Abstract
Epithelial to Mesenchymal Transition (EMT) in cancer is a process that allows cancer cells to detach from neighboring cells, become mobile and metastasize and shares many signaling pathways with development. Several molecular mechanisms which regulate oncogenic properties in neoplastic cells such as proliferation, resistance to apoptosis and angiogenesis through transcription factors or other mediators are also regulators of EMT. These pathways and downstream transcription factors are, in their turn, regulated by ubiquitination and the Ubiquitin–Proteasome System (UPS). Ubiquitination, the covalent link of the small 76-amino acid protein ubiquitin to target proteins, serves as a signal for protein degradation by the proteasome or for other outcomes such as endocytosis, degradation by the lysosome or directing these proteins to specific cellular compartments. This review discusses aspects of the regulation of EMT by ubiquitination and the UPS and underlines its complexity focusing on transcription and transcription factors regulating EMT and are being regulated by ubiquitination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A process termed Epithelial to Mesenchymal Transition (EMT) permits detachment of an epithelial cell belonging to an epithelial membrane from its neighboring cells in order to transverse the dissolving basement membrane and become motile. Cell motility during EMT is facilitated by the loss of critical adhesion molecules and junctional complexes which provide adhesion between adjacent epithelial cells [1]. Other changes happen concomitantly such as acquisition of a fibroblast-like shape, downregulation of epithelial markers and upregulation of mesenchymal markers. The reverse process, Mesenchymal to Epithelial transition (MET), happens when the fibroblast-like cell regains epithelial characteristics and establishes adhesions with adjacent cells. EMT and MET are processes that physiologically take place during development or injury healing. Developmental EMT gives rise to the three embryonal layers of differentiation—ectoderm, mesoderm and endoderm—and further to different tissues and cell types in complex but ordered patterns and is an integral process of the differentiation to the various cell types which are regulated in time and space. An example of developmental EMT takes place during gastrulation when the epiblast layer produces a midline invagination called the primitive streak, from which cells are mobilized by undergoing an EMT and produce the mesoderm and endoderm [2]. Other examples can be found in implantation and somitogenesis. In many instances, two or more rounds of EMT/MET are necessary for the final differentiation of specialized cells in developing organs.
A significant part of the phenotypic differences between various cell types of a multicellular organism are due to transcriptional and post-transcriptional differences in the regulation of various cellular proteins as the genetic content (DNA sequences) of each cell is the same for a given individual. Post-translational modifications such as phosphorylation, hydroxylation, acetylation and ubiquitination can regulate conformation, function, localization and turnover of cell proteins impacting on cell morphology, activity and cellular communication in the multicellular organism. Ubiquitination is the covalent attachment of molecules of the small 76-amino acid protein ubiquitin to a target protein which is then marked for destruction by the proteasome or the lysosome, transportation to specific compartments, endocytosis or participation in a range of processes [3, 4]. Ubiquitination, along with other post-translational modifications of proteins, is a regulated process where multiple regulators cooperate for its execution. Many signal transducers and transcription factors involved in EMT are regulated by ubiquitination and the Ubiquitin–Proteasome System (UPS). In addition, ubiquitination and the UPS have a general role on the execution of transcription signaling through modification of histones [5] which also impacts EMT. In this review, the role of ubiquitination and the UPS in the regulation of transcription factors of EMT and their transcription function will be discussed. Transduction cascades involved in the regulation of these transcription factors will not be discussed in detail.
EMT in cancer
Three processes are using EMT to enable cell movement. In development, normal embryonal cells use EMT to position themselves appropriately and to obtain different specific morphologies and functions present in the multicellular organism. In adult tissue injury repair, EMT mobilizes adjacent epithelial cells to heal open wounds but may also lead to fibrosis. Finally, EMT enables tissue invasion and metastatization of malignant epithelial cells. These three EMT types have recently been termed types 1, 2 and 3, respectively [6]. During cancer-associated EMT or type 3 EMT, epithelial cancer cells acquire the ability to detach from their initial site, pass through the dismantled basement membrane into adjacent tissues and even metastasize to distant sites. In vitro and in vivo criteria for EMT confirmation have been established [7] and fall into two broad categories (Fig. 1). On one hand, they include upregulation of specific mesenchymal proteins and downregulation of epithelial-associated proteins. On the other hand, they describe general properties of the transformed cell. Mesenchymal proteins induced in EMT include S100A4 [also called FSP1 (Fibroblast-Specific Protein 1)], vimentin, type I collagen and its receptor kinase DDR2 (Discoidin Domain Receptor tyrosine kinase 2), cadherin N and OB, transcription factors SNAIL1 and 2, ZEB1 and 2 and TWIST, and nuclear localization of β-catenin. Downregulated epithelial proteins include E-cadherin, ZO-1 (Zona Occludens 1), cytokeratins, claudins, occludins and basement membrane components collagen IV and laminin 1 [7]. General cell properties induced by the transition include a change in morphology with the acquisition of spindle shape, loss of epithelial cell polarity and stress fiber redistribution, resistance to apoptosis induction and enhanced migratory capability. Resistance to apoptosis in neoplastic cells undergoing EMT is accompanied by the acquisition of a stem cell phenotype [8] also associated with drug resistance. For the in vivo experimental confirmation of EMT, the introduction of a cell reporter construct in epithelial cells that subsequently continues to be expressed in resulting mesenchymal cells has been proposed. Despite initial debate stemming largely from difficulty to define it in vivo [9], EMT is increasingly accepted as a hallmark capability of cancer cells that promotes invasion and metastasis [10]. Its acceptance has been helped by the realization that EMT in a cancer cell may not be complete, and only part of the EMT markers may be expressed in each instance [7]. An example is collective migration during which cells detach from the epithelial site, acquire mesenchymal properties but move en block without losing adhesions between them [11]. Neoplastic cells that have undergone EMT retain the capability to undergo Mesenchymal to Epithelial reverting Transition (MErT) when in the appropriate environment in the metastatic site, and this plasticity makes them elusive but may concomitantly constitute a significant advantage for their survival at metastatic foci. The acceptance of EMT as intrinsic to the malignant process has been further aided by the discovery that beyond specific EMT-inducing factors a multitude of general cancer-regulating pathways are also important EMT regulators.
Disruption of intercellular adhesions is at the center stage of the process of EMT. The main intercellular connecting molecule at adherens junction is E-cadherin, which turnover is regulated by ubiquitination. At adherens junctions, E-cadherin molecules span the cytoplasmic membrane and make homotypic contacts with E-cadherin molecules of neighboring cells [12, 13]. Their cytoplasmic domain binds together with α-catenin, p120 catenin and β-catenin with the actin cytoskeleton. E-cadherin molecules are continuously incorporated in adherens junctions and removed by clathrin-mediated endocytosis, being in a dynamic equilibrium [14]. Endocytosis of E-cadherin is effectuated after phosphorylation by c-src kinase followed by ubiquitination with the aid of c-cbl family E3 ligase Hakai [15]. p120 catenin prevents E-cadherin endocytosis by masking Hakai interaction sites in the juxtamembrane domain of E-cadherin. In contrast, during EMT of cancer, E-cadherin is downregulated through both transcriptional suppression of its gene (see below) and increased degradation after endocytosis. Growth factors and TGFβ signaling promote ubiquitinated E-cadherin degradation by directing early endosomes to late endosomes and lysosomes instead of recycling to the cell surface [16, 17]. TGFβ signaling, in addition to promoting EMT through the canonical SMAD pathway, directly phosphorylates Partitioning defective 6 (PAR6) resulting in recruitment of E3 ligase Smurf1 and ubiquitination followed by proteasome degradation of GTPase RhoA and, finally, promoting tight junction dissolution and cell polarity loss [18]. Thus, signaling from the cell surface leads concomitantly to a program of direct junction dissolution and a program of transcriptional repression of junction components. Both programs contribute to EMT phenotype characterized by motility and loss of cell polarity [19]. The UPS is an integral part of both junction dissolution programs regulation.
