical nitric oxide (NO), coupled with the up-regulated enzyme nitric oxide synthase (NOS) which produces NO, are activities which impact tumor growth. Previously, we adapted four human breast cancer cell lines: BT-20, Hs578T, T-47D, and MCF-7 to elevated concentrations of nitric oxide (or high NO [HNO]). This was accomplished by exposing the cell lines to increasing levels of an NO donor over time. Significantly, the HNO cell lines grew faster than did each respective (“PARENT”) cell line even in the absence of NO donor-supplemented media. This was evident despite each “parent” being morphologically equivalent to the HNO adapted cell line. Herein, we characterize the HNO cells and their biological attributes against those of the parent cells. Pairs of HNO/parent cell lines were then analyzed using a number of key cellular activity criteria including: cell cycle distribution, DNA ploidy, response to DNA damage, UV radiation response, X-ray radiation response, and the expression of significant cellular enzymes. Other key enzyme activities studied were NOS, p53, and glutathione S-transferase-pi (GST-pi) expression. HNO cells were typified by a far more aggressive pattern of growth and resistance to various treatments than the corresponding parent cells. This was evidenced by a higher S-phase percentage, variable radioresistance, and up-regulated GST-pi and p53. Taken collectively, this data provides evidence that cancer cells subjected to HNO concentrations become resistant to free radicals such as NO via up-regulated cellular defense mechanisms, including p53 and GST-pi. The adaptation to NO may explain how tumor cells acquire a more aggressive tumor phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The treatment regimes most often offered to patients for breast cancer include surgery, radiation therapy, chemotherapy, and biologicals (monoclonal antibodies). Breast cancer ranks second among leading causes of cancer deaths for women in the US, with about 230,000 new cases and over 39,500 deaths estimated for 2011 [1]. Worldwide, cancer arising in the breast is the predominant form of the neoplastic disease among women, with 1.38 million cases diagnosed and 458,400 deaths during 2008 [2]. Regardless of how patients present (e.g., with varying degrees of tumor size, stage, histological grade, and hormone receptor status), there is a broad array of disease outcomes from these morphologically identical tumors. It has been proposed that these varying outcomes are the result of variations in the tumor microenvironment [3–5]. The tumor microenvironment may further cause genetic alterations that affect tumor evolution and growth patterns, which ultimately impacts responses to therapeutic regimes and altering outcomes [3].
The free radical nitric oxide (NO) is thought to be an important element in this tumor microenvironment. It is well known that there is a varying concentration of NO in tumors [6]. At lower concentrations, it is beneficial being anti-inflammatory and having a tumor suppressive function. At high levels, however, it can become mutagenic and lead to tumor formation [6, 7]. NO, when produced by nitric oxide synthesis (NOS) in low concentrations, can be a highly effective hypoxic radiosensitizer; it has been shown to inhibit damage to DNA caused by radiation [8]. Significantly increased levels of NO and elevated expression of NOS, however, is associated with tumor formation. Whether antagonistic or beneficial, the role NO plays in any particular microenvironment is a dichotomy and dependent upon the concentration level present.
Three enzymatic isoforms of NOS are known to exist: inducible (iNOS), endothelial (eNOS), and neural (nNOS) [9]. In prior work, our laboratory explored expression levels of NOS and nitrotyrosine in various carcinomas [9–12]. Work by other reserachers has demonstrated that NO is overexpressed in multiple cancer types [13–19]. In vitro studies have alluded to there being NO concentration levels, which when reached, result in phenotypic changes in tumors [20].
Concurrent in vivo work also supports the premise that NO is involved in tumor formation. NO is produced by various iso-enzymes of NOS, and as NOS expression increases, so do other NO related metabolic constitutions such as nitrotryosine. These NO byproducts appear in a variety of human tumors including breast cancer [3, 9, 10, 16, 18, 19, 21], and in tumor-infiltrating macrophages, endothelial cells, and/or tumor cells [22, 23]. By example, in a large (n = 161) study of breast tumors, 61 % were found to express iNOS, and within that group, iNOS up-regulation was a marker for tumor progression. A subgroup within the iNOS-expressing tumors demonstrated a positive correlation between lymph node status and tumor size. Importantly, for those breast cancer patients found testing positive, versus negative, for iNOS, there was a statistically significant difference in 5-year survival outcomes: 67.1 % for those positive and 84.8 % testing negative [24].
The role of NO in tumor growth and development is not completely understood. To further clarify NO behavior patterns we created a model cell line system within our laboratory using a variety of breast cancer cell lines that were adapted to NO. These cells were gradually adapted to high concentrations of NO over a prolonged period of months [7]. Two significant results were observed: (1) the original “parent” cell lines adapted to the high nitric oxide (HNO), and (2) the resultant HNO adapted cell lines grew faster and more aggressively than did each respective “parent” cell line. This was found to be the case even when the HNO cells were later grown in media not augmented with a free radical donor (NO). The current study reported herein compares and characterizes four human breast adenocarcinoma cell lines by examining their biological behavior profiles and underlying physiological states. It creates a venue to analyze cellular response to variations in concentration of free radicals such as NO, with an expectation of a more complete comprehension of the relationship between cellular mechanisms, tumor growth and development, and therapeutic outcome.
