Abstract
Background
hTERT contains a high density of minisatellites, of which rare alleles of hTERT-VNTR2-2nd have been reported to be associated with prostate cancer. This shows an association between VNTR and cancer, but this repeat sequence is likely to be associated with genomic instability. Therefore, we investigated the effects of hTERT-VNTR2-2nd on gastrointestinal cancer and the relationship between repeated sequence and chromosome instability.
Methods
A case–control study was performed using DNA from 818 cancer-free controls, 539 cases with gastric cancer, 275 cases with colon cancer and 274 cases with rectal cancer. To determine whether minisatellites affect gene expression, expression levels were examined using TERT-reporter vectors in cell lines. In addition, the length of the hTERT-VNTR2-2nd alleles were determined in blood and cancer tissues from 107 gastric cancers, 112 colon cancers and 76 rectal cancers patients to determine whether the repeat sequence was associated with genomic instability during cancer development.
Results
No statistically significant association between hTERT-VNTR2-2nd and risk of gastrointestinal cancer was detected. However, it has been shown that VNTRs inserted into the enhancer region can regulate the expression of TERT in gastrointestinal cancer cells. Moreover, hTERT-VNTR2-2nd was analyzed in matched blood and cancer tissue from patients with gastrointestinal cancer and in seven among 294 subjects, and hTERT-VNTR2-2nd was found to be rearranged.
Conclusions
We suggest that minisatellites are associated with genomic instability in cancer and that the hTERT-VNTRs region may increase hTERT expression in gastrointestinal cancer cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ends of eukaryotic chromosomes undergo degradation and end-to-end fusion problem during cell division (Blackburn 2000). To protect the coding region from this event, telomeres that are non-coding repeat region are specialized at the ends of eukaryotic chromosomes (Blackburn 2001). Maintaining chromosome terminal length by telomerase is necessary for immortal proliferation in stem cells or most malignancies (Shay and Bacchetti 1997). Template-containing RNA components (TR) and human telomerase reverse transcriptase (hTERT) are major components for chromosome end synthesis (Jiang et al. 1999; Morales et al. 1999), of which the telomerase catalytic subunit, hTERT, has been found to be abnormal in many human cancers (Ito et al. 1998; Kanaya et al. 1998; Kyo et al. 1998, 1999; Takakura et al. 1998).
In previous studies, we identified five variable number tandem repeat (VNTR, mini-satellite) regions present in the intron region of hTERT, of which rare alleles of hTERT-VNTR 2-2nd were associated with prostate cancer (Kim et al. 2003; Leem et al. 2002). The region of hTERT-VNTR2-2nd found that the repeat unit was 61-bp long, with 44 repeated VNTRs being the most common allele (Yoon et al. 2010a). In particular, 28 and 37 repeated VNTR alleles were detected only in a group of prostate cancer patients, and the rare hTERT-VNTR2-2nd allele containing these cancer specific alleles was statistically associated with prostate cancer risk (Yoon et al. 2010a). And we confirmed that the hTERT-VNTR2-2nd allele region could influence the expression of hTERT in prostate cancer (Yoon et al. 2010a). In addition, rearrangement occurrence in the VNTR6-1 and VNTR6-2 regions present in intron six was confirmed by comparison of normal and cancerous tissues (Leem et al. 2002). Our previous study demonstrated the relationship between minisatellite variants and several cancers (Ahn et al. 2009; Jeong et al. 2007; Seol et al. 2008; Yoon et al. 2008, 2010b), and also revealed that VNTR alleles influences gene expression (Kwon et al. 2010; Yoon et al. 2010a).
In this study, we investigated the allelic variation and genetic rearrangement of hTERT-VNTR2-2nd in gastrointestinal cancer and their association with hTERT expression. A case control study using a PCR-based method was performed and compared the allelic distribution of hTERT-VNTR2-2nd in DNA samples from 818 cancer-free controls and 1088 patients with gastrointestinal cancer. In sequence, we identified the effect of the VNTRs on the transcriptional levels of hTERT-promoter driven reporter gene in gastrointestinal cancer cell lines. In addition, the length of the hTERT-VNTR2-2nd allele was determined in the blood and cancer tissue of gastrointestinal cancer patients to determine whether the repeat sequence was associated with genomic instability during cancer development. Here, we report that hTERT-VNTR2-2nd alleles may be related to genomic instability and regulation of expression level of hTERT in gastrointestinal cancer.