Ubiquitination and the Ubiquitin–Proteasome System (UPS)
Ubiquitination refers to the attachment of the 76-amino acid protein ubiquitin to a target protein. It is established through an amide bond between the carboxylic acid of the terminal glycine of an activated ubiquitin molecule and the ε-amine of a lysine residue in the target protein. Ubiquitination takes place with a series of enzymatic reactions executed by three types of enzymes. The first step involves E1 or ubiquitin-activating enzyme which loads an ubiquitin molecule in an ATP-dependent manner onto a second enzyme, ubiquitin-conjugating enzyme or E2. E2-linked ubiquitin is subsequently transferred to a target protein by a third type of enzymes called ubiquitin ligases or E3 [20]. Human genome encodes for two E1 enzymes (UBA1 and UBA6), about 30 to 40 E2 enzymes and probably about 600 E3 ligases [21, 22].
E3 ligases belong to two families characterized by specific domains, RING (Really Interesting New Gene) family and HECT (Homologous to Human Papilloma Virus E6 Carboxyterminal domain) family which differ in their catalytic mode but both execute ubiquitin ligation to the target protein. A third type of E3s, U-box ligases, can be considered a subfamily of RING E3 ligases, U-box domain being an atypical RING domain. RING domains of E3 ligases constitute the interactive surface with the ubiquitin-conjugating enzyme E2 bound to ubiquitin. Some E3s are single polypeptides that possess both the RING E2-binding domain and the substrate-binding domain, while other E3s represent complexes of several distinct proteins, one of which is the RING domain E2-binding protein. Another binds the target (substrate) protein to be ubiquitinated, while often, a third peptide serves as a linker between them [23]. HECT ligases are constituted by various amino terminal domains, while their carboxy-terminus is occupied by a HECT domain first identified and named after E3 ligase E6-AP (Human Papilloma Virus E6-Associated Protein). HECT domain has two subdomains, one of which binds the E2 ubiquitin-conjugating enzyme and the other binds the substrate protein.
RING type E3s comprise about 95% of human E3s, while HECT type E3s are less abundant and count 28 members in human genome [24]. Similar to other post-translational modifications, ubiquitination is reversible, and there exist five families of deubiquitinizing enzymes that perform this reaction and preserve cellular ubiquitin reserves and reverse inappropriate ubiquitination [25]. Deubiquitinases attack the isopeptide bond between the carboxy terminal glycine of ubiquitin and the ε-amino group of a lysine of another ubiquitin molecule or of a target protein. In some instances such as in transcription, it is the sequence of ubiquitination/deubiquitination that is necessary for normal function.
Ubiquitin molecule has seven lysine residues at positions 6, 11 27, 29, 33, 48 and 63. Ubiquitination of each of these lysine residues has signaling potential. The number of ubiquitin molecules attached also encodes for different outcomes [26]. A target protein may become monoubiquitinated (a single ubiquitin molecule attached), multiubiquitinated (one ubiquitin molecule attached in several different lysine residues) or polyubiquitinated (a chain of ubiquitins attached in the same lysine residue). Lysine 48 ubiquitin chains of at least four molecules are the trigger for recognition of the target protein by the proteasome and subsequent degradation [26]. Occasionally, lysine 6- and 11-mediated ubiquitin chains have been observed to signal for target protein proteasome degradation. Lysine 63-mediated ubiquitin attachment leads less often to proteasome degradation but serves mostly as a signal for autophagy-mediated proteolysis. Moreover, it serves nonproteolytic functions including DNA repair and receptor kinases endocytosis. Other processes requiring ubiquitination are DNA transcription and DNA damage tolerance.
The proteasome is a hollow cylinder multiprotein structure of 2.5 MDa comprised of a core particle (CP or 20S proteasome) covered in one or both sides by a regulatory particle (RP or 19S proteasome). RP is comprised of a lid and a base subcomplex and functions in ubiquitinated protein recognition, unfolding of the proteins, deubiquitination which allows ubiquitin molecules to be recycled and delivery of the target proteins to the CP [20]. The different subunits of RP possess specific activities to accomplish all these functions. Three subunits of the base subcomplex possess ubiquitin recognition domains that allow them to recognize polyubiquitin chains. Subunit Rpn11 (S13 in mammals) of the lid subcomplex is a deubiquitinase and recycles ubiquitin from proteins that had been recognized. The 19S base subcomplex is made up of six ATPases and three other peptides. ATPases belong to the AAA (ATPases Associated with various cellular Activities) family and are able to hydrolyze all four nucleotide triphosphates and to alter the conformation of proteasome substrate protein, preventing their aggregation before they enter the CP to be degraded [20].
CP is made of four rings of seven-member proteins each that are stacked one on top of the other. The two identical peripheral rings are called α rings (with subunits α1 to 7), and also, the two identical central rings are called β rings (with subunits β1 to 7) [27]. The proteasome possesses three enzymatic activities, a trypsin-like (postbasic residues cleavage) activity, a chymotrypsin-like (posthydrophobic residues cleavage) activity and a postglutamyl (caspase-like or postacidic residues cleavage) activity that resides in subunits β1, β2 and β5, respectively, and degrade target proteins producing fragments of four to 14 amino acids.
EMT transcription network and the UPS
Receiving cues not only from the environment but also through self-sufficient signals, neoplastic cells acquire EMT through activation of several oncogenic transcription factors (that will be referred to as intermediate transcription factors) and, in their turn, regulate and are regulated by transcription factors of the core EMT machinery such as SNAIL family regulators, ZEB and TWIST. A discussion of these transcription factors with emphasis on the interception with the UPS follows. Ubiquitin-like proteins such as SUMO (Small Ubiquitin-like Modifier) and NEDD8 (Neural precursor cell Expressed and Developmentally Downregulated protein 8) also regulate many of these transcription factors but they will not be discussed.
Intermediate transcription factors
NF-κB represents a family of five transcription factors that are important for both inflammation/immunity and carcinogenesis [28]. NF-κB has been associated with chemotherapy resistance in various cancers. It is a downstream target of several signal pathways among which is AKT kinase, an EMT inducer and drug resistance mediator that phosphorylates NF-κB-activating kinase IKK. IKK phosphorylates the NF-κB inhibitor I-κB which is then ubiquitinated by E3 ligase βTrCP for proteasome degradation. NF-κB is activated by several other pathways including TNFα [29, 30]. In addition to inducing genes that inhibit apoptosis (e.g., BCL-2, BCL-XL and A1) and promote proliferation (e.g., Cyclin D1 and C-myc), NF-κB induces genes of the core EMT program such as SNAIL and SLUG, TWIST and ZEB [31, 32]. SNAIL contributes to EMT by suppressing transcription of adhesion proteins E-cadherin, claudins and occludins as well as tumor suppressor RKIP (Raf kinase inhibitory protein) which is an inhibitor of NF-κB (Fig. 2). In this way, a feed-forward loop is established in which NF-κB induces SNAIL which suppresses RKIP preventing it from inhibiting NF-κB [33]. SNAIL is also upregulated by NF-κB in a post-translational manner, in which NF-κB inhibits SNAIL phosphorylation by GSK3β, thus, preventing its subsequent ubiquitination by E3 ligase βTRCP and proteasome degradation [34]. SLUG (also known as SNAIL2) suppresses expression of E-cadherin, claudins and occludins, while ZEB proteins suppress expression of E-cadherin and zona occludens protein ZO-1. NF-κB is a critical regulator of bHLH (basic helix-loop-helix) transcription factor TWIST which regulates several hundred genes. A feed-forward loop is present in this case, too, as TWIST activates transcription of kinase AKT2, a NF-κB activator [35, 36].