Experimental procedures
Cell lines and cell adaptation
Four human breast adenocarcinoma cell lines were used for this study: Hs578T, BT-20, MCF-7, and T-47D. The Hs578T cell line was obtained from Abbott Laboratories (Abbott Park, IL, USA). All other cell lines were acquired from American Type Culture Collection (Manassas, Virginia, USA). T-47D was grown in RPMI-1640 media. MCF-7 and BT-20 were each grown in MEM media. Hs578T was grown in DMEM media (Invitrogen, Carlsbad, CA).
These four cell lines have previously been reported on by our group as to how they were adapted to grow in comparatively high levels of NO [7]. To summarize, the cell adaptation process was conducted over a period of time by exposing parent cell lines to an NO donor, (Z)-1-[2-(2-aminoethyl)-N-(2-amminoethyl) amino] diazen-1-ium-1,2-diolate (DETA-NONOate) (Sigma Life Sciences, St. Louis, MO, USA), at an initial 50 μM concentration. This was increased in increments of 25 μM up to a point which was lethal to each parent cell line (a concentration of 600 μM). The newly adapted HNO cell lines were designated as BT-20-HNO, Hs578T-HNO, MCF-7-HNO, and T-47D-HNO, respectively. Each media type was augmented with 10 % fetal calf serum (which was inactivated at 56 °C for 30 min), 100 μg/ml streptomycin, 2 mM l-glutamine, a 2.5 μg/ml Amphotericin B solution, and 100 U/ml penicillin. In addition, the MEM media was supplemented with 1 mM sodium pyruvate and 100 mM MEM nonessential amino acids (CellGro, Inc., Manassas, VA, USA). All cell lines were maintained in a humidified incubator at 37 °C and a 5 % CO2 concentration.
Fluorescence-activated cell sorting (FACS) analysis
Parent and HNO cell line samples were collected and preserved in ice-cold 70 % ethanol in preparation for FACS analysis. The suspension of cells was washed with phosphate buffered saline (PBS) two times and then suspended in 50 μl of a 100 μg/ml ribonuclease solution (Sigma-Aldrich Corp., St. Louis, MO, USA) in PBS for 5 min, followed by 200 μl of a 50 μg/ml solution of propidium iodide in PBS. This solution was added to each cell line. A cell cycle profile was created for each cell line using an EPICS Elite ESP flow cytometer/cell sorter (Beckman Coulter Inc., Fullerton, CA, USA), and excitation was accomplished through use of a 15 mW air-cooled 488 nm argon ion laser. The propidium iodide emission (λ max = 620 nm) was then captured with the use of a 610-nm long-pass filter. Two independent trials were run for each cell line, and 20,000–50,000 cells were analyzed for each experiment for each cell line. The mean was recorded for each trial; results from a representative trial are reported in the Results section.
Immunoblotting of cell line samples
Western blots were run to determine levels of expression for iNOS, eNOS, p53, and GST-pi in parent and respective HNO cell lines for BT-20, Hs578T, MCF-7, and T-47D. Cellular disruption was accomplished with a 1× sodium dodecyl sulfate (SDS) buffer solution (62.5 mM Tris–HCl, pH 6.8, 2 % SDS, 10 % glycerol, 50 mM DTT and 0.01 % bromophenol blue). SDS-polyacrylamide gel electrophoresis was used for separation of proteins, which were then transferred onto a nitrocellulose membrane. Nonspecific binding was prevented by blocking with 5 % nonfat dry milk in tris-buffered saline with tween (TBST; 10 mM Tris–Cl, pH 7.5, 0.15 M NaCl, and 0.05 % Tween 20) kept overnight at 4 °C, and the membranes were then incubated with anti-p53 antibody (Thermo Fisher Scientific Inc., Waltham, MA, USA; Catalog #P53 ab-8), anti-iNOS antibody (Abcam Inc., Cambridge, MA, USA; Catalog #ab3523), anti-eNOS antibody (Cell Signaling Technology, Inc., Danvers, MA; Catalog# 9572), or anti-GSTpi antibody (Thermo Fisher Scientific Inc.; Catalog #RB-050-A1) in TBST containing 5 % nonfat dry milk for 2 h at 22 °C. The membranes were then washed with TBST and incubated, alternately with horseradish peroxidase-conjugated goat anti-mouse secondary antibody, or horseradish peroxidase-conjugated goat anti-rabbit secondary antibody for 1 h. After washing the membranes, protein bands were visualized using enhanced chemiluminescent substrate (Thermo Fisher Scientific Inc.; SuperSignal West Pico Chemiluminescent Substrate). The blot was exposed to X-ray film for various time periods, and subsequently the film was developed. An earlier method [25] was used to record the protein band densities in Adobe Photoshop V7.0 (Adobe Systems Inc., San Jose, CA, USA). β-Actin was then used as a comparative control, and the expression level was normalized against it. Each experiment was repeated in triplicate, and data are presented as the average expression level ± standard error (SE), with error bars representing variations of less than 5 %. The densities of individual bands were added together in cell lines where multiple bands were observed for an antibody. It is also of note that both the mutant and wild-type forms of p53 interact with the anti-p53 antibody used in this study.