Materials and methods
Preparation of case–control study and genomic DNA from peripheral blood lymphocytes and cancer tissues
We performed a case–control study with genomic DNA from the 818 cancer-free controls and 539 cases with gastric cancer, 275 cases with colon cancer and 274 cases with rectal cancer (Table 1). The controls consist of similar proportion of sex and age range to the cases (Table 1). A total of 818 individuals in the control group with no personal history of cancers or current cancer were recruited and completed an interview. Cases with gastrointestinal cancer and controls were recruited from three different hospitals in two different cities. The bioethics committees of Dong-A University Hospital, Inje University Paik Hospital and Chungbuk National University Hospital approved the study plan and procedures: [Dong-A University Hospital (#IRB-07-10-7; Busan, Korea), Inje University Paik Hospital (#IRB11-011) and the Chungbuk National University Hospital (#IRB-2006-1; Cheongju, Korea)].
For PCR experiments, genomic DNA was isolated from 400 µL whole blood, using a Blood DNA Mini Kit (Qiagen, CA). A total of 295 cancerous tissues and their respective non-cancerous tissues were obtained from patients with gastrointestinal cancer and were immediately frozen in liquid nitrogen. Cancer and normal cells were laser captured and microdissected using a Pix Cell II LCM system and stained by the HistoGen LCM Frozen Section Staining Kit (Arturus, USA). Malignant cells were captured and their genomic DNA was isolated by using the PicoPure DNA extraction kit (Arturus, USA) (Jeong et al. 2007).
PCR analysis of hTERT-VNTR2-2nd
Primer sequences used for amplification of hTERT-VNTR2-2nd are as follows: hTERT-VNTR2-2nd forward 5′-TGGGAGCATCACTCACAGGA, hTERT-VNTR2-2nd reverse 5′-GGAACACAGCCAACCCCTTA (Leem et al. 2002). PCR analysis of human DNA samples was performed using the Go Taq polymerase (Promega, USA) with 100 ng genomic DNA. Genomic DNA was amplified using primers under the following standard PCR conditions: 50 mM KCl, 10 mM Tris–HCl, pH 9.0, 3 mM MgCl2, 0.2 mM dTTP, dCTP, dGTP and dATP in a final volume of 30 µl. Cycle conditions were 94 °C for 2 min, then 30 cycles consisting of 45 s at 94 °C, then 2 min 30 s at 69 °C in a 9700 Thermalcycler (Perkin-Elmer, CA, USA). The last elongation step was extended to 7 min at 72 °C. PCR products were analyzed by gel electrophoresis (1 V/cm) in TAE buffer through 0.8% agarose gel.
Cells and luciferase assay
Human cell lines were examined for the effect of the hTERT-VNTR2-2nd on hTERT expression: 293T (human embryonic kidney cell line; obtained from Korean Cell Line Bank (KCLB), South Korea), MCF7 (breast cancer cell line; obtained from KCLB, South Korea), AGS (gastric cancer cell line; obtained from KCLB, South Korea) and HCT116 (colon cancer cell line; obtained from KCLB, South Korea). For the luciferase assay, cells (1 × 105) were seeded in 12-well plates, cultured overnight and transfected with the hTERT promoter–luciferase plasmids (0.5 µg per well; constructed as described in (Yoon et al. 2010a)) by use of FuGENE6 transfection reagent (Roche Diagnostics, USA) and the ratio of DNA to FuGENE6 was 1:3 (Yoon et al. 2010a). The cells were analyzed using a dual-luciferase reporter assay system (Promega) 48 h after completion of the transfection procedure. The Firefly luciferase activity was normalized according to Renilla luciferase activity and expressed as relative luciferase units to reflect the promoter activity. Triplicate transfections per each construct were examined for one experiment, and final results were calculated by four independently repeated experiments.