In addition to the points outlined above, signal transduction pathways culminating in NF-κB activation are regulated by ubiquitination with degradation or nondegradational outcomes in multiple points [37, 38] that will not be further detailed here. In the transcription level, NF-κB function is also regulated by ubiquitination and the UPS through the availability and modification of cofactors. The family of I-κB regulators is comprised of several members including canonical members I-κBα, I-κBβ and I-κBγ and noncanonical members BCL3, I-κBζ, I-κBη and I-κBNS which act as cofactors in NF-κB transcription regulation [39]. Among these NF-κB cofactors acting in the nucleus during transcription, BCL-3 functions either as a coactivator or a corepressor and is UPS regulated [40, 41]. Ligase TBLR1 (Transducer β-like related) mediates a GSK3-independent BCL-3 ubiquitination and degradation, while an unidentified ligase mediates a GSK3-dependent degradation [42, 43]. GSK3 phosphorylation-dependent degradation is not mediated by ligase Fbw7 [43] which is an enzyme acting on phosphodegron sites (ubiquitinating lysines after phosphorylation of an amino acid nearby). Whether this function is performed by ligase βTRCP which also ubiquitinates other family members has not been reported but it is worth investigating. TBLR1 and the related ligase TBL1 are general transcription de-repressors as they ubiquitinate and facilitate degradation of the NCoR/SMRT and the CtBP1/2 corepressor complexes [44].
Transcription factor HIF (Hypoxia Inducible Factor), a heterodimer of HIF-1α or HIF-2α and HIF-1β (also called ARNT, Aryl-hydrocarbon receptor nuclear translocator) are key mediators of the cellular response to hypoxic conditions [45]. In normoxic conditions, HIF-α subunits are hydroxylated by dioxygenases of the PHD (prolyl-4-hydroxylase domain) family (hydroxylating a proline residue) and FIH1 (Factor Inhibiting HIF1, hydroxylating an aspartine residue), become substrates for ubiquitination by E3 ligase VHL (Von Hippel Lindau) and are proteasome degraded. In contrast, in hypoxia, prolyl hydroxylases are inhibited, and HIF-α is stabilized and can heterodimerize with constitutive unit HIF-1β for the execution of their transcriptional program of more than 100 genes by binding to HREs (HIF Response Elements) on DNA with the consensus sequence RCGTG (where R is one of the pyrimidines). In addition, HIF has HREs-independent regulatory functions by interactions with other pathways such as Ras, TGFβ, NOTCH and C-myc [46–48]. HIF promotes EMT, in most, but not all, settings [49], by induction of transcription factors SNAIL, SLUG, ZEB1 and 2 and TWIST. HIF also induces NF-κB activity and both knockdown of VHL or HIF mutations leading to resistance to VHL-mediated degradation result in NF-κB upregulation [50].
C-myc is a transcription factor of the basic helix-loop-helix leucine zipper family implicated in neoplastic transformation but also in stem cell maintenance. It takes part in the pathogenic translocation t (8;14) of Burkitt lymphoma. It binds DNA as a heterodimer with protein Max. The heterodimer recognizes the so-called E box sequence CACGTG and leads to the recruitment of cofactors and the core transcription machinery. C-myc is considered in general a weak transcriptional activator and is also a transcriptional repressor for some target genes [51]. In neoplasia, it leads to proliferation, although it can also act as a promoter of apoptosis by upregulating p14ARF, a p53 activator. As a result, C-myc transformation effects are favored in cells that have a nonfunctional p14ARF/MDM2/p53 axis. In addition, p14ARF inhibits C-myc directly in a negative feedback loop. Ubiquitination of C-myc with the help of E3 ligase SKP2 complex promotes both C-myc transcriptional function and turnover, although ubiquitination is not required for transcription [51]. It is probable that, as proposed for other transcription factors, C-myc ubiquitination and subsequent degradation promote transcription by allowing the recruitment of new C-myc molecules to access the DNA-binding site if activating signals persist in order for transcription to continue. FBWX7 ligase promotes C-myc degradation independently of promoter binding but dependent on previous phosphorylation [52]. In contrast, phosphorylation of C-myc by IKK kinases at Serine 62 protects C-myc from ubiquitination and degradation [53].
C-myc overexpression promotes EMT in breast cancer cells in vitro and in vivo [54]. These cells obtain a fibroblast-like configuration and downregulate E-cadherin. E-cadherin transcriptional suppressor SNAIL is upregulated both by increased transcription and decreased ubiquitination and proteasome degradation after C-myc transfection [55]. C-myc cooperates with TGFβ in SNAIL transcriptional upregulation [56]. An additional way by which C-myc influences E-cadherin is through induction of microRNA miR-9 which suppresses E-cadherin mRNA translation [57]. α-Catenin, another component of adherens junctions, is also a target of miR-9 (Fig. 3). Dissolution of adherens junctions leads to β-catenin release from it and, depending on the state of the GSK3β/Axin/APC destruction complex, initiation of transcription. C-myc is a β-catenin target gene and, thus, a feed-forward loop is established [58].
c-myc induction of EMT by induction of SNAIL and of miRNA miR-9 leading to suppression of both E-cadherin and α-catenin. Resulting adherens junction resolution frees β-catenin which induces c-myc in a feed-forward loop. Proteins in ovals are directly UPS-regulated. Arrows denote activation and inverse T signs denote inhibition
ETS (E26 transformation specific) transcription factor family with 27 human members is implicated in a wide range of cancers. Up to 80% of prostate cancers harbor translocations between the promoter of androgen receptor (AR)-regulated serine protease TMPRSS2 and one of ETS family genes, most commonly ERG (Ets Related Gene), in a way that ETS transcription factors come to the proximity of the AR-regulated promoter and are upregulated by androgens in prostate tissue where AR signaling is robust [59]. In Ewing sarcoma, a pathognomonic translocation is almost invariably present which brings an ETS family member, most commonly FLI1 (Friend leukemia virus integration 1) in this case, in proximity of EWS (Ewing Sarcoma) gene [60]. FLI1 is upregulated under the influence of EWS. The ERG transcription program in TMPRSS2-ERG1-bearing prostate cancer activates Wnt signaling by upregulating FRIZZLED-4, other pathway proteins [61] and C-myc signaling [62] and promotes EMT with repression of E-cadherin and active β1-integrin [61]. PEA3 (Polyomavirus Enhancer Activator 3), another ETS family transcription factor, is activated by EGFR signaling and leads to transcriptional repression of miR-125a, a repressor of ARID3B (AT-rich interactive domain 3B) in ovarian cancer [63]. As a result, ARID3B, a mesenchymal specification promoter [64] is upregulated. Additionally, PEA3 promotes expression of SNAIL and matrix metalloproteases MMP9 and MMP14 contributing to cell migration [65, 66].
ETS family members ETV1, ETV4 and ETV5 which participate in prostate cancer translocations as alternative partners of TMPRSS2 instead of ERG are regulated by UPS with the aid of RING E3 ligase COP1 (Constitutive Photomorphogenic 1) [67]. Truncated forms produced by the translocations lack the critical domain leading to proteasome-mediated degradation and are more stable, a fact that may contribute to their tumorigenicity [67].
p53, a well-known tumor suppressing transcription factor sensing DNA damage and mediating either apoptosis or cell cycle arrest, has a role in EMT prevention. DNA damage response involves activation of kinases such as ATM and ATR which then activate checkpoint kinases CHK1 and 2 (Checkpoint kinases 1 and 2) [68]. These kinases, in their turn, phosphorylate p53 leading to its stabilization and activation. When the cell is not under stress, p53 is unstable because it is ubiquitinated by E3 ligase MDM2 (and other ligases) and degraded by the proteasome. When activated in response to DNA damage, p53 executes a transcriptional program leading, depending on post-translational modifications and coactivators available to either cell cycle arrest which gives time for DNA repair or to apoptosis if damage is sensed to be irreversible. NF-κB regulator BCL-3 is an inhibitor of p53 transcription activity [69], and given that BCL-3 is regulated by the UPS, it represents an additional mode of p53 regulation by the system.
Subcellular localization (nuclear export) of p53 is regulated by monoubiquitination with the aid of NEDD4 family member WWP1 [70]. p53 family members p63 and p73 are also regulated by NEDD4 ligases [71, 72].
