Abstract
Quantitative real-time PCR (qRT-PCR) is a standard method to measure gene expression in function exploring. Accurate and reproducible data of qRT-PCR requires appropriate reference genes, which are stably expressed under different experimental conditions. However, no housekeeping genes were validated as internal controls for qRT-PCR in Sinonovacula constricta. In this study, we classified the transcriptome data of two tissues for Vibrio infection and Cd2+ stress into ten clusters based on the gene expression patterns. Among them, cluster 5 had the most stable gene expression patterns regardless of tissues and treatments as the database for candidate reference genes. A total of 55 orthologs of classical housekeeping genes in the clam transcriptome were annotated. Combined the expression profiles and housekeeping genes in S. constricta, we chose eight candidate reference genes and validated their expression in Vibrio-infected samples and different tissues by qRT-PCR. Their expression stability was analyzed by three different algorithms geNorm, NormFinder and BestKeeper. Although the rank of the eight candidate reference genes is different in different treatments using different software, RS9 could be the best reference genes for normalization of qRT-PCR expression data in S. constricta under various treatments considering the above analysis. Meanwhile, the ranking of genes based on the CV values of transcriptomic data was similar to the validation results. This study provides for the first time a list of suitable reference genes for S. constricta and a valuable resource for further studies of clam immune defense systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The razor clam (Sinonovacula constricta) is an importantly economic bivalve species, and distributes in intertidal zones and estuarine waters along the coast of West Pacific Ocean. In China, it is a member of four important and traditional shellfish with breeding history for hundreds of years. Meanwhile, owing to its benthic lifestyle and strong adaptability, it is a main species cultivated with crustacean for polyculture. Because of its commercial value and ecological niche, molecular and genetic researches in S. constricta have been increasing (Niu et al. 2017; Peng et al. 2017; Ran et al. 2017; Zhao et al. 2017).
Gene expression analysis is an important and useful procedure in our understanding of gene function, pathways and networks underlying cellular and biological processes. Although there are available technologies to evaluate gene expression, such as northern blotting, microarray and next-generation sequencing, quantitative real-time PCR (qRT-PCR) is still a common method, even used for the validation of sequencing results, owing to its high sensitivity and convenience (Bustin et al. 2005; Kubista et al. 2006). However, the normalization of qRT-PCR data is a prerequisite before accounting for variations between samples, which can minimize the non-specific variations obtained from RNA quality, the efficiencies of reverse transcription and quantity of samples (Guénin et al. 2009). To assure the accuracy of qRT-PCR results among tissues and over time, the appropriate selection of the reference genes is crucial. Housekeeping genes are constitutive genes that express for the maintenance of basic cellular functions and should remain constant under different tissues and different physiological conditions (Thellin et al. 1999; Dheda et al. 2004). Hence, the internal reference controls of qRT-PCR mostly are housekeeping genes. Actually, various studies have reported housekeeping genes used for reference control expressed variably under different tissues, developmental stages, and different treatments (Stürzenbaum and Kille 2001). In previous studies, it was validated that the expression levels of the same gene normalized by the different reference genes varied greatly, even acquired opposite results (Niu et al. 2015; Koramutla et al. 2016). These findings showed that the choice of reliable reference gene is essential for accurate normalization of target gene expression levels. It’s necessary to validate the stable housekeeping genes in the given organisms and experiments.
The identification of optimal reference genes for qRT-PCR has been carried out in aquatic organisms, such as fish (Ingerslev et al. 2006; Filby and Tyler 2007; Zheng and Sun 2011), shrimp (Dhar et al. 2009; Valenzuela-Castillo et al. 2017) and mollusc (Cubero-Leon et al. 2012; Du et al. 2013; Song et al. 2017; Volland et al. 2017). Unfortunately, qRT-PCR normalization in immunology studies using non-model organism, such as bivalves, frequently with not validated genes or with genes previously validated under different experimental conditions or other species (Volland et al. 2017).
The razor clam is a promising studied species among the bivalves. For decreasing the damage by disease, its immune response to pathogen attracts widespread attention. A number of commonly used housekeeping genes in bivalves, such as β-actin (ACT), elongation factor 1-α (EF1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 18S and tubulin (TUB) are reported in previous studies (Meistertzheim et al. 2007; Mateo et al. 2010; Li et al. 2011; Liu et al. 2014a, b). In addition to these genes, we selected housekeeping genes from the stable expression of the high-throughout data from S. constricta treated by Cd2+ and Vibrio parahaemolyticus (Wang et al. 2016; Zhao et al. 2017) as candidate reference genes. In this study, we validated eight candidate housekeeping genes associated with five target tissues and Vibrio infection in S. constricta to identify the most stable internal controls for normalization of qRT-PCR data in response to pathogen.
Materials and methods
Selection of candidate reference genes and primers design
Based on the RNA-seq data of 12 libraries for the gill and hepatopancreas of the razor clam under Cd2+ stress (Wang et al. 2016) and Vibrio infection (Zhao et al. 2017), genes with similar expression levels were classified to ten different clusters. Candidate reference genes were selected from clusters with stable expression trends and were the orthologs of the housekeeping genes of Crassostrea gigas (Du et al. 2013). According to the FPKM values of the candidate reference genes in the 12 transcriptome, the coefficient of variation (CV) of these genes were calculated in Microsoft Excel for the analysis of expression stability (Robinson et al. 2010). CV was calculated as SD/mean. According to the CV value and commonly used internal controls, six candidate genes were selected. In addition, β-actin (ACT) and tubulin (TUB), used commonly in bivalves, were added as candidate reference genes (Volland et al. 2017) (Glyceraldehyde-3-phosphate dehydrogenase is a common housekeeping gene in most of animals, but there is no orthologs in the transcriptome of S. constricta). The cDNA sequences of these selected housekeeping genes were obtained from our RNA-seq transcriptome dataset of razor clam. The qRT-PCR primers were designed by Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/) with the melting temperature between 59 and 61 °C (Table 1). Lysozyme (F: AACTATTGGCTGGACTGTGGCTC; R: GACATTAGAACATCCTGGCTGG) and C1q-domain-containing protein (C1qDC) (F: AATGCTTACAACTCCCACGCCG; R: CCACGCTTTGCTGTCACTACTG) were chosen to validate the applicability of the reference genes.
Sample collection
Adult clams (S. constricta) with an average body weight of 10 g, average body length of 5.5 cm and average shell width of 2.25 cm were purchased from a commercial clam fishery (Ningbo, Zhejiang, China) in June 2017. The clams were held in aerated 40 l tanks, with filtered seawater at a salinity of 20‰ and a temperature of 16 °C. After accommodation for three days, all the clams was exposed to V. parahaemolyticus with the final concentration of 107 CFU/mL. The gill tissues from five individuals were merged as one sample and the samples were collected at 0, 6, 12, 24 and 48 h, respectively. The tissues were dissected and immediately frozen in liquid nitrogen, and stored at − 80 °C for later use. Each time point had three biological replicates.
Total RNA extraction and cDNA synthesis
The gill tissues from five individuals with the same treatments were homogeneously mixed. Total RNA was extracted using RNAiso Plus (TaKaRa, Tokyo, Japan) according to manufacturer’s instructions. RNA integrity was confirmed by gel electrophoresis with the proportion of the ribosomal bands as a ratio of 2.0. The quantity and quality of the total RNA were measured by a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The samples with A260/A280 and A260/A230 ratios greater than 1.8 were used for cDNA synthesis. Total RNA was diluted with nuclease-free water and 1 µg RNA of each sample was reverse transcribed using a PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa).
qRT-PCR analysis
Real-time PCR reactions were performed using an ABI 7500 with 7500 software v2.0.1 (Applied Biosystems). The 20 µl PCR mixture contained 10 µl of PowerUp SYBR Green Master Mix (Applied Bfiosystems), 0.8 µl of 10 µM forward primer, 0.8 µl of 10 µM reverse primer, 1 µl of cDNA template and 7.4 µl of nuclease-free water. Cycling conditions were an initial UDG activation step at 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 15 s at 95 °C, 1 min at 60 °C and then a melting curve stage after the cycling stage. For each reference gene, no-template reactions were run as negative PCR controls. To calculate the gene-specific PCR efficiency, standard curves were generated from dilution series of cDNA template for each primer pair.
Analysis of gene expression stability
Gene expression stability was evaluated by geNorm (qbase+, version 3.1) (Hellemans et al. 2007), NormFinder (version 0.953) (Andersen et al. 2004) and BestKeeper (version 1) (Pfaffl et al. 2004). The raw expression CT values for the eight selected genes in the 15 vibrio-infected samples and 15 tissues samples were exported into an Excel sheet. RefFinder (http://150.216.56.64/referencegene.php?type=reference) includes geNorm, Normfinder, BestKeeper and delta CT that can be used to rank the stability order of reference genes stability generated from each statistical algorithm.
Results
Selection of candidate reference genes
We identified ten different clusters with various gene expression patterns (Fig. 1). There were 631 to 4091 genes in each cluster. Gene expression patterns of clusters 1, 2, 3, 4 and 6 (Fig. 1a–d, f) were highly variable between gills and hepatopancreas, while the gene expression changed with the different treatments in the last four clusters (Fig. 1g–j). Only genes in cluster 5 (Fig. 1e) showed the most stable expression trend. Therefore, 2061 unigenes in cluster 5 were the dataset for selecting candidate reference genes. Based on the published 317 housekeeping genes in C. gigas (Du et al. 2013), a total of 55 orthologs of S. constricta housekeeping genes were identified (Supplementary Table S1). There were 11 housekeeping genes in cluster 5 among the 55 orthologs. To measure the stability of these gene expression, we calculated their CV values based on the FPKM values across 12 transcriptomes (Table 2). The first nine genes in Table 2 except 18S rRNA and Methionyl-tRNA synthetase had lower CV values. According to the lower CV values and commonly used reference genes, we chose six housekeeping genes from these 11 genes as candidate internal controls, including RS9, RL12, RL13, EIF3, EF1 and 18S.
Clusters of genes in 12 transcriptome of two tissues in response to Vibrio and Cd2+ in S. constricta. control group post 12 h in the gill (G12N) and in the hepatopancreas (L12N), control group post 48 h in the gill (G48N) and in the hepatopancreas (L48N), vibrio treatment post 12 h in the gill (G12B) and in the hepatopancreas (L12B), vibrio treatment post 48 h in the gill (G48B) and in the hepatopancreas (L48B), Cd2+ treatment post 12 h in the gill (G12Cd) and in the hepatopancreas (L12Cd), and Cd2+ treatment post 12 h in the gill (G48Cd) and in the hepatopancreas (L48Cd)
qRT-PCR specificity and efficiency
A total of eight candidate genes were selected to normalize the gene expression levels in S. constricta using qRT-PCR. The specificity of the primers was confirmed using a melting curve analysis by single peak melting curves of the qPCR products without primer-dimer formation (Supplementary Figure S1). The amplification efficiencies of primer pairs ranged from 95.6% of RL13 to 104.4% of ACT (Table 1), and the standard curves displayed highly linear correlation coefficients (R2), which were ranged between 0.921 and 0.998.
Analysis of gene transcription stability
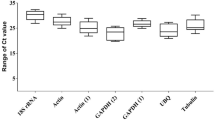
We tested eight candidate housekeeping genes for expression stability in vibrio infected razor clams at five different time points (0, 6, 12, 24 and 48 h) and five different tissues, including gill, hepatopancreas, siphon, hemocytes and foot from five individuals with three biological replicates. The raw data of qRT-PCR was performed using boxplot analysis (Fig. 2) and listed in the supplementary Table S2 and Table S3. The results showed 18S was the most abundant housekeeping gene with the lowest mean CT value of 9 and 13 in both vibrio-infected samples and tissues, respectively. EIF3 was the least expressed housekeeping gene with the highest mean CT value of 23 in vibrio-infected samples. The results also revealed that genes were characterized by the smaller variation in transcript levels in vibrio-infected samples than in tissues. Among them, EIF3 gene had the smallest variation in transcript levels either in vibrio-infected samples or in tissues. In addition, RS9, RL12, RL13, EIF3 and EF1 had relatively stable expression across all the tissues and vibrio-treated samples tested.
In this study, we used three algorithms to select better-suited housekeeping genes for higher-accuracy stability rankings. The results in different algorithms differed, even in the most appropriate reference genes (Fig. 3; Table 2). But the first four or five stably expressed genes showed little variation in each software. In addition, TUB and 18S were identified to be the least stable expressed gene in vibrio-infected samples and tissues, respectively. Among vibrio-infected samples, EIF3 and RS9 were the most stable reference genes analyzed by geNorm and NormFinder (Fig. 3a). In addition, all candidate reference genes could be considered stable for vibrio-infected samples according to geNorm stability values (M) below 0.5 (Hellemans et al. 2007). In tissue samples, geNorm identified RS9 and EF1 to be the most stable reference genes, followed by RL13, EIF3, RL12, ACT, TUB and ultimately 18S. Except TUB and 18S, the stability values (M) of other candidate housekeeping genes were below 0.5 (Fig. 3b). It showed that five genes selected from the stable expression data of transcriptome except 18S were validated to express stably. NormFinder identified RL13 as the most stable reference gene tested, followed by RL12, ACT, EF1, RS9, EIF3, TUB and 18S. BestKeeper calculates the standard deviation (SD) to identify the stable reference genes with the SD value inversely proportional to the stability of expression. As Table 3 shown, EF1 is the most stably expressed in vibrio-infected condition, while EIF3 is the most stably expressed in tissues. Similar to the results of geNorm, genes in vibrio-infected samples expressed more stable than those in tissues based on the SD distribution in the two groups. According to the integration of four algorithms’ results by RefFinder (Fig. 4), EIF3, RS9 and ACT were the three most stable reference genes in vibrio-infected conditions, while RL13, RS9, EF1 and EIF3 were the four most stable reference genes in five different tissues of S. constricta.
Determination of the optimal number of reference genes
In regards to determine the optimum number of reference genes, geNorm used a pairwise number variation Vn/Vn + 1 analysis. A suggested cut-off value for the pairwise variation is 0.15. The result below it suggesting that the inclusion of an additional reference gene is not required (Vandesompele et al. 2002). In vibrio-infected samples, the V2/3 value was 0.063 suggesting that EIF3 and RS9 (on the basis of the M value) were enough for normalization (Fig. 5). Compared to V2/3 value, the V3/4 value was a little lower, which indicated the inclusion of a third reference gene could improve slightly stability of normalization. In tissues, the V2/3 value was 0.138 suggesting that RS9 and EF1 were enough for normalization. BestKeeper does not provide a measurement for the optimal number of reference genes. We identified the three genes with the highest correlation coefficient (r) as the minimal number of reference genes according to the authors’ advice (Pfaffl et al. 2004).
Validation of the reference genes
Analysis of the above algorithms indicated RS9 as the top ranked reference gene in terms of stability in gene-expression for tissues and Vibrio-infected treatments. And EIF3 expressed more stably under Vibrio infection. In contrary, TUB and 18S were found to be the least stable. Therefore, we chose i-type lysozyme and C1qDC to validate the applicability of these reference genes for the normalization, either as single or in combination in response to Vibrio infection at different time points. The results showed the expression patterns of lysozyme and C1qDC differed normalizing by the most and least stable reference genes (Fig. 6). For lysozyme, the expression showed higher consistency compared to normalization by all the reference genes except the 12 h-treatment group. It showed significant variation when TUB and 18S were as normalizer compared to normalization by RS9 and EIF3 and their combinations in the 12 h-treatment group (Fig. 6a) At 48 h, the expression profile normalized by 18S showed significant variation. As the qRT-PCR data of C1qDC shown, TUB was the least stable reference gene as normalizer in the 12 h-treatment group and 48 h-treatment group (Fig. 6b).
Variation in lysozyme and C1qDC gene-expression data normalized by different reference genes and their combinations. Lysozyme (a) and C1qDC (b) expression pattern under Vibrio challenge at different time points. The five kinds of bars indicate the two genes’ expression levels normalized by different reference genes (RS9, EIF3, TUB and 18S) and their combination, respectively
Discussion
Quantitative real-time PCR is a popular technology to measure the expression levels of target genes under all kinds of experimental conditions, including different tissues, different treatments or different developmental stage, on account of its high sensitivity, specificity and cost-efficiency (Ginzinger 2002). Vibrio sp. is commonly detected in marine and estuarine environments (Thompson et al. 2004) and is one of the main pathogens to bivalves and is responsible for massive mortality, including S. constricta. Hence, exploring the expression patterns of immune-related genes is imperative. However, the accuracy of gene expression levels by qRT-PCR needs to be normalized with an internal control. The internal control should have a relatively stable expression level regardless of experimental variations. There are little information about S. constricta housekeeping genes and no study to examine the suitability of potential housekeeping genes for qRT-PCR in S. constricta. In this study, we clustered gene expression data of 12 transcriptome into ten clusters based on similar expression patterns. About half of these clusters showed the expression patterns had significant tissue specificity. The perfect internal control gene should show stable expression regardless of experimental variations. For this purpose, the cluster 5 was chose to be the candidate database with the stable gene expression patterns in spite of different tissues and different stresses. We identified 55 orthologs of housekeeping genes in S. constricta blasting to those of C. gigas, and combined the CV values to select six candidate reference genes. In addition, two genes commonly used as internal control genes in Mollusca were added in the list of candidate reference genes. These eight genes were validated the expression stabilities by qRT-PCR in both vibrio-infected samples and different tissues of S. constricta.
We selected three algorithms (geNorm, NormFinder and BestKeeper) to evaluate the stability of the expression levels of eight candidate reference genes. Under vibrio infected conditions, EIF3 was the most stable internal control genes followed by RS9 based on the consistent results of the three methods. The pairwise variation analysis performed by geNorm indicated that EIF3 and RS9 were enough for normalization. Among the five different tissues, the results from different methods differed. The integrative results by RefFinder showed that RL13 was the most stable internal control genes followed by RS9. The pairwise variation analysis indicated that RS9 and EF1 were enough for normalization. According to the conclusion that genes with M ≤ 0.5 calculated by geNorm have high reference target stability, all the genes tested could be considered stable in vibrio infected conditions, and all the genes except 18S expressed stably among tissues. Almost all the genes with stable expression may result from the filter criterion that the reference genes selected from the gills and hepatopancreas transcriptome in response to Vibrio infection with stable expression. Meanwhile, we used the CV values to measure the stability of genes selected relying on the transcriptome data. The ranking of these genes based on the transcriptome data was similar to that validated in qRT-PCR, especially the least stable gene, 18S. It indicated that the cluster analysis of transcriptome combining the CV ranking is an important reference for candidate internal controls selection used in qRT-PCR.
It was apparent that the best ranked reference gene for one treatment was not applicable to the other treatments. For experimental convenient, it was imperative to identify the reference genes that show acceptable stability in expression across various treatments. RS9 was the most stable reference gene in both vibrio-infected samples and different tissues based on the results. It belongs to small subunit ribosomal proteins, and takes part in ribosome biogenesis in all cell types, as a common internal control for qRT-PCR in human cells (Aychek et al. 2008). Ribosomal protein-encoding genes are widely used as internal controls in humans and other animals, even plants (Hsiao et al. 2001; Thorrez et al. 2008; Barsalobres-Cavallari et al. 2009). In this study, another two ribosomal proteins, RL12 and RL13 were also tested for the stable expression, which belong to large subunit ribosomal proteins. RL13 had the most stable expression among different tissues based on the data of Normfinder and RefFinder, which was found to be one of the best reference genes in mustard aphid (Koramutla et al. 2016). RL12 had lower stability either in different tissues or under vibrio challenge. This result may reflect their different functions in protein synthesis. In marine bivalves, the stability and suitability of the ribosomal protein-encoding genes as reference genes has been validated in the previous studies. RL7 and RS18 were found to be the most stably expressed genes during the development of Crassostrea gigas larvae by OsHV-1 infection (Du et al. 2013). RS18 gene was also found to be the most stably expressed gene in Mya arenaria after Vibrio challenge (Mateo et al. 2010). 40S ribosomal protein s20 (RPS20) was validated to express most stably in Ruditapes philippinarum hemocytes in response to copper stress (Volland et al. 2017). Our study validated that a certain ribosomal protein gene can express stably under environmental challenge again.
EIF3 was the most stable gene under vibrio infection, which is required for several steps in the initiation of protein synthesis, targets a subset of mRNA participating kinds of cellular processes (Masutani et al. 2007; Lee et al. 2015). It was validated that EIF3 was not regulated by experimental conditions in animals (Kouadjo et al. 2007; Zhang et al. 2012) and plants (Shi et al. 2012). However, the expression of EIF3 was variable depending on the different tissues in this study, indicating that the protein synthesis is different among the tissues.
EF1, ACT, 18S and TUB are common reference genes with validation or without validation in papers of bivalves (Volland et al. 2017) and used in many organisms, including animals and plants. ACT and 18S genes were the most common internal controls for qRT-PCR in S. constricta under a range of experimental conditions including different tissues, pathogen infection and ocean acidification (Li et al. 2011; Niu et al. 2015; Peng et al. 2017). ACT expressed at moderately abundant levels in all the tissues and encodes a ubiquitous cytoskeleton protein. At the present study, ACT showed variable in different tissues and more stable in gills under vibrio infection at different time points. Although ACT is one of the first and often used reference genes, it has been showed to vary considerably and be unsuitable as internal control to normalize the gene expression analysis in some cases (Selvey et al. 2001). The 18S is a component of the small subunit of eukaryotic ribosomes (40S) involved in the translation process (Boujedidi et al. 2012), which was used for a reference gene in many previous studies (Zhang et al. 2011; Banni et al. 2014). However, in this study, the 18S gene was the least stable gene across the two datasets. It has also been found inappropriate for normalization of qRT-PCR analysis in C. gigas (Du et al. 2013), R. philippinarum (Volland et al. 2017) and Rapana venosa (Song et al. 2017). In S. constricta, the expression of 18S should be validated before it is used for the internal control for qRT-PCR. EF1 is a member of the G-protein family and plays a key role in protein translation (Browne and Proud 2002), which is frequently used in C. gigas as the reference gene, and is the most stable reference gene in hemocytes of flat oyster Ostrea edulis (Morga et al. 2010) and in gametogenesis of Mytilus edulis (Cubero-Leon et al. 2012). The stability of expression of EF1 in razor clam remains unknown. Based on the results, EF1 has a moderately stable expression in different tissues and vibrio-infected samples of S. constricta. TUB is another least stable candidate gene in our study, which plays a crucial role in cell structural maintenance. TUB was found to be the most stable gene in development stages of Hippoglossus hippoglossus (Fernandes et al. 2008) and in goat (Costa et al. 2012). Taken together, it indicates that candidate reference genes differ among species or treatments.
To verify the actual utility of validated reference genes in this study, the expression profile of lysozyme and C1qDC (MF289989) was conducted in the razor clam under Vibrio infection at different time-points. Lysozyme and complement component play important roles in protecting animals against bacteria pathogens (Callewaert and Michiels 2010; Cui et al. 2018). Their mRNA expressions were induced following bacterial challenges in other mollusk (Ren et al. 2012; Bathige et al. 2013; Liu et al. 2014a, b). Here, the expressions of the two genes were remarkably expressed at 12 h after infection by Vibrio normalized by the four selected reference genes. However, the least stable genes TUB and 18S showed significant variation compared with the most stable genes RS9 and EIF3. Even in the result of C1qDC expression pattern at 48 h after infection normalized by 18S showed a very different profile than others. Therefore, these results further proved the importance of choosing the appropriate reference genes.
Conclusions
There were just 18S and β-actin commonly used for normalization of qRT-PCR data in S. constricta without validation. We combined the expression patterns of transcriptome to select candidate housekeeping genes, which suggested that the cluster analysis of transcriptome combining the CV values of FPKM of candidate reference genes is an important reference. In addition, three algorithms were used for the decision of the most appropriate internal control, and RS9 was identified as the recommended reference gene for qRT-PCR analysis under Vibrio infection and in different tissues. RL13 and EIF3 were the most stable reference genes in different tissues and vibrio-infected samples, respectively. The results of this study provide the primary reference to select the internal controls for qRT-PCR in S. constricta, and will aid further studies of clam immune response.
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Aychek T, Miller K, Sagi-Assif O, Levy-Nissenbaum O, Israeli-Amit M, Pasmanik-Chor M, Jacob-Hirsch J, Amariglio N, Rechavi G, Witz IP (2008) E-selectin regulates gene expression in metastatic colorectal carcinoma cells and enhances HMGB1 release. Int J Cancer 123:1741–1750
Banni M, Hajer A, Sforzini S, Oliveri C, Boussetta H, Viarengo A (2014) Transcriptional expression levels and biochemical markers of oxidative stress in Mytilus galloprovincialis exposed to nickel and heat stress. Comp Biochem Phy C 160:23–29
Barsalobres-Cavallari CF, Severino FE, Maluf MP, Maia IG (2009) Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Mol Biol 10:1
Bathige SDNK., Umasuthan N, Kasthuri SR, Whang I, Lim BS, Nam BH, Lee J (2013) A bifunctional invertebrate-type lysozyme from the disk abalone, Haliotis discus discus: genome organization, transcriptional profiling and biological activities of recombinant protein. Dev Comp Immunol 41:282–294
Boujedidi H, Bouchet-Delbos L, Cassard-Doulcier AM, Njiké-Nakseu M, Maitre S, Prévot S, Dagher I, Agostini H, Voican CS, Emilie D (2012) Housekeeping gene variability in the liver of alcoholic patients. Alcohol Clin Exp Res 36:258–266
Browne GJ, Proud CG (2002) Regulation of peptide-chain elongation in mammalian cells. FEBS J 269:5360–5368
Bustin S, Benes V, Nolan T, Pfaffl M (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34:597–601
Callewaert L, Michiels CW (2010) Lysozymes in the animal kingdom. J Biosci 35:127–160
Costa J, Passos M, Leitão C, Vasconcelos G, Saraiva M, Figueiredo J, Van den Hurk R, Silva J (2012) Levels of mRNA for bone morphogenetic proteins, their receptors and SMADs in goat ovarian follicles grown in vivo and in vitro. Reprod Fert Develop 24:723–732
Cubero-Leon E, Ciocan CM, Minier C, Rotchell JM (2012) Reference gene selection for qPCR in mussel, Mytilus edulis, during gametogenesis and exogenous estrogen exposure. Environ Sci Pollut R 19:2728–2733
Cui Y, Wei Z, Shen Y, Li C, Shao Y, Zhang W, Zhao X (2018) A novel C1q-domain-containing protein from razor clam Sinonovacula constricta mediates G-bacterial agglutination as a pattern recognition receptor. Dev Comp Immunol 79:166–174
Dhar AK, Bowers RM, Licon KS, Veazey G, Read B (2009) Validation of reference genes for quantitative measurement of immune gene expression in shrimp. Mol Immunol 46:1688–1695
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112–119
Du Y, Zhang L, Xu F, Huang B, Zhang G, Li L (2013) Validation of housekeeping genes as internal controls for studying gene expression during Pacific oyster (Crassostrea gigas) development by quantitative real-time PCR. Fish Shellfish Immunol 34:939–945
Fernandes JM, Mommens M, Hagen Ø, Babiak I, Solberg C (2008) Selection of suitable reference genes for real-time PCR studies of Atlantic halibut development. Comp Biochem Phy B 150:23–32
Filby AL, Tyler CR (2007) Appropriate’housekeeping’genes for use in expression profiling the effects of environmental estrogens in fish. BMC Mol Biol 8:10
Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30:503–512
Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19
Hsiao L-L, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, Dillon W, Lee KF, Clark KE, Haverty P (2001) A compendium of gene expression in normal human tissues. Physiol Genom 7:97–104
Ingerslev H-C, Pettersen EF, Jakobsen RA, Petersen CB, Wergeland HI (2006) Expression profiling and validation of reference gene candidates in immune relevant tissues and cells from Atlantic salmon (Salmo salar L.). Mol Immunol 43:1194–1201
Koramutla MK, Aminedi R, Bhattacharya R (2016) Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci Rep 6:25883
Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J (2007) Housekeeping and tissue-specific genes in mouse tissues. BMC Genom 8:127
Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L (2006) The real-time polymerase chain reaction. Mol Aspects Med 27:95–125
Lee AS, Kranzusch PJ, Cate JH (2015) eIF3 targets cell proliferation mRNAs for translational activation or repression. Nature 522:111
Li C, Li H, Su X, Li T (2011) Identification and characterization of a clam ferritin from Sinonovacula constricta. Fish Shellfish Immunol 30:1147–1151
Liu N, Pan L, Gong X, Tao Y, Hu Y, Miao J (2014a) Effects of benzo (a) pyrene on differentially expressed genes and haemocyte parameters of the clam Venerupis philippinarum. Ecotoxicology 23:122–132
Liu HH, Xiang LX, Shao JZ (2014b) A novel C1q-domain-containing (C1qDC) protein from Mytilus coruscus with the transcriptional analysis against marine pathogens and heavy metals. Dev Comp Immunol 44:70–75
Masutani M, Sonenberg N, Yokoyama S, Imataka H (2007) Reconstitution reveals the functional core of mammalian eIF3. EMBO J 26:3373–3383
Mateo DR, Greenwood SJ, Araya MT, Berthe FC, Johnson GR, Siah A (2010) Differential gene expression of γ-actin, Toll-like receptor 2 (TLR-2) and interleukin-1 receptor-associated kinase 4 (IRAK-4) in Mya arenaria haemocytes induced by in vivo infections with two Vibrio splendidus strains. Dev Comp Immunol 34:710–714
Meistertzheim AL, Tanguy A, Moraga D, Thébault MT (2007) Identification of differentially expressed genes of the Pacific