RNA isolation and reverse transcription PCR
Total RNA was isolated from cancer cell lines using QIAGEN RNeasy Mini Kit. For the reverse transcription reaction, a mixture of total RNA (3 µg), an oligo(dT)20 (50 nM), and 10 mM dNTP was incubated at 65 °C for 5 min. A mixture of 1X RT buffer, 25 nM MgCl2, 0.1 M DTT and RNaseOUT™ was incubated at 42° C for 2 min. After incubation at 42 °C for 2 min, Invitrogen SuperScript III (200 U/µl) was added. This mixture was then incubated at 42 °C for 50 min followed by 70 °C for 15 min. RNase H (2 U/µl) was then added and incubated at 37 °C for 20 min in a 9700 Thermocycler (Perkin-Elmer) with the following primer sequences (exon 2): hTERT forward 5′-AGTGACCGTGGTTTCTGTGTGGTG-3′ and hTERT reverse 5′-GCCTGGAACCCAGAAAGATGGTCT-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward 5′-CCCTTCATTGACCTCAACTACATGG-3′ and GAPDH reverse 5′-CCTGCTTCACCACCTTCTTGATGTC-3′. PCR products containing hTERT and GAPDH were amplified using gene specific primers under standard PCR conditions: 50 mM KCl, 10 mM Tris–HCl, pH 9.0, 3.0 mM MgCl2, 0.2 mM dTTP, dCTP, dGTP, and dATP in a final volume of 40 µl. Thermocycling conditions were as follows: one cycle of initial denaturation for 2 min at 94 °C, 30 cycles of 30 s at 94 °C, annealing for 15 s at 62 °C and extension for 45 s at 72 °C, followed by a final 7 min extension at 72 °C in a 9700 Thermocycler. Cosmo G Taq DNA polymerase was used for amplification. PCR products were analyzed by gel electrophoresis (1 V/cm) in TAE buffer through 1.2% agarose gel.
Statistical analysis
Regression analyses were performed to determine the odds ratios (ORs) in the association between control and case groups. ORs were estimated using the natural logarithm and its standard error. Where relevant, we used a Chi squared test with 1° of freedom to assess statistical significance. Differences were considered significant for confidence intervals (CIs) of 95%. All tests were two-sided, with p < 0.05 being considered statistically significant. Statistical analyses were performed using MS Excel with CHITEST and R statistical software (v2.5.1, www.r-project.org) with chisq.test for the calculation of Chi squared values.
Results
Analysis of hTERT-VNTR2-2nd allelic polymorphism in controls and cases with gastrointestinal cancer
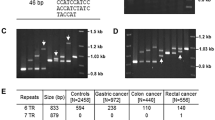
Using the PCR amplification with diagnostic primers on human genomic DNA samples isolated from 818 cancer-free unrelated individuals (412 males and 397 females), we analyzed the degree of polymorphism within the hTERT-VNTR2-2nd (Table 1). A similar distribution of allelic frequency of hTERT-VNTR2-2nd was revealed in the male and female controls (Table 2). The five alleles of hTERT-VNTR2-2nd ranged from 2605 to 2910 bp in length and contained 39–44 copies of the repeat unit, with 44 copies being present in the most common allele (total controls, 61.31%; male controls, 61.05%; female controls, 61.59%) (Table 2) (Yoon et al. 2010a). Seven different genotypes with the five alleles were found for hTERT-VN
TR2-2nd in controls (Fig. 1a).
Allele typing at hTERT-VNTR2-2nd in cancer-free controls and gastrointestinal cancer patients. Electrophoretic patterns of PCR products of hTERT-VNTR2-2nd in controls and patients. Seven haplotype patterns of hTERT-VNTR2-2nd were detected in DNA from 818 cancer-free controls (a) and 539 gastric cancer (b). Six haplotype patterns were detected in DNA from 275 colon cancer patients (c). Seven haplotype patterns were detected in DNA from 274 rectal cancer patients (d). Three rare haplotypes were detected in DNA from patients with gastric cancer (37/44) and rectal cancer (35/44, 42/50). The first and last lanes correspond to a 1-kb marker (M)
We analyzed the frequency and distribution of the polymorphic hTERT-VNTR2-2nd alleles between controls and cancer cases for evaluation of a correlation between hTERT-VNTR2-2nd variants and gastrointestinal cancer. To compare DNA obtained from the 818 controls and from 539 patients with gastric cancer (335 males and 204 females), 275 patients with colon cancer (148 males and 127 females) and 274 patients with rectal cancer (177 males and 97 females), we conducted a case–control study (Table 2). hTERT-VNTR2-2nd had seven haploid types in controls (Fig. 1a), seven haploid types in gastric cancer patients (Fig. 1b), six haploid types in colon cancer patients (Fig. 1c), seven haploid types in rectal cancer patients (Fig. 1d). Although there was no statistically significant difference between controls and cases, three rare alleles (35, 37 and 50 repeats) were detected in gastric and rectal cancer (Table 2, Fig. 1).