X-ray radiation exposure growth assays
An intensity-modulated radiation therapy (IMRT) treatment plan was designed for both HNO and parent cell lines consisting of differing doses of radiation ranging from 0 to 28 Gy. Cells placed into 96-well microtiter plates (100 μl/well) in the appropriate media (e.g., standard media for parent cells; media supplemented with 600 μM DETA-NONOate for HNO cells), and upon reaching ~50 % confluency, the cells were irradiated. Three hours before exposure to the radiation, an additional 150 μl of media was added to each well, bringing the total volume per well to 250 μl.
A computed tomography scan (PQ5000; Philips Medical Systems Inc., Andover, MA, USA) was conducted on the microtiter plates placed into a specially designed phantom able to hold tissue culture plates [26]. An IMRT treatment plan, consisting of two ~235-cm3 cuboidal planning target volumes (PTVs), was created using an Eclipse treatment planning system (Varian Corp., Palo Alto, CA, USA). The PTVs were located at either end of the plate. Each remaining plate area was divided into four rows of parent cells and four rows of the respective HNO cells. The phantom was placed on the patient treatment table and irradiated at 1, 2, 5, 10, 14, or 28 Gy using 6 MV photons delivered by a clinical linear accelerator (2100CD; Varian Corp). A total of six, equally spaced beam angles were used (52°), ranging from 28° to 336°. Each plate then received two paired doses generated as consistently as permissible. The paired dosage strengths varied: 1 and 2 Gy, 5 and 10 Gy, or 14 and 28 Gy. A fixed ratio of 2:1 was maintained between the PTVs. The duration of the experiment was about 60 min, which included a complete trip to and from the IMRT facility. An additional control plate was created and brought to the IMRT facility, but was not exposed to radiation. After exposure to the radiation, the plates were placed in an incubator for another 96 h at 37 °C.
Cell viability was determined using the diphenylamine (DPA) assay, which colorimetrically measures the quantity of DNA within cells [27]. The reagents for the DPA assay were obtained from Sigma-Aldrich. After the 96-h incubation period the media was removed from all wells. A 1:5 mixture of chilled acetaldehyde (0.16 % in water)/perchloric acid (20 % vol/vol) was added to the cells (60 μl), followed by a 4 % DPA solution in glacial acetic acid (100 μl). After incubating for 24 h at 37 °C, the absorbance of each well was measured with a spectrophotometer (SpectraMax® Plus 384; Molecular Devices, Inc., Sunnyvale, CA, USA) and read at 595 nm. At least two independent trials were conducted for each of the cell lines, and a minimum of eight replicate microtiter wells were tested for each dose. Each trial was normalized against the mean absorbance readings of the untreated control cells. A representative trial is presented in the Results section; values are presented as the mean normalized absorbance ± SE of the mean (SEM).
Ultraviolet (UV) exposure growth assays
Both UV and X-ray radiation are well recognized to cause damage to DNA. Appropriate media types were placed into 96-well microtiter plates (i.e., standard media for parent cells; media supplemented with 600 μM DETA-NONOate for HNO cells) which were then seeded (50 μl) with the respective parent and HNO cells. The plates were incubated overnight in a CO2 incubator and allowed to reach ~70 % confluency. The cells were exposed to a UV germicidal light (254 nm, 13.4 W UV output; plate positioned ~51 cm from light source) for 0, 2, 4, 6, 8, or 10 min in a sterile biological hood. The lid of the microtiter plate was removed to provide complete irradiation of the cells. After the UV exposure cycle was completed, 100 μl of fresh media was added to each well, bringing the total volume in each well to 150 μl. The plates were incubated for an additional 72 h at 37 °C.
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay was used to measure cell proliferation/viability. The MTT assay was completed by removing the media from all wells and adding 100 μl of 2 mg/ml MTT (Sigma-Aldrich Corp.) in PBS to each well, which was then allowed to incubate for 5 h at 37 °C. The MTT was removed from all cells, leaving purple formazan crystals, which were then dissolved in 100 μl of DMSO. The absorbance of each well was read with a spectrophotometer at 540 nm (SpectraMax® Plus 384; Molecular Devices, Inc.). A minimum of three independent trials were carried out for each cell line; a minimum of eight replicate wells were established for each experimental condition, per cell line. Data was normalized against the mean absorbance readings of the untreated control cells. A representative trial is presented in the Results section; values are presented as the mean normalized absorbance ± SEM.