Two deubiquitinating enzymes are participating in p53 regulation. USP10 (Ubiquitin Specific Protease 10) is mainly located in the cytoplasm and reverses p53 monoubiquitination which allows nuclear re-entry. In contrast, HAUSP (Herpes virus-Associated Ubiquitin Specific Protease, alternatively named USP7) is active in the nucleus-stabilizing p53 [73]. HAUSP is also a deubiquitinase for MDM2, and thus, the final result of its action on MDM2/p53 axis is complex and may depend on its concentration [74].
As mentioned, beyond its paramount role in cell cycle inhibition and apoptosis, p53 is an EMT suppressor (Fig. 4). This function is mediated through induction of microRNAs of the miR-200 and miR-192 families which then suppress translation of ZEB1 and 2 [75, 76]. In addition to ZEB1 and 2, SNAIL, SLUG and TWIST are upregulated in pancreatic acinar cells when p53 is knocked out and cells undergo EMT [77]. p53 downregulates SLUG through promotion of its mdm2-mediated ubiquitination and proteasome degradation which leads to E-cadherin expression [78]. CDK inhibitor p21, a p53 transcription target, has been found to decrease EMT of breast cancer cells induced by Ras and C-myc (Fig. 4) [79]. In contrast, cancer-associated mutant p53 promotes SNAIL, SLUG and TWIST induction and EMT [80–82]. In addition, mutant p53 promotes EMT by interfering with the function of metastasis suppressor p63 [83, 84]. This is because mutant p53 retains the ability to be phosphorylated and interact with isopropyl isomerase PIN1, thus, allowing for p63 sequestration and inhibition. Mutant p53 promotes with an unknown mechanism nuclear accumulation and activity of NF-κB in response to TNFα stimulation [85]. In contrast, mutant p53 lose the ability to act as a suppressor of hyaluronan receptor CD44 transcription, thus, allowing this protein to promote EMT [86, 87].
EMT core transcription factors and the UPS
Transcription factors activated through various EMT pathways cooperate to induce a set of other transcription regulators that directly suppress the epithelial phenotype and promote mesenchymal phenotype by dissolving intercellular junctions through downregulation of E-cadherin and other junctional components and by upregulating mesenchymal markers such as N-cadherin and vimentin [88]. These core EMT transcription factors include SNAIL1 and SLUG (also named SNAIL2), ZEB1 and 2, TWIST1 and 2 and E12/ E47. They cooperate with the intermediate transcription factors which are direct targets of EMT-inducing pathways.
SNAIL family transcription regulators are upregulated by growth factor receptor signaling, TGFβ family, WNT, Hedgehog, NF-κB, NOTCH, G-coupled protein receptors and Interleukin-6 (IL-6) (Fig. 5) [89–92]. IL-6 upregulates SNAIL through activation of STAT3 (Signal Transducer and Activator of Transcription 3) and promotes the metastatic potential of head and neck carcinoma cells in a xenograft model [92]. Reciprocally, SNAIL induces WNT signaling through WNT gene expression [93].
SNAIL, in cooperation with SMAD3 and 4, downregulates tight junction component CAR (Coxsackie and Adenovirus Receptor), claudins, occludin and E-cadherin [94]. Corticosteroids interfere with SNAIL binding to E-cadherin promoter, inhibit TGFβ-induced EMT and enhance E-cadherin transcription [95]. SNAIL is regulated by ubiquitination-mediated by ligases MDM2 and βTRCP subsequent to phosphorylation by kinase GSK3β, and thus, its stability is UPS-dependent [90, 96].
ZEB1 is regulated by NOTCH signaling, and silencing NOTCH by siRNA has led to ZEB1 downregulation in pancreatic cancer cells leading to MET and reversal of gemcitabine resistance [90]. ZEB2 is upregulated by NF-κB, TGFβ, ETS and HIF signaling directly through respective binding sites in its promoter and by Hedgehog signaling indirectly through TGFβ [97]. In the same study, it was found that besides E-cadherin, ZEB2 suppresses cyclin D1 and telomerase (Fig. 6). Through the suppression of these proteins, it promotes cell cycle arrest and senescence, respectively. Other important targets of ZEB1 and 2 transcription suppression are miRs of the miR-200 family [98]. These miRs represent a family of five members and target ZEB1 and 2 in a feedback loop in which ZEBs promote EMT, and miR-200 s promote the reverse process, MET, by repressing ZEBs. miR-200 family member miR-200a also targets β-catenin, another way of suppressing EMT [99]. ZEB1 suppresses another miR, miR-203, which is a suppressor of stemness genes SOX2 and KLF4 and, thus, promotes the stem cell phenotype [100] and establishes a link between this phenotype and EMT. A link of stemness and EMT with antineoplastic drugs resistance has been recognized [101].
Transcription modulators, ZEBs, are found in the center of regulation by multiple intermediate EMT transcription factors and lead to EMT induction but also promote cell cycle arrest, cell senescence and stem cell phenotype. Proteins in ovals are directly UPS-regulated. Arrows denote activation and inverse T signs denote inhibition
ZEB1 as well as SLUG has been found to be upregulated by E3 ligase cullin7/FBXW8 complex, resulting in downregulation of E-cadherin and increased invasion of human trophoblastic cell lines [102]. RNA interference for cullin7 reversed E-cadherin suppression and decreased migration of these cells.
TWIST bHLH transcription factors play a role in development and promote cancer EMT. They are upregulated by NF-κB and HIF and contribute to E-cadherin downregulation. TWIST further promote EMT by upregulating miRNA miR-10b which is a repressor of transcription factor HOXD10, finally resulting in upregulation of RHOC [103, 104]. HOXD10 is a homeobox-type factor which represses RHOC. RHOC, a Rho family GTPase, is a promoter of cell motility and is upregulated in various cancer types [105]. The hyaluronan receptor CD44, a stem cell marker activates kinase c-SRC and promotes TWIST-mediated upregulation of RHOC providing a link between EMT and the stem cell phenotype [104]. Another study associates TWIST-induced EMT and stem cell phenotype with activation of β-catenin and AKT by this transcription modulator [36]. In addition, TWIST stimulates IL-6 production and STAT3 activation in breast cancer cells which display enhanced invasiveness [106]. As mentioned, STAT3 promotes expression of SNAIL [92] and, as a result, a feed-forward mechanism promoting EMT is established.
TWIST factors have been found to be proteasome substrates at least during apoptosis [107]. Class I bHLH transcription factors E12/E47 stimulate transcription of miR-495 which is a suppressor of E-cadherin and additionally promotes stem cell phenotype [108]. E12/E47 and inhibitor Id are also proteasome regulated [109, 110].
EMT core transcription factors, similar to intermediate transcription factors important for EMT described in the previous section, have roles in development and have been hijacked by cancer. It is probable that adult tissue stem cells maintain part of embryonal stem cell characteristics that favor long-term survival and renewal potential and are primed for EMT if additional signals arrive, explaining the connection between stemness, inhibition of apoptosis and EMT. It is important to note that different EMT transcription factors may contribute to different aspects of the EMT phenotype, and only their coordinate action under specific conditions produce EMT. For example, as discussed, the role of SNAIL in intercellular junction dissolution by E-cadherin downregulation needs to be complemented with the action of TWIST for motility promotion by RHOC upregulation.
Ubiquitin–Proteasome System’s (UPS) role in transcription
The UPS has an important role in the regulation of the transcription process in general and, through this role, has an additional influence in the transcription factors of EMT function and activity. Ubiquitination of proteins taking part in transcription leads to their proteasomal degradation, directly to other outcomes or to nondegradational outcomes followed by proteasome degradation after a required ubiquitin chain has been added to an initial monoubiquitination [111].