For analysis of susceptibility for cancer according to rare alleles of hTERT-VNTR2-2nd, each hTERT-VNTR2-2nd allele was divided into two groups (common and rare alleles) based on their frequency in the controls. Rare alleles were grouped through threshold frequency, ≤ 1%. The six alleles of hTERT-VNTR2-2nd were grouped as three common alleles (42, 43 and 44 repeats) and five rare alleles (35, 37, 39, 40 and 50 repeats) (Table 2; bold represents the rare alleles). Consistent with comparison of allele frequency between controls and cases, these data showed no significant association between rare alleles and OR of gastrointestinal cancer (data not shown).
Regulation of the VNTR polymorphism on the hTERT promoter within gastrointestinal cancer
To investigate whether hTERT-VNTR2-2nd alleles affect hTERT expression levels, we used reporter vectors which contained the hTERT promoter, luciferase gene and four different sizes of hTERT-VNTR2-2nd alleles were published in our study (Fig. 1a) (Yoon et al. 2010a). We analyzed VNTRs carry between 37 and 44 copies of the 61 bp repeats unit, and they were positioned in the enhancer region of the pGL3-Basic vector with hTERT core promoter which was published (Yoon et al. 2010a) in the promoter region. Transfection of control vector (pBT304) and vectors with each VNTR repeat variant (37, 39, 42, 44 copies) was used for estimation of expression level in 293T, MCF7, AGS and HCT116 cells. Expression of hTERT was checked by RT-PCR through each cell line before the luciferase assay began. RT-PCR analysis revealed that 293T, MCF7, AGS and HCT116 cells expressed the hTERT gene (Fig. 2a). Then, the luciferase activity between the control pBT304 and pBT304 with each VNTR repeat did not show statistically significant difference in 293T and MCF7 cells (Fig. 2b, c). AGS and HCT116 cells were also transiently transfected with pBT304 or pBT304 + each length of VNTR. Interestingly, as shown in Fig. 2b, luciferase assays revealed that each VNTRs stimulated the activity of the hTERT promoter, representing approximately 1.3–3.7-fold increases in AGS and HCT116, respectively (Fig. 2c). Although we could not find a statistically significant difference based upon VNTR length, we identified a potential of hTERT-VNTR2-2nd within hTERT gene expression regulation. Furthermore, when we used the pBT304 with 2443 bp of unrelated genomic region, there was no significant difference in luciferase activity (Fig. 2b, c).