Single cell gel electrophoresis (CometAssay™)
DNA damage taking place in both the parent and HNO cell lines was measured with the use of a CometAssay™ kit (Trevigen, Inc., Gaithersburg, MD, USA). Cell lines were grown as before using standard media for parent cells, and media supplemented with 600 μM DETA-NONOate for HNO cells. All cells were harvested and then counted with the use of a hemocytometer, adjusted to a total concentration of 1×105 cells/ml. Cells were resuspended in PBS and then immediately mixed in 10-μl aliquots of the cell suspension and combined with 100 μl of 1 % low melting point agarose (Sigma-Aldrich) at 37°C. The solution was dispensed (75 μl) onto a CometSlide™ and then placed in cold lysing buffer (2.5 mM NaCl, 100 mM Na2EDTA, 10 mM Tris, 1 % N-lauryl sarcosine sodium salt, pH 10) and kept at 4°C for 1 h. It was rinsed carefully with tris-borate buffer (10.8 g Tris, 5.5 g boric acid, and 0.93 g Na2EDTA in 1 l dH2O) twice and placed on a horizontal gel electrophoresis platform covered in tris-borate buffer. Slides were exposed to 13 V for 10 min, then quickly immersed into 70 % ethanol, removed, and allowed to air-dry. Using 100 μl of a silver staining solution, the slides were then fixed, stained and cover-slipped. The resulting Comet tails were viewed using a Reichert Microstar IV microscope (Reichert, Inc., Depew, NY, USA) at 400X, and images were captured with the Dazzle Multimedia software package (Pinnacle Systems, Inc., Mountain View, CA, USA). The distance from the center of the cell nucleus to the tip of the tail was used to measure the tail lengths in pixel units. One independent trial was conducted, and a minimum of 19 Comet tails were measured for each slide. Values are reported as the mean Comet tail length ± SEM.
Gene Chip analysis
A comparative analysis was conducted on all four parent and HNO cell line pairs using a “QuantArray 3.0” (Perkin-Elmer, Boston, MA) system. All of the data was collected and analyzed using a R 2.9 Windows compatible platform, and the statistical analytic package “Limma 2.16” [28] was used to assess gene expression levels. Background correction was conducted consistent with other efforts [29] in which the “Normexp” background correction methodology was used, with an “Offset” threshold value of 50. Background correction was performed with weights of control spots positioned at 0. Normalization was realized by using “loess” and “scale” methods of “Normalize Within Arrays” and “Normalize Between Arrays” for within array and between array methodologies, respectively. Details of the procedures used herein may be found in [30]. “QuantArray” was used to control quality levels, and only those spots found acceptable by the software were included for signal transformations. Finally, normalized signals were used to determine the importance of differentiated expression levels with the use of linear modeling systems through “lmFit” and Bayes statistics achieved with the use of “eBayes,” “topTable,” as previously reported [31].
Results
A number of comparative experiments were conducted to characterize and determine the biological properties of the original parent cell lines and each respective adapted HNO cell line.
Cell cycle distribution profiles for both parent and HNO cell lines were determined using FACS analysis of propidium iodide stained cells (Table 1). Results for each parent/HNO cell line pair show the parent cell line exhibited a higher percentage of cells in the G1 phase, with the HNO cell line having a greater percentage in the S phase. For the four cell line pairs analyzed, no aneuploidy or aneuploidy modifications were found between the parent/HNO cell lines.
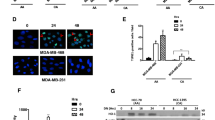
Western blot analysis was used to determine the expression level of p53 for each of the parent/HNO cell line pairs (Fig. 1). The results demonstrated significant up-regulation for the adapted HNO cell lines versus parent cell lines in the BT-20-HNO and Hs578T-HNO (approximately 3-fold and 5-fold increases, respectively). High expression levels were observed in both the MCF-7-HNO cell line and the parent MCF-7 cells; T-47D-HNO and T-47D parent cells exhibited significantly lower levels of expression.
Comparative cell line survivability against exposure to X-ray and UV radiation of parent versus HNO cell lines was done using growth curve assays. Results from treatment with X-ray radiation (Fig. 2) indicate that Hs578T-HNO cells exhibited greater viability than did the respective parent cells at the higher doses tested. Little to no difference was observed between parent and HNO cell line pairs for the other three cell lines pairs (MCF-7, BT-20, and T-47D). Results of the UV radiation studies (Fig. 3) indicate that all four cell line pairs showed responses to UV radiation exposure, with responses being particularly sensitive to exposure duration; typically, the more lengthy the exposure period, the greater the toxicity level.
The degree of damage to DNA from exposure to HNO levels in the four parent/HNO cell line pairs was determined through use of single cell gel electrophoresis, or CometAssay™, and compared to the respective parent cell lines. Results appear in Fig. 4 and demonstrate that Hs578T-HNO had significantly shorter Comet tails than the parent cell line. The other cell lines showed little difference between the cell line pairs.
Expression of the two isoenzymes iNOS and eNOS were determined through Western blot analysis. Results appear in Figs. 5 and 6. The results indicate that iNOS is highly elevated in both the BT-20-HNO and Hs578T-HNO cell lines as compared to each respective parent cell line, but decreased in MCF-7-HNO and T-47D-HNO as compared to their respective parent cell lines (Fig. 5). Both the BT-20-HNO and MCF-7-HNO cell lines were found to have increased expression of eNOS, while Hs578T-HNO and T-47D-HNO had decreased expression when compared to their corresponding parent cell lines (Fig. 6).