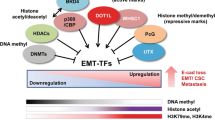
Ubiquitination of transcription machinery components and histones both play parts in the transcription process. In many instances, ubiquitination of transcription factors and cofactors signals for transcription complex assembly followed by polyubiquitination which interrupts transcription initiation phase. Coactivators bound to activated transcription factors recruit histone acetyltransferases such as CBP (CREB Binding Protein)/p300 and p/CAF (p300/CBP-Associated Factor) and histone arginine methyltransferases such as CARM1 (Coactivator-associated Arginine Methyltransferase-1) and PRMT-1 (Protein Arginine Methyltransferase-1) [112]. These enzymes promote histone acetylation and methylation that opens nucleosomes in order for the transcription complex to obtain access to transcription factor-binding sequences in target promoters. The signal for histone methylation is provided by sequential histone monoubiquitination and deubiquitination [113–115]. Histones H2A and H2B are part of the nucleosome octamer comprised of pairs of histones H2A, H2B, H3 and H4 wrapped around 147 bases of DNA. They are both ubiquitinated in mammalian cells with a greater percentage (10%) of H2A being ubiquitinated than H2B of which only 1% is ubiquitinated at any given time [116, 117]. Ubiquitination of the two histones appears to have opposing roles in transcription. H2B ubiquitination on lysine 123 promotes H3 methylation at lysine 4 (H3K4) and promotes transcription, while H2A ubiquitination has the reverse effect on H3K4 methylation and represses transcription. The 19S regulatory part of the proteasome is involved in histone ubiquitination [118, 119]. In addition, RING domain-containing E3 ligase hPIRH2 (human p53-induced ring-containing H2) has a role in histone acetylation. It binds transcription factors such as nuclear receptors and promotes suppression of histone deacetylase 1 (HDAC1), thereby stabilizing histones in the acetylated state [120]. Only the wild type hPIRH2 which is able to downregulate HDAC1 retains the ability to promote transcription, while a mutant not able to repress HDAC1 is also unable to promote transcription [120].
Concomitant with histone modifications that facilitate nucleosome dissociation from the promoter transcription initiation site and transcription machinery binding [121, 122], ubiquitination of corepressors CtBP1/2 and NCoR/SMRT leads to their proteasome degradation-releasing transcriptional repression in order for the transcription complex to bind DNA [44]. Many transcription factors such as nuclear receptors undergo ubiquitination after DNA binding [123, 124]. In parallel, a molecular complex called mediator is recruited and helps recruit, in its turn, RNA polymerase II to begin transcription [125]. By this time, ubiquitin ligases complete the attachment of at least four ubiquitin molecules to transcription factor molecules which can now be recognized by the proteasome for degradation. E3 ligase activity possessing components of the general transcription machinery may facilitate this ubiquitination [126]. A protein called TSG101 (Tumor Susceptibility Gene 101) transiently protects some transcription factors such as the AR from polyubiquitination in order to complete their function on transcription initiation before degradation [127]. Proteasomal degradation contributes to time regulation of transcription as it allows for its prompt termination if no new transcription factor molecules bind the promoter.
Ubiquitination and the UPS are, thus, contributing to the regulation of several steps of transcription such as initiation complex assembly, histone modifications and transcription elongation. Whether these multiple regulations of transcription take place in transcriptional regulation by EMT-involved transcription factors has not been specifically reported but this is likely at least for the themes that concern general transcription components and universal histone modifications. Indeed, as mentioned in previous sections, core EMT transcription factors are ubiquitination targets for proteasomal degradation, and it remains plausible that the mechanism of ubiquitination-mediated transcription function termination is at play for these factors, too. In addition, ubiquitination-mediated modulation of chromatin landscape may proceed through modification of transcription cofactors to influence EMT. For example, ligase CHIP (Carboxyl terminus of Hsp70-Interacting Protein) has been found to inhibit coactivator SRC-3 (Steroid hormone Receptor Co-activator 3) and, thus, interfere with transcription activation of TWIST and β-catenin and transcription of mesenchymal marker vimentin [128, 129].
E3 ligases regulating transcription factors of EMT
RING and HECT family E3 ligases participate in EMT regulation. RING E3s are more abundant and represent critical regulators of several transcription factors involved in EMT (Fig. 7). E3 ligases also regulate other carcinogenesis processes through the same pathways involved in EMT signaling and through further substrates that are involved in carcinogenesis beyond EMT. SCF (Skp1/Cullin/F-box) E3 ligases represent a subfamily of RING-type ligases and several members are involved in EMT. Their structural organization groups several proteins and includes a cullin molecule which is the scaffold protein of the complex, an F-box protein that associates through a Skp (S phase kinase-associated protein) protein with cullin and binds the substrate to be ubiquitinated and a ROC (also called RBX) RING finger protein that links the complex through cullin with the ubiquitin-loaded E2 enzyme [130]. The assembly of the ligase complex depends on cullin neddylation, the association with the ubiquitin-like protein NEDD8, which opens cullin configuration and facilitates E2 enzyme association [131].
βTRCP, a RING-type E3 ligase of the SCF subfamily, regulates EMT through its involvement in WNT, Hedgehog and NF-κB cascades and in SNAIL degradation. βTRCP recognizes phosphorylated substrates and, thus, the process of target destruction is tightly regulated by at least one and sometimes two steps of phosphorylation followed by ubiquitination. In the NF-κB pathway, for example, βTRCP ubiquitinates multiple substrates including inhibitors I-κBα, I-κBβ and I-κBγ as well as the precursor NF-κB forms p105 and p100. Inhibitors I-κB need to be phosphorylated by kinases IKK in order to be recognized by βTRCP for ubiquitination and then proteasome degradation. Ubiquitination of p105 and p100 leads to partial cleavage by the proteasome which produces the active factors p50 and p52, respectively. SNAIL transcription factor is also a target for βTRCP ubiquitination [132]. As a result, βTRCP has both promoting and inhibiting roles in EMT.
SKP2 (S-phase kinase-associated protein 2) is the F-box component of another SCF family RING E3 ligase and is involved in carcinogenesis processes through regulation of Cyclin-Dependent Kinase inhibitors p27 and p21 [133]. SKP2 regulates EMT by promoting C-myc transcription. It ubiquitinates C-myc acting as a cofactor for its transcription activity [134].
FBW7 (F-box and WD repeat domain-containing 7, also designated FBXW7 or hCDC4) is another SCF E3 ligase regulating EMT through its role in degradation of transcription factor C-myc. C-myc ubiquitination by FBW7 requires the phosphorylation of serine at position 62 by MAPK followed by the phosphorylation of threonine at position 58 by GSK3. These phosphorylations create the recognition site for FBW7 binding to C-myc [135]. C-myc is a transcriptional target of NOTCH and, thus, it is also regulated indirectly by FBW7 as the ligase targets both NOTCH and presenelin, a component of its activating enzyme γ-secretase [136].
FBW7 mutations are synergistic with p53 mutations in cancer induction in experimental models, given that p53 represents a safeguard mechanism of unopposed C-myc activity. This is a developmentally preserved mechanism. In hematopoiesis, for example, FBW7 preserves hematopoietic stem cells quiescence, while its deletion results in transient cell growth due to C-myc and cyclin E (another FBW7 target) [137] overactivity but, finally, to stem cell exhaustion due to p53-induced apoptosis. Analogously, in carcinogenesis, concomitant FBW7 and p53 mutations would lead to unopposed EMT.
An additional member of the SCF family of ligases regulating EMT pathways is Cullin 7/FBXW8 [138]. SKP1 participates in the ligase complex associating Cullin 7 with FBXW8, and ROC1 is the E2 recognizing unit. As mentioned, Cullin 7/FBXW8 plays a role in EMT by participating in degrading ZEB 1 and SLUG [102]. In addition, a Cullin 7 ligase complex is further implicated in EMT regulation by interfering with p53 function in a degradation-independent manner [139]. This complex associates with p53 and impairs its transcriptional activity without exhibiting an E3 ligase activity towards it [139]. The mechanism of the interference of Cullin 7 with p53 function is unknown but may involve retention of p53 in the cytoplasm. This mechanism has been shown for another E3 ligase, PARC, which retains p53 in the cytoplasm [140].