Effect of allelic variation of hTERT-VNTR2-2nd in hTERT-promoter activity. ahTERT mRNA expression in 293T, MCF7, AGS and HCT116 cells were confirmed by RT-PCR. b The effect of VNTR regions on hTERT gene expression were analyzed by the luciferase reporter system. 293T, MCF7, AGS and HCT116 cell lines were transfected with 6 different plasmids. c The average value of the promoter activity and p value were schematized. Two rare alleles TR (37 and 39 copies) and two common alleles TR (42 and 44 copies) were inserted in the pBT304 plasmid: pBT304 + TR37, pBT304 + TR39, pBT304 + TR42, and pBT304 + TR44. The pBT304 + 2443 plasmid was made by insertion of an irrelevant 2443-bp fragment instead of TR
Analysis of minisatellite instability in gastrointestinal cancer tissues
hTERT, which located at the subtelomeric region at 15p3.3, contains a high density of minisatellites that may play a role in its chromosomal instability (Yoon et al. 2010a). This possibility was examined by comparing the polymorphic alleles of hTERT-VNTR2-2nd minisatellites in the blood and cancer tissues from 107 gastric cancers, 112 colon cancers and 76 rectal cancers patients. These cancer tissues were not included in determination of rare alleles for each minisatellite because of their small sample size (Fig. 3). In DNA obtained from both blood and cancer tissue of patients with gastrointestinal cancer, there were four cases of small deletions or loss of heterozygosity (LOH) in hTERT-VNTR2-2nd in DNA obtained from cancer tissues (Fig. 3a). Among the 107 gastric cancer patients, the frequency of rearrangement was 3.74% (4/107) in hTERT-VNTR2-2nd (Fig. 3a). The frequencies of rearrangement of hTERT-VNTR2-2nd in colon and rectal cancer tissues were 0.89% (1/112) in colon cancer tissues and 2.63% (2/76) in rectal cancer tissues (Fig. 3b, c).
Rearrangement of hTERT-VNTR2-2nd in cancer patient tissue. Instability of hTERT-VNTR2-2nd in paired peripheral blood and cancer tissue from gastric (a), colon (b) and rectal (c) cancer patients were detected. Cancer tissue samples are indicated by asterisks and rearrangements in cancer tissues are indicated by arrows. The first and last lanes correspond to a 1-kb marker (M)
Discussion
In particular, if the gene is located in the terminal region of the chromosome and contains high density repeats such as VNTR, certain VNTR alleles present in the gene may be involved in rearrangement or susceptibility to cancer (Ahn et al. 2009; Jeong et al. 2007; Kwon et al. 2010; Seol et al. 2008; Yoon et al. 2008, 2010a, b). hTERT, located at the end of chromosome 5, has these characteristics, genetic mutations involving important carcinogenic pathways have been identified and are associated with susceptibility to cancer (Carpentier et al. 2007). Telomeres roles as a cap for the protection of the DNA from exonuclease degradation and recombination events at telomeric regions could lead to genomic instability (Blackburn 2001). Therefore, we were interested in rearrangement and susceptibility in cancer of specific VNTR regions for the hTERT.
We previously reported that the short rare hTERT-VNTR2-2nd alleles are correlated with prostate cancer susceptibility when compared with common alleles (Yoon et al. 2010a). In this study, we examined association with rare alleles for gastrointestinal cancer, but no significant differences were found. In addition, previous results showed a significant difference in prostate cancer, while no significant difference was found in breast cancer, a female cancer (data not shown). These results suggest that the cancer susceptibility of rare hTERT-VNTR2-2nd alleles vary according to the cancer type and is not applied in gastrointestinal cancer.
And then we hypothesized that the hTERT-VNTR2-2nd polymorphism which affects the expression of hTERT in gastrointestinal cancer. One of the characteristics of gastric cancer cells is telomerase expression and telomerase activity is frequently up-regulated by a variety of mechanisms during gastric cancer development (He et al. 2010). In most embryonic and adult tissues, the RNA component of telomerase is expressed (Feng et al. 1995); on the contrary, expression of hTERT is highly regulated and correlates with telomerase activity (Kilian et al. 1997; Meyerson et al. 1997; Nakamura et al. 1997).
We also analyzed the putative binding sites for the transcription factors, estrogen receptor 1 (ER/ESR1; 5), v-Erb1 (5 sites) and NF-kappaB (3 sites) in the hTERT-VNTR2-2nd (Yoon et al. 2010a). Several study showed that cell and tissue level in gastrointestinal cancer revealed the presence of estrogen receptor a (ERa) and estrogen receptor b (ERb) (Messa et al. 2000; Singh et al. 1997; Takano et al. 2002). Furthermore, poor prognosis of gastric cancer patients is related with expression of ERa and the absence of ERb expression, and both of ERa and ERb could use as a marker for gastric cancer (Xu et al. 2010). In addition, NF-kappaB is constitutively activated in gastric carcinoma tissues and NF-kappaB activation is correlated with clinicopathological features of tumor aggression of gastric carcinoma (Sasaki et al. 2001). Coincidence with luciferase assay, this analysis suggests that hTERT-VNTR2-2nd is a potential region for regulation of hTERT gene expression even though it located in non-coding region, intron.