The free radical protective mechanism provided by glutathione-S-transferase-pi (GST-pi) expression was determined via Western blot analysis. The results appear in Fig. 7 and demonstrated GST-pi was elevated in both Hs578T-HNO and MCF-7-HNO, relative to their parent cell lines. BT-20 had significant expression in both the parent and the HNO cell lines, while T-47D had little expression in both the parent and HNO cell lines.
Gene chip analysis on the four breast cancer cell line pairs showed a wide range of gene types being up-regulated and down-regulated. Table 2 lists the top ten and bottom ten genes that were most up-regulated or down-regulated for each of the four breast cancer call line pairs. In brief, for the BT-20 parent/BT-20-HNO cell line pair, the ALPI, PLSCR2, GAPDHS, KCNQ4, FCN1, AC215219.3-2, STXBP4, PRR19, RP11-196D18.5/RP11-196D18.4/AC012005.5/AC006328.1/SEPT14, and CHCHD7 genes were up-regulated, and KCNK18, HMGB3, EIF5A2, ADNP, RAET1E, KIR2DL2, HTR1F, TRIB2, LENG3, and UBE2V2 were down-regulated. For the Hs578T parent/Hs578T-HNO cell line pair, the SPTBN1, IL1F7, HSPA2, ACADVL, FCRL1, SMARCA5, SNAPC1, SLC35A4, VPS33B, and WFDC5 were up-regulated, and INO80E, RP11-49 G10.8, COMMD9, RP11-64P14.7/OR1Q1, NRG2, PIGP, KLHL13, EIF4A3, TM7SF3, and FMO6P were down-regulated. For the MCF-7 parent/MCF-7-HNO and T-47D parent/T-47D-HNO cell line pairs, DTX4, MUC1, FOSB, COL5A2, SASH1, CAMK2N1, KLK8, ZFP36L1, EGFR, and MED13L, and STXBP4, DSCAML1, SNAPC1, ESM1, PCDH10, PLA2G3, CCL8, UHRF1BP1, CDC23, UBE2Z being up-regulated, and C14orf147, TTRAP, EIF4A2, C5orf5, SC4MOL, IDI1, RP3-527 F8.2, RP4-640 H8.2, C11orf55, and SFRS7, PKP1, RP11-420 G6.4, EMX2, THUMPD3, MYO18A, C1orf2, RP3-402 G11.5, FIZ1, and MGEA5 being down-regulated, respectively.
Discussion
NO is well recognized as a robust radiosensitizer of mammalian cells experiencing hypoxia. NO donors have been used in a variety of studies and demonstrated a synergistic enhancement for radiosensitivity, in both in vivo and in vitro settings [32–39]. It has also been shown that the cytotoxic impact of radiation is significantly increased through up-regulated NOS in tumor cells [34].
However, a dichotomy exists between enhanced radiosensitization and conditions where elevated levels of NO and up-regulated NOS are contributors to tumor progression. The relationship between NOS expression and tumor growth is evident in the literature from studies with a variety of cancer types including: (1) human oral squamous cell carcinoma [10, 40], (2) gastric carcinoma [41], (3) lymph node metastases, and most significantly for this current work, (4) breast cancer, to a point that it is a universal property for human solid tumors [7, 15–17, 42–45].
As indicated in our prior works, a number of tumor cell lines have been adapted to HNO concentration levels including those from lung [11, 46–48], head and neck [3, 49–52], and breast [7], suggesting that NO adaptation is also a universal property. In an attempt to further elucidate the impact of extended exposure from NO on underlying biological systems, we herein characterize the four newly created breast adenocarcinoma HNO cell lines [7], along with each respective parent cell line.
In the harsh NO environment, all four rapidly proliferating HNO cell lines exhibited a greater percentage of cells in the S-phase of the cell cycle than their corresponding parent cell lines (Table 1). This result, together with the previously reported faster growth rates of the HNO cells relative to the parent cells [7], suggests the HNO cells are programmed for a more proliferative state than their corresponding parent cells. This finding correlates with the previously reported MTT growth curves which showed that the HNO adapted cells grow faster than the parent cell lines [7].
While cell cycle distribution was relatively consistent across the four HNO cell lines, p53 expression (Fig. 1) was not: there was a significant increase in BT-20-HNO and Hs578T-HNO compared to their respective parent cell lines. While p53 was detected in MCF-7 parent/MCF-7-HNO and T-47D parent/T-47D-HNO cells, slightly higher expression levels were observed in the parent MCF-7 and T-47D cells suggesting that a p53-independent adaptation process was utilized in MCF-7 and T-47D. In contrast, the lack of p53 expression observed in the BT-20 and Hs578T parent cells, combined with the elevated levels of p53 detected in the BT-20-HNO and Hs578T-HNO cells, suggests that the p53 pathway may be involved in the HNO adaptation of BT-20 and Hs578T.