VHL is the substrate-recognizing component of a crucial ligase that regulates hypoxia responses through transcription factor HIF. VHL plays a role in EMT both through HIF regulation and through regulation of other substrates [141]. The complex of the RING ligase in which VHL participates also includes the scaffold protein Cullin 2, Elongin that links cullin 2 to VHL and the E2-conjugating enzyme-binding RING protein RBX1. Beyond HIF, other interacting partners of VHL complex that affect EMT include several proteins that regulate adhesion and the extracellular matrix such as fibronectin, hydroxylated collagen IV and microtubules. Renal tight junctions are maintained by VHL action through upregulation of claudin 1 and occludin [142].
MDM2 (Mouse double minute 2) is a RING-type E3 ligase well-known as a negative regulator of p53. Together with p53, MDM2 (also known as HDM2 in humans) forms a regulatory feedback loop functioning in DNA damage (or other stress) response. MDM2 physiologic action restrains p53 activity and, being a target gene of p53 transcription, terminates p53 activity when DNA damage is repaired. The pathway is also active and important in development [143]. Mdm2 knockout mice are embryonic lethal due to massive apoptosis except if p53 is also knocked out. In oncogenesis, dysregulated activity of MDM2 ligase may restrain physiologic p53 response and promote neoplastic transformation.
MDM2 is composed of 491 amino acids and contains an amino terminal p53-binding domain; centrally, an acidic domain and a zinc finger next to it; and a carboxy terminal RING domain [144]. A MDM2-related protein, MDM4 (also called MDMX or HDM4 in humans), has the same domain organization as MDM2 but lacks ligase activity. As a result, although it can bind p53 and inhibit its transcriptional activity, it cannot promote its proteasome degradation [145]. Nevertheless, MDM4 binds MDM2, and it can modulate positively or negatively MDM2 ligase activity in different settings [144, 146]. MDM2 recognizes other proteins in addition to p53 for ubiquitination. An important MDM2 substrate for EMT is, as mentioned in a previous section, transcription regulator SNAIL [78]. In this way, MDM2 may both promote EMT by ubiquitinating p53 for degradation and inhibit EMT by promotion of SNAIL degradation.
Transcription of MDM2 is upregulated by several pathways inducing EMT such as TGFβ and receptor tyrosine kinases pathways [147]. SMAD2- and SMAD3-binding sites have been identified in one of the two alternative promoters of MDM2 gene [148]. Activation of transcription factor AP-1 as well as ETS family factors downstream of the Ras/Raf pathway activates MDM2 transcription through distinct sites in the same promoter [149, 150]. In addition, MEK–ERK signaling downstream of Ras upregulates MDM2 post-transcriptionally by promoting nuclear export of its mRNA [151]. Thus, MDM2 is induced by several EMT-inducing transcription factors that may override the decrease of MDM2 production secondary to p53 suppression required in the EMT process. Furthermore, the PI3K/AKT pathway activated downstream of various receptors post-translationally activates MDM2 through phosphorylation.
Perspectives and conclusion
Although the above discussion of UPS regulation of EMT-related transcription is not meant to be exhaustive, a glimpse of its complexity becomes evident. This discussion reveals also the close relationship of EMT with other carcinogenesis processes which share essentially the same signaling and transcription factors. Several transcription factors well-known for their implication in malignancy such as NF-κB, HIF and C-myc are primary players in EMT, while the foremost tumor suppressor p53 is concomitantly an important EMT suppressor. Deeper inside EMT regulation, a layer of core transcription factors cooperate with intermediate transcription factors for direct changes leading to the execution of EMT with dissolution of adhesions and mesenchymal marker induction.
The recent revelation that EMT phenotype is associated with stem cell phenotype [8] and drug resistance [101] has implications for understanding carcinogenesis but also overcoming this resistance. It argues for a common signaling matrix that originates and serves development and is hijacked by the neoplastic process. An additional argument is offered by the connection of EMT signaling with asymmetric cell division, characteristic of normal and cancer stem cells. Asymmetric cell division is regulated by factors playing roles in EMT such as TGFβ and p53 [18, 152], and as a result, it enters the list of the processes with a close relationship and common regulation with EMT. The UPS, as a regulator of these factors, enters the equation in multiple knots. Thus, a therapeutic intervention could be introduced in several levels. For the time being, the only antineoplastic drug in clinical use directly affecting the UPS, proteasome inhibitor bortezomib, acts in a point where specificity is precluded. One could imagine that inhibition of other parts of the system such as individual ligases could offer therapeutic specificity dependent on molecular lesions of individual carcinomas. For example, ubiquitination of p53 by MDM2 could be a target of therapeutic intervention in tumors that harbor wild type p53 [153]. Several inhibitors of MDM2/p53 interaction such as cis-imidazoline compound nutlins as well as thiobenzodiazepine compounds are under investigation [154, 155]. In contrast, use of inhibitors of MDM2 E3 ligase activity could be more complicated given that, as mentioned, MDM2 have other ubiquitination targets such as SLUG, stabilization of which would have EMT-promoting effects. This example illustrates the subtleties that the transfer of molecular interventions to the clinical arena may hold. Nevertheless, a therapeutic intervention that successfully inhibits EMT could have a significant impact inhibiting not only metastasis but also parallel processes of carcinogenesis served by the same pathways.
References
Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nature Rev Mol Cell Biol. 2006;7:131–42.
Acloque H, Adams MS, Fishwick K, et al. Epithelial–mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–49.
Strieter ER, Korasick DA. Unraveling the complexity of ubiquitin signaling. ACS Chem Biol. 2012;7:52–63.
Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–8.
Shukla A, Chaurasia P, Bhaumik SR. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci. 2009;66:1419–33.
Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. J Clin Invest. 2009;119:1420–8.
Zeisberg M, Neilson EG. Biomarkers for epithelial–mesenchymal transitions. J Clin Invest. 2009;119:1429–37.
Mani SA, Guo W, Liao MJ, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15.
Tarin D. The fallacy of epithelial–mesenchymal transition in neoplasia. Cancer Res. 2005;65:5996–6001.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Micalizzi DS, Farabaugh SM, Ford HL. Epithelial–mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–34.
Harris TJC, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11:502–14.
Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899.
Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907–17.
Fujita Y, Krause G, Scheffner M, et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 2002;4:222–31.
Palacios F, Tushir JS, Fujita Y, D’Souza-Schorey C (2005) Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell–cell adhesion during Epithelia to Mesenchymal Transitions. 25: 389–402
Janda E, Nevolo M, Lehmann K, et al. Raf plus TGFβ-dependent EMT is initiated by endocytosis and lysosomal degradation of E-cadherin. Oncogene. 2006;25:7117–30.
Ozdamar B, Bose R, Barrios-Rodiles M, et al. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–9.
Viloria-Petit AM, Wrana JL. The TGFβ–Par6 polarity pathway. Linking the Par complex to EMT and breast cancer progression. Cell Cycle. 2010;9:623–4.
Voutsadakis IA (2010) Ubiquitin, ubiquitination and the ubiquitin–proteasome system in cancer. Atlas Genet Cytogen Oncol Haematol. URL:// Atlas GeneticsOncology.org/Deep/UbiquitinCancerID20083.httml
Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signaling pathways. Nature Rev Mol Cell Biol. 2009;10:319–31.
van Wijk SJL, Timmers HTM. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24:981–93.
Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434.
Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–406.
Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207.
Voutsadakis IA. Pathogenesis of colorectal carcinoma and therapeutic implications: the roles of the ubiquitin–proteasome system and Cox-2. J Cell Mol Med. 2007;11:252–85.
Wolf DH, Hilt W. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim Biophys Acta. 2004;1695:19–31.
Wertz IE, Dixit VM. Signaling to NF-κB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2:a003350.
Wu Y, Zhou BP. TNF-α/NF-κB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102:639–44.
Min C, Eddy SF, Sherr DH, Sonenshein GE. NF-κB and epithelial to mesenchymal transition of cancer. J Cell Biochem. 2008;104:733–44.
Pham CG, Bubici C, Zazzeroni F, et al. Upregulation of Twist-1 by NF-κB blocks cytotoxicity induced by chemotherapeutic drugs. Mol Cell Biol. 2007;27:3920–35.