It has been suggested that cancer cell development accelerates the accumulation of genetic variation, causing genomic instability, which plays an important role in cancer development and progression (Lengauer et al. 1998). In addition, hTERT, located at the end of the chromosome, contains a high-density minisatellite that may play an important role in chromosome instability (Yoon et al. 2010a). We examined the hTERT-VNTR2-2nd in genomic DNA from blood and cancer tissue derived from 295 patients with gastrointestinal cancer and detected seven cases with small deletions or LOH in hTERT-VNTR2-2nd (Fig. 3); frequency of rearrangement was 3.9%. This frequency is higher than in the previously known minisatellite region of H-ras (Krontiris et al. 1985).
Thus, these results strongly suggest that hTERT-VNTR2-2nd alleles may be associated with chromosomal instability in cancer. Based on these findings, further investigations, such as large-scale epidemiological studies of the association between minisatellite and cancer risk, could provide a useful reference for understanding gene function and complex genomic characteristics.
References
Ahn EK, Kim WJ, Kwon JA, Choi PJ, Kim WJ, Sunwoo Y, Heo J, Leem SH (2009) Variants of MUC5B minisatellites and the susceptibility of bladder cancer. DNA Cell Biol 28:169–176
Blackburn EH (2000) The end of the (DNA) line. Nat Struct Biol 7:847–850
Blackburn EH (2001) Switching and signaling at the telomere. Cell 106:661–673
Carpentier C, Lejeune J, Gros F, Everhard S, Marie Y, Kaloshi G, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Sanson M (2007) Association of telomerase gene hTERT polymorphism and malignant gliomas. J Neurooncol 84:249–253
Feng J, Funk W, Wang S, Weinrich S, Avilion A, Chiu C, Adams R, Chang E, Allsopp R, Yu J et al (1995) The RNA component of human telomerase. Science 269:1236–1241
He XL, Qiao Q, Ge NJ, Nan J, Shen SQ, Wang ZZ, Yang YF, Bao GQ (2010) Irradiation-induced telomerase activity and gastric cancer risk: a case-control analysis in a Chinese Han population. BMC Cancer 10:9
Ito H, Kyo S, Kanaya T, Takakura M, Inoue M, Namiki M (1998) Expression of human telomerase subunits and correlation with telomerase activity in urothelial cancer. Clin Cancer Res 4:1603–1608
Jeong YH, Kim MC, Ahn EK, Seol SY, Do EJ, Choi HJ, Chu IS, Kim WJ, Kim WJ, Sunwoo Y et al (2007) Rare exonic minisatellite alleles in muc2 influence susceptibility to gastric carcinoma. PLoS One 2:10
Jiang X-R, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar AG, Wahl GM, Tlsty TD et al (1999) Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet 21:111–114
Kanaya T, Kyo S, Takakura M, Ito H, Namiki M, Inoue M (1998) hTERT is a critical determinant of telomerase activity in renal-cell carcinoma. Int J Cancer 78:539–543
Kilian A, Bowtell DDL, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet 6:2011–2019
Kim JH, Leem SH, Sunwoo Y, Kouprina N (2003) Separation of long-range human TERT gene haplotypes by transformation-associated recombination cloning in yeast. Oncogene 22:2452–2456
Krontiris TG, DiMartino NA, Colb M, Parkinson DR (1985) Unique allelic restriction fragments of the human Ha-ras locus in leukocyte and tumour DNAs of cancer patients. Nature 313:369–374
Kwon JA, Lee SY, Ahn EK, Seol SY, Kim MC, Kim SJ, Kim SI, Chu IS, Leem SH (2010) Short Rare MUC6 minisatellites-5 alleles influence susceptibility to gastric carcinoma by regulating gene expression. Hum Mutat 31:942–949
Kyo S, Takakura M, Tanaka M, Kanaya T, Inoue M (1998) Telomerase activity in cervical cancer is quantitatively distinct from that in its precursor lesions. Int J Cancer 79:66–70
Kyo S, Kanaya T, Takakura M, Tanaka M, Yamashita A, Inoue H, Inoue M (1999) Expression of human telomerase subunits in ovarian malignant borderline and benign tumors. Int J Cancer 80:804–809