Both X-ray and UV radiation are energy sources known to induce DNA damage. It is well known that X-ray radiation reacts with water in the body to produce free radicals that can in turn cause DNA damage, while UV photons can react directly with DNA, generating potentially toxic photoproducts (Thymidine — Thymidine dimers) that interfere with DNA replication. In all four of the breast tumor cell line pairs studied herein, the adapted HNO lines were more resistant to UV irradiation than its corresponding parent cell line at longer exposure times (Fig. 3). While Hs578T-HNO was comparably radioresistant relative to its corresponding parent cell line (Fig. 2), the other HNO adapted cell lines tested all had little or no resistance to radiation when compared to the respective parent cell lines.
The Comet assays (Fig. 4) indicate that for Hs578T a greater amount of DNA damage was present in the parent cell lines relative to the HNO cell line. The other three cell lines showed no appreciable differences between the parent and the HNO adapted cell lines. The X-ray, UV, and Comet assay results of the Hs578T-HNO cell line, along with the loss of p53 (a known activator of DNA repair enzymes) expression observed between the Hs578T parent and HNO cell lines, indicate an up-regulation of p53, and possibly other DNA repair enzymes as well, occurred during the adaptation of the Hs578T cell line. Similar up-regulation of p53 was observed in the BT-20-HNO cells, suggesting this enzyme was also involved in the adaptation of the BT-20 cells. In contrast, while both MCF-7-HNO and T-47D-HNO cells had p53 expression, it was not up-regulated when compared to their parent cell lines. This finding suggests that perhaps other DNA repair enzymes not studied herein were responsible for the cell survival during the adaptation process.
In addition to the DNA repair enzymes, cancer cells are also known to utilize GST-pi as a universal free radical protective mechanism. We have previously shown that together with the expression of eNOS, the expression of GST-pi also increases as carcinogenesis progresses in head and neck squamous cell carcinomas [3, 53]. The expression/amplification of GST-pi has been linked to chemotherapy resistance [54] and to decreased survival after neoadjuvant treatment [55], which at first may seem counterintuitive until one factors in that many chemotherapeutic agents work through free radicals. The GST-pi isoform in particular has been found to prevent cellular damage by detoxifying and eliminating electrophilic and redox molecular species generated during oxidative stress [56]. In this study we found that GST-pi was expressed in three of the breast cancer cell lines studied, and that the HNO derived cells for these three cell lines expressed more GST-pi than their respective parent cell line (Fig. 5). We also found that the expression of iNOS, rather than eNOS, was high in these tumors. Both of these results are consistent with previously reported studies of GST-pi [57] and NOS [21] expression in tongue tumor carcinomas. These data support the idea that tumor cells, regardless of the site of origin, are capable of producing NO by using either of the NOS isoenzymes. Regardless of the isoform, GST-pi and various DNA repair mechanisms are up-regulated in response to the increase free radicals encountered in the microenvironment. It is yet to be determined which of the DNA repair systems predict which patients will have a particular outcome. This line of investigation would merit further study.
The gene chip analysis of the parent and HNO adapted cells show a wide array of gene products being up- or down-regulated, in response to being adapted to NO. Surprisingly, there was no clear pattern across the cell lines based solely on the genes that had expression level changes. This lack of uniformity suggests that there are multiple ways to modulate the cellular machinery to compensate for the adaptive pressures of increased NO.
In this study, we set out to provide an initial survey of the cellular and molecular makeup of four breast cancer cell lines that had been adapted to HNO. In the two accompanying papers [58, 59]we further assess the biology of HNO adapted cells as they relate to both clinical specimens with respect to mitochondrial mutations and how these findings related to the BT-20 parent/BT-20-HNO cell line model reported herein. Clearly, there is still a comparatively large amount of work that needs to be done to fully understand the mechanisms of how cells adapt to increasing NO. Unlocking these mechanisms will lead to new treatment approaches for not only breast cancers, but also many other solid tumors.
References
American Cancer Society Inc., Breast Cancer Facts & Figures 2011–2012. Copyright 2011 [Accessed 27 March 2012]; Available from: http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-030975.pdf.
Jemal A, et al. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61(2):69–90.
Bentz BG, et al. Nitric oxide synthase type 3 is increased in squamous hyperplasia, dysplasia, and squamous cell carcinoma of the head and neck. Ann Otol Rhinol Laryngol. 1999;108(8):781–7.
Nathan C, Xie QW. Nitric oxide synthases: roles, tolls, and controls. Cell. 1994;78(6):915–8.
Singh S, et al. The impact of intracellular generation of nitric oxide on the radiation response of human tumor cells. Radiat Res. 2009;171(5):572–80.
Paradise WA, Vesper BJ, Goel A, Waltonen JD, Altman KW, Haines III GK, Radosevich JA. Nitric oxide: perspectives and emerging studies of a well known cytotoxin. Int J Mol Sci. 2010;11:2715–45.
Vesper BJ, et al. Long-term adaptation of breast tumor cell lines to high concentrations of nitric oxide. Tumor Biol. 2010;31(4):267–75.