Chua HL, Bhat-Nakshatri P, Clare SE, et al. NF-κB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–24.
Vonach C, Viola K, Giessrigl B, et al. NF-κB mediates the 12(S)-HETE-induced endothelial to mesenchymal transition of lymphendothelial cells during the intravasation of breast carcinoma cells. Br J Cancer. 2011;105:263–71.
Bachelder RE, Yoon S-O, Franci C, et al. Glycogen synthase kinase-3 is an endogenous inhibitor of Snail transcription: implications for the epithelial–mesenchymal transition. J Cell Biol. 2005;168:29–33.
Cheng GZ, Chan J, Wang Q, et al. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67:1979–87.
Li J, Zhou BP. Activation of β-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer. 2011;11:49.
Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30:383–91.
Landström M. The TAK1-TRAF6 signalling pathway. Int J Biochem Cell Biol. 2010;42:585–9.
Espinosa L, Bigas A, Mulero MC. Alternative nuclear functions for NF-κB family members. Am J Cancer Res. 2011;1:446–59.
Dechend R, Hirano F, Lehmann K, et al. The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene. 1999;18:3316–23.
Keutgens A, Shostak K, Close P, et al. The repressing function of the oncoprotein BCL-3 requires CtBP, while its polyubiquitination and degradation involve the E3 ligase TBLR1. Mol Cell Biol. 2010;30:4006–21.
Viatour P, Dejardin E, Warnier M, et al. GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol Cell. 2004;16:35–45.
Keutgens A, Zhou X, Shostak K, et al. BCL-3 degradation involves its polyubiquitination through a FBW7-independent pathway and its binding to the proteasome subunit PSMB1. J Biol Chem. 2010;285:25831–40.
Perissi V, Scafoglio C, Zhang J, et al. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell. 2008;29:755–66.
Haase VH. Oxygen regulates epithelial-to-mesenchymal transition: insights into molecular mechanisms and relevance to disease. Kidney Int. 2009;76:492–9.
Kim WY, Perera S, Zhou B, et al. HIF2α cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–70.
Chen J, Imanaka N, Chen J, Griffin JD. Hypoxia potentiates Notch signalling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer. 2010;102:351–60.
Xing F, Okuda H, Watabe M, et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene. 2011;30:4075–86.
Scortegagna M, Martin RJ, Kladney RD, et al. Hypoxia-inducible factor-1α suppresses squamous carcinogenic progression and epithelial–mesenchymal transition. Cancer Res. 2009;69:2638–46.
Pantuck AJ, An J, Liu H, Rettig MB. NF-κB-dependent plasticity of the epithelial to mesenchymal transition induced by Von Hippel–Lindau inactivation in renal cell carcinomas. Cancer Res. 2010;70:752–61.
Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242–52.
Yada M, Hatakeyama S, Kamura T, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–25.
Yeh P-Y, Lu Y-S, Ou D-L, Cheng A-L. IκB kinases increase Myc protein stability and enhance progression of breast cancer cells. Mol Cancer. 2011;10:53.
Trimboli AJ, Fukino K, de Bruin A, et al. Direct evidence for epithelial–mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–45.
Cho KB, Cho MK, Lee WY, Kang KW. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 2010;293:230–9.
Smith AP, Verrecchia A, Fagà G, et al. A positive role for Myc in TGFβ-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene. 2009;28:422–30.
Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biol. 2010;12:247–56.
Khew-Goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nature Cell Biol. 2010;12:209–11.
Turner DP, Watson DK. ETS transcription factors: oncogenes and tumor suppressor genes as therapeutic targets for prostate cancer. Expert Rev Anticancer Ther. 2008;8:33–42.
Kovar H. Context matters: the hen or egg problem in Ewing’s sarcoma. Semin Cancer Biol. 2005;15:189–96.
Gupta S, Iljin K, Sara H, et al. FZD4 as a mediator of ERG oncogene-induced WNT signalling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–45.
Sun C, Dobi A, Mohamed A, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–53.
Cowden Dahl KD, Dahl R, Kruichak JN, Hudson LG. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia. 2009;11:1208–15.
Takebe A, Era T, Okada M, et al. Microarray analysis of PDGFRα+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev Biol. 2006;293:25–37.
Cowden Dahl KD, Zeineldin R, Hudson LG. PEA3 is necessary for optimal epidermal growth factor receptor-stimulated matrix metalloproteinase expression and invasion of ovarian tumor cells. Mol Cancer Res. 2007;5:413–21.
Yuen H-F, Chan Y-K, Grills C, et al. Polyomavirus enhancer activator 3 protein promotes breast cancer metastatic progression through Snail-induced epithelial–mesenchymal transition. J Pathol. 2011;224:78–89.
Vitari AC, Leong KG, Newton K, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–6.
Vousden KH, Prives C. Blinded by the SteLight: the growing complexity of p53. Cell. 2009;137:413–31.
Kashatus D, Cogwell P, Baldwin AS. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 2006;20:225–35.
Laine A, Ronai Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene. 2007;26:1477–83.
Miyazaki K, Ozaki T, Kato C, et al. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem Biophys Res Commun. 2003;308:106–13.
Melino G, Knight RA, Cesareni G. Degradation of p63 by Itch. Cell Cycle. 2006;5:1735–9.
Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–36.
Masuya D, Huang C, Liu D, et al. The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J Pathol. 2006;208:724–32.
Chang C-J, Chao C-H, Xia W, et al. p53 regulates epithelial–mesenchymal transition and stem cell properties through modulating miRNAs. Nature Cell Biol. 2011;13:317.
Kim T, Veronese A, Pichiorri F, et al. p53 regulates epithelial–mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–83.
Pinho AV, Rooman I, Real FX. p53-dependent regulation of growth, epithelial–mesenchymal transition and stemness in normal pancreatic epithelial cells. Cell Cycle. 2011;10:1312–21.
Wang S-P, Wang W-L, Chang Y-L, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nature Cell Biol. 2009;11:694–704.
Liu M, Casimiro MC, Wang C, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci USA. 2009;106:19035–9.
Zhang Y, Yan W, Chen X. Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J Biol Chem. 2011;286:16218–28.
Kogan-Sakin I, Tabach Y, Buganim Y, et al. Mutant p53R175H upregulates Twist1 expression and promotes epithelial–mesenchymal transition in immortalized prostate cells. Cell Death Diff. 2011;18:271–81.
Ohashi S, Natsuizaka M, Wong GS, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–84.
Girardini JE, Napoli M, Piazza S, et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91.
Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Diff. 2011;18:1487–99.
Weisz L, Damalas A, Liontos M, et al. Mutant p53 enhances nuclear factor κB activation by tumor necrosis factor α in cancer cells. Cancer Res. 2007;67:2396–401.
Jiang Z, Jones R, Liu JC, et al. RB1 and p53 at the crossroad of EMT and triple-negative breast cancer. Cell Cycle. 2011;10:1563–70.
Godar S, Ince TA, Bell GW, et al. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73.
Gemmill RM, Roche J, Potiron VA, et al. ZEB1-responsive genes in non-small cell lung cancer. Cancer Lett. 2011;300:66–78.
De Craene B, van Roy F, Berx G. Unraveling signalling cascades for the Snail family of transcription factors. Cell Signalling. 2005;17:535–47.
Wang Z, Li Y, Kong D, et al. Acquisition of epithelial–mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the Notch signaling pathway. Cancer Res. 2009;69:2400–7.
Bagnato A, Rosanò L. Epithelial–mesenchymal transition in ovarian cancer progression: a crucial role for the endothelin axis. Cells Tissues Organs. 2007;185:85–94.
Yadav A, Kumar B, Datta J, et al. IL-6 promotes head and neck tumor metastasis by inducing epithelial–mesenchymal transition via the JAK–STAT3–SNAIL signaling pathway. Mol Cancer Res. 2011;9:1658–67.
Stemmer V, de Craene B, Berx G, Behrens J. Snail promotes Wnt target gene expression and interacts with β-catenin. Oncogene. 2008;27:5075–80.
Vincent T, Neve EPA, Johnson JR, et al. A SNAIL1–SMAD3/4 transcriptional repressor complex promotes TGF-β mediated epithelial–mesenchymal transition. Nature Cell Biol. 2009;11:943–50.
Zhang L, Lei W, Wang X, et al. Glucocorticoid induces mesenchymal-to-epithelial transition and inhibits TGF-β1-induced epithelial-to-mesenchymal transition and cell migration. FEBS Lett. 2010;584:4646–54.
Yook JI, Li X-Y, Ota I, et al. A Wnt-Axin2-GSK3β cascade regulates Snail1 activity in breast cancer cells. Nature Cell Biol. 2006;8:1398–14.
Katoh M, Katoh M. Integrative genomic analyses of ZEB2: transcriptional regulation of ZEB2 based on SMADs, ETS1, HIF1α, POU/OCT, and NF-κB. Int J Oncol. 2009;34:1737–42.
Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop-a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–7.
Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391:535–41.
Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biol. 2009;11:1487–95.
Voutsadakis IA. Molecular predictors of gemcitabine response in pancreatic cancer. World J Gastrointest Oncol. 2011;3:153–64.
Fu J, Lv X, Lin H, et al. Ubiquitin ligase Cullin 7 induces epithelial–mesenchymal transition in human choriocarcinoma cells. J Biol Chem. 2010;285:10870–9.
Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8.
Burguignon LYW, Wong G, Earle C, et al. Hyaluronan–CD44 interaction promotes c-Src-mediated Twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J Biol Chem. 2010;285:36721–35.
Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91–8.
Sullivan NJ, Sasser AK, Axel AE, et al. Interleukin-6 induces an epithelial–mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009;28:2940–7.
Demontis S, Rigo C, Piccinin S, et al. Twist is substrate for caspase cleavage and proteasome-mediated degradation. Cell Death Diff. 2006;13:335–45.
Hwang-Verslues WW, Chang P-H, Wei P-C, et al. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–74.
Sun L, Trausch-Azar JS, Ciechanover A, Schwartz AL. E2A protein degradation by the ubiquitin–proteasome system is stage-dependent during muscle differentiation. Oncogene. 2007;26:441–8.
Sun L, Trausch-Azar JS, Ciechanover A, Schwartz AL. Ubiquitin–proteasome-mediated degradation, intracellular localization, and protein synthesis of MyoD and Id1 during muscle differentiation. J Biol Chem. 2005;280:26448–56.
Bhat KP, Greer SF. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim Biophys Acta. 2011;1809:150–5.
Jenster G, Spencer TE, Burcin MM, et al. Steroid receptor induction of gene transcription: a two-step model. Proc Natl Acad Sci USA. 1997;94:7879–84.
Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–40.
Dover J, Schneider J, Tawiah-Boateng MA, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–71.
Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8.
Higashi M, Inoue S, Ito T. Core histone H2A ubiquitylation and transcriptional regulation. Exp Cell Res. 2010;316:2707–12.
Chandrasekharan MB, Huang F, Sun Z-W. Histone H2B ubiquitination and beyond. Epigenetics. 2010;5:460–8.
Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitination in transcriptional control: a FACT-finding mission. Genes Dev. 2007;21:737–43.
Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004;13:435–42.
Logan IR, Gaughan L, McCracken SRC, et al. Human PIRH2 enhances androgen receptor signalling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol Cell Biol. 2006;26:6502–10.
Boeger H, Bushnell DA, Davis R, et al. Structural basis of eukaryotic gene transcription. FEBS Lett. 2005;579:899–903.
Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–73.
Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26.
Ramamoorthy S, Nawaz Z. E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl Recept Signal. 2008;6:e006.
Vijayvargia R, May MS, Fondell JD. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 2007;67:4034–41.
Conaway RC, Brower CS, Conaway JW. Emerging roles of ubiquitin in transcriptional regulation. Science. 2002;296:1254–8.
Burgdorf S, Leister P, Scheidtmann KH. TSG101 interacts with apoptosis-antagonizing transcription factor and enhances androgen receptor-mediated transcription by promoting its monoubiquitination. J Biol Chem. 2004;279:17524–34.
Kajiro M, Hirota R, Nakajima Y, et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol. 2009;11:312–9.
Patterson C, Ronnebaum S. Breast cancer quality control. Nat Cell Biol. 2009;11:239–41.
Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nature Rev Cancer. 2011;11:629–43.
Soucy TA, Dick LR, Smith PG, et al. The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer. 2010;1:708–16.
Kim S-E, Yoon J-Y, Jeong W-J, et al. H-Ras is degraded by Wnt/β-catenin signaling via β-TrCP-mediated polyubiquitination. J Cell Sci. 2009;122:842–8.
Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nature Rev Cancer. 2008;8:438–49.
von der Lehr N, Johansson S, Wu S, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–200.
Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature Rev Cancer. 2008;8:83–93.
O’Neal J, Grim J, Strack P, et al. (2005) FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–24.
Minella AC, Welcker M, Clurman BE. Ras activity regulates cyclin E degradation by the Fbw7 pathway. Proc Natl Acad Sci USA. 2005;102:9649–54.
Sarikas A, Xu X, Field LJ, Pan Z-Q. The cullin7 E3 ligase: a novel player in growth control. Cell Cycle. 2008;7:3154–61.
Jung P, Verdoodt B, Bailey A, et al. Induction of cullin 7 by DNA damage attenuates p53 function. Proc Natl Acad Sci USA. 2007;104:11388–93.
Nikolaev AY, Li M, Puskas N, et al. Parc: a cytoplasmic anchor for p53. Cell. 2003;112:29–40.
Kaelin Jr WG. The von Hippel–Lindau tumour suppressor protein: O2 sensing and cancer. Nature Rev Cancer. 2008;8:865–73.
Harten SK, Shukla D, Barod R, et al. Regulation of renal epithelial tight junctions by the von Hippel–Lindau tumor suppressor gene involves occludin and claudin 1 and is independent of E-cadherin. Mol Biol Cell. 2009;20:1089–101.
Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in MDM4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nature. 2001;29:92–5.
Perry ME. The regulation of the p53-mediated stress response by MDM2 and MDM4. Cold Spring Harb Perspect Biol. 2010;2:a000968.
Marine J-C, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Diff. 2010;17:93–102.
Barboza JA, Iwakuma T, Terzian T, et al. MDM2 and MDM4 loss regulates distinct p53 activities. Mol Cancer Res. 2008;6:947–54.
Manfredi JJ. The Mdm2–p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580–9.
Araki S, Eitel JA, Batuello CN, et al. TGF-β1-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. J Clin Invest. 2010;120:290–302.
Ries S, Biederer C, Woods D, et al. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103:321–30.
Phelps M, Darley M, Primrose JN, Blaydes JP. p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor α-positive breast cancer cells. Cancer Res. 2003;63:2616–23.
Phelps M, Phillips A, Darley M, Blaydes JP. MEK–ERK signalling controls Hdm2 oncoprotein expression by regulating hdm2 mRNA export to the cytoplasm. J Biol Chem. 2005;280:16651–8.
Cicalese A, Bonizzi G, Pasi CE, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–95.
Allende-Vega N, Saville MK. Targeting the ubiquitin–proteasome system to activate wild-type p53 for cancer therapy. Sem Cancer Biol. 2010;20:29–39.
Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31.
Zhuang C, Miao Z, Zhu L, et al. Synthesis and biological evaluation of thio-benzodiazepines as novel small molecule inhibitors of the p53–MDM2 protein–protein interaction. Eur J Med Chem. 2011;46:5654–61.
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Voutsadakis, I.A. Ubiquitination and the Ubiquitin–Proteasome System as regulators of transcription and transcription factorsin epithelial mesenchymal transition of cancer. Tumor Biol. 33, 897–910 (2012). https://doi.org/10.1007/s13277-012-0355-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0355-x