Leem SH, Londono-Vallejo JA, Kim JH, Bui H, Tubacher E, Solomon G, Park JE, Horikawa I, Kouprina N, Barrett JC et al (2002) The human telomerase gene: complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene 21:769–777
Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396:643–649
Messa C, Russo F, Pricci M, Di Leo A (2000) Epidermal growth factor and 17 beta-estradiol effects on proliferation of a human gastric cancer cell line (AGS). Scand J Gastroenterol 35:753–758
Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q et al (1997) <em > hEST2 </em > the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785–795
Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, Shay JW (1999) Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet 21:115–118
Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955–959
Sasaki N, Morisaki T, Hashizume K, Yao T, Tsuneyoshi M, Noshiro H, Nakamura K, Yamanaka T, Uchiyama A, Tanaka M et al (2001) Nuclear factor-kappa B p65 (RelA) transcription factor is constitutively activated in human gastric carcinoma tissue. Clin Cancer Res 7:4136–4142
Seol SY, Lee SY, Kim YD, Do EJ, Kwon JA, Kim SI, Chu IS, Leem SH (2008) Minisatellite polymorphisms of the SLC6A19: susceptibility in hypertension. Biochem Biophys Res Commun 374:714–719
Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33:787–791
Singh S, Poulsom R, Wright NA, Sheppard MC, Langman MJ (1997) Differential expression of oestrogen receptor and oestrogen inducible genes in gastric mucosa and cancer. Gut 40:516–520
Takakura M, Kyo S, Kanaya T, Tanaka M, Inoue M (1998) Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res 58:1558–1561
Takano N, Iizuka N, Hazama S, Yoshino S, Tangoku A, Oka M (2002) Expression of estrogen receptor-alpha and -beta mRNAs in human gastric cancer. Cancer Lett 176:129–135
Xu CY, Guo JL, Jiang ZN, Xie SD, Shen JG, Shen JY, Wang LB (2010) Prognostic role of estrogen receptor a and estrogen receptor beta in gastric cancer. Ann Surg Oncol 17:2503–2509
Yoon YH, Seol SY, Heo J, Chung CN, Park IH, Leem SH (2008) Analysis of VNTRs in the solute carrier family 6, member 18 (SLC6A18) and lack of association with hypertension. DNA Cell Biol 27:559–567
Yoon SL, Jung SI, Do EJ, Lee SR, Lee SY, Chu IS, Kim WJ, Jung J, Kim CS, Cheon SH et al (2010a) Short rare hTERT-VNTR2-2(nd) alleles are associated with prostate cancer susceptibility and influence gene expression. BMC Cancer 10:10
Yoon SL, Kim DC, Cho SH, Lee SY, Chu IS, Heo J, Leem SH (2010b) Susceptibility for breast cancer in young patients with short rare minisatellite alleles of BORIS. BMB Rep 43:698–703
Acknowledgements
We gratefully acknowledge patients and their caregivers for their willing participation in this project and for consenting to the use of information obtained from the study.
Funding
This work was supported by the Dong-A University research fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Jeong-Ah Kwon, Mi-So Jeong, Se-Lyun Yoon, Jeong-Yeon Mun, Min-Hye Kim, Gi-Eun Yang, Seong-Hwan Park, Jin-Woong Chung, Yung Hyun Choi, Hee-Jae Cha and Sun-Hee Leem declare that they have no competing interests.
Ethical approval
This study was conducted with informed written consent from participants and after approval by the bioethics committees of Dong-A University Hospital (#IRB-07-10-7; Busan, Korea), Inje University Paik Hospital (#IRB11-011) and the Chungbuk National University Hospital (#IRB-2006-1; Cheongju, Korea).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kwon, JA., Jeong, MS., Yoon, SL. et al. The hTERT-VNTR2-2nd alleles are involved in genomic stability in gastrointestinal cancer. Genes Genom 41, 1517–1525 (2019). https://doi.org/10.1007/s13258-019-00882-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-019-00882-y