Bentz BG, et al. Nitrosative stress induces DNA strand breaks but not caspase mediated apoptosis in a lung cancer cell line. J Carcinog. 2004;3(1):16.
Bentz BG, et al. Cytoplasmic localization of endothelial constitutive nitric oxide synthase in endometrial carcinomas. Tumor Biol. 1997;18(5):290–300.
Bentz BG, et al. Endothelial constitutive nitric oxide synthase (ecNOS) localization in normal and neoplastic salivary tissue. Head Neck. 1998;20(4):304–9.
Bentz BG, et al. The kinetics and redox state of nitric oxide determine the biological consequences in lung adenocarcinoma. Tumor Biol. 2007;28(6):301–11.
Bentz BG, et al. The yin and yang of nitric oxide: reflections on the physiology and pathophysiology of NO. Head Neck. 2000;22(1):71–83.
Keydar I, et al. Establishment and characterization of a cell line of human breast carcinoma origin. Eur J Cancer. 1979;15(5):659–70.
Beevi SS, Rasheed MH, Geetha A. Evidence of oxidative and nitrosative stress in patients with cervical squamous cell carcinoma. Clin Chim Acta. 2007;375(1–2):119–23.
Loibl S, et al. Expression of endothelial and inducible nitric oxide synthase in benign and malignant lesions of the breast and measurement of nitric oxide using electron paramagnetic resonance spectroscopy. Cancer. 2002;95(6):1191–8.
Duenas-Gonzalez A, et al. Expression of inducible nitric oxide synthase in breast cancer correlates with metastatic disease. Mod Pathol. 1997;10(7):645–9.
Tschugguel W, et al. Expression of inducible nitric oxide synthase in human breast cancer depends on tumor grade. Breast Cancer Res Treat. 1999;56(2):145–51.
Cobbs CS, et al. Expression of nitric oxide synthase in human central nervous system tumors. Cancer Res. 1994;55(4):727–30.
Chandra R, et al. Expression of nitric oxide synthase type 3 in reflux-induced esophageal lesions. Otolaryngol. Head Neck Surg. 2001;124(4):442–7.
Ambs S, et al. Frequent nitric oxide synthase-2 expression in human colon adenomas: implication for tumor angiogenesis and colon cancer progression. Cancer Res. 1998;58(2):334–41.
Chen YK, Hsue SS, Lin LM. Increased expression of inducible nitric oxide synthase for human buccal squamous-cell carcinomas: immunohistochemical, reverse transcription-polymerase chain reaction (RT-PCR) and in situ RT-PCR studies. Head Neck. 2002;24(10):925–32.
Hibbs JB, et al. Nitric oxide: a cytotoxic activated macrophage effector molecule. Biochem Biophys Res Commun. 1988;157(1):87–94.
Hibbs JB, Vavrin A, Taintor RR. l-Arginine is required for expression of the activated macrophage effector mechanism causing selective metabolic inhibition in target cells. J Immunol. 1987;138(2):550–65.
Loibl S, et al. The role of early expression of inducible nitric oxide synthase in human breast cancer. Eur J Cancer. 2005;41(2):265–71.
Miller, L.P. Quantifying western blots without expensive commercial quantification software. Copyright 2007. [Accessed 20 May 2010]; Available from: http://www.lukemiller.org/journal/2007/08/quantifying-western-blots-.
Altman MB, et al. Characterization of a novel phantom for three-dimensional in vitro cell experiments. Phys Med Biol. 2009;54(5):N75–82.
Natarajan N, et al. Adaptation of the diphenylamine (DPA) assay to a 96-well plate tissue culture format and comparison with the MTT assay. BioTechniques. 1994;17(1):166–71.
Smyth, G.K., et al., Limma: Linear Models for Microarray Data. Bioinformatics and computationalbBiology solutions using R and bioconductor, 2005: 397–420.
Ritchie ME, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23(20):2700–7.
Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–73.
Smyth, G.K., Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol, 2004. 3: Article3.
Blackett NM, et al. Radiation modifying effect of the free radical norpseudopelletierene-N-oxyl on normal bone marrow stem cells in vitro and in vivo. Radiat Res. 1974;58(3):361–72.
Bristow, R.G. and R.P. Hill, Molecular and cellular basis of radiotherapy. In I.F. Tannock and R.P. Hill, editors. The basic science of oncology. 1998, McGraw-Hill: New York. p. 95–321.
Chen HHW, et al. Increased expression of nitric oxide synthase and cyclooxygenase-2 is associated with poor survival in cervical cancer treated with radiotherapy. Int J Radiation Oncology Biol Phys. 2005;63(4):1093–100.
Cooke BC, Fielden EM, Johnson M. Polyfunctional radiosensitizers: I. Effects of a nitroxyl biradical on the survival of mammalian cells in vitro. Radiat Res. 1976;65(1):152–62.
De Ridder M, et al. Hypoxic tumor cell radiosensitization through nitric oxide. Nitric Oxide. 2008;19(2):164–9.
George R, et al. Audio-visual biofeedback for respiratory-gated radiotherapy: impact of audio instruction and audio-visual biofeedback on respiratory-gated radiotherapy. Int J Radiat Oncol Biol Phys. 2006;65(3):924–33.
Griffin RJ, et al. Radiosensitization of hypoxic tumor cells in vitro by nitric oxide. Int J Radiat Oncol Biol Phys. 1996;36(2):377–83.
Hogle WP. Radiation therapy in the treatment of breast cancer. Semin Oncol Nurs. 2007;23(1):20–8.
American Cancer Society, I. Oral Cancer. Copyright 2008 [Accessed 17 June 2012]; Available from: http://www.cancer.org/downloads/PRO/OralCancer.pdf.
Wang YZ, et al. Expression of nitric oxide synthase in human gastric carcinoma and its relation to p53, PCNA. World J Gastroenterol. 2005;11(1):46–50.
Glynn, S., et al., iNOS expression is associated with basal-like breast cancer phenotype and predicts poor survival in ERneg breast cancer. Nitric Oxide, 2007. 17(Supp. 1): S17.
Nakamura Y, et al. Nitric oxide in breast cancer: induction of vascular endothelial growth factor-C and correlation with metastasis and poor prognosis. Clin Cancer Res. 2006;12(4):1201–7.
Thomsen LL, et al. Nitric oxide synthase in human breast cancer. Br J Cancer. 1995;72(1):41–4.
Bulut AS, et al. Significance of inducible nitric oxide synthase expression in benign and malignant breast epithelium: an immunohistochemical study of 151 cases. Virchows Arch. 2005;447(1):24–30.
Masri FA, et al. Abnormalities in nitric oxide and its derivatives in lung cancer. Am J Respir Crit Care Med. 2005;172(5):597–605.
Lee TW, et al. Differential expression of inducible nitric oxide synthase and peroxisome proliferator-activated receptor gamma in non-small cell lung carcinoma. Eur J Cancer. 2003;39(9):1296–301.
Liu CY, et al. Increased level of exhaled nitric oxide and up-regulation of inducible nitric oxide synthase in patients with primary lung cancer. Br J Cancer. 1998;78(4):534–41.
Rosbe KW, et al. Immunohistochemical characterization of nitric oxide synthase activity in squamous cell carcinoma of the head and neck. otolaryngol. Head Neck Surg. 1995;113(5):541–9.
Yarmolyuk Y, et al. A model system for dtudying nitric oxide in tumors: high nitric oxide-adapted head and neck squamous cell lines. Tumor Biol. 2011;32(1):77–85.
Bentz BG, et al. Nitric oxide and apoptosis during human head and neck squamous cell carcinoma development. Am J Otolaryngol. 2002;23(1):4–11.
Gavilanes J, et al. Nitric oxide synthase activity in human squamous cell carcinoma of the head and neck. Laryngoscope. 1999;109(1):148–52.
Bentz BG, Haines III GK, Radosevich JA. Glutathione S-transferase pi in squamous cell carcinoma of the head and neck. Laryngoscope. 2000;110(10 Pt 1):1642–7.
Cullen KJ, et al. Glutathione S-transferase pi amplification is associated with cisplatin resistance in head and neck squamous cell carcinoma cell lines and primary tumors. Cancer Res. 2003;63(23):8097–102.
Shiga H, et al. Prognostic value of p53, glutathione S-transferase pi, and thymidylate synthase for neoadjuvant cisplatin-based chemotherapy in head and neck cancer. Clin Cancer Res. 1999;5(12):4097–104.
Dourado DF, Fernandes A, Ramos MJ. Mammalian cytosolic glutathione transferases. Curr Protein Pept Sci. 2008;9(4):325–37.
Mulder TP, et al. Glutatione S-transferases and glutathione in human head and neck cancer. Carcinogenesis. 1995;16(3):619–24.
De Vitto, et al. Part II. Mitochondrial mutational status of high nitric oxide adapted cell line BT-20 (BT-20-HNO) as it relates to human primary breast tumors. Tumor Biol. 2012. doi:10.1007/s13277-012-0555-4.
De Vitto, et al. Part III. Molecular changes induced by high nitric oxide adaptation in human breast cancer cell line BT-20 (BT-20-HNO): a switch from aerobic to anaerobic. Tumor Biol. 2012. doi:10.1007/s13277-012-0564-3.
Acknowledgments
The authors wish to thank the UIC Flow Cytometry facility (Drs. Karen Hagen and Jewel Graves) for their assistance with the FACS study, and the UIC Core Genomics Facility (Dr. Zarema Arbieva and Mr. Oleksiy Karpenko for their assistance with the gene chip study. This work was supported by a Veterans Affairs merit review grant (J.A.R.).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vesper, B.J., Onul, A., Haines, G.K. et al. Part I. Molecular and cellular characterization of high nitric oxide-adapted human breast adenocarcinoma cell lines. Tumor Biol. 34, 203–214 (2013). https://doi.org/10.1007/s13277-012-0530-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0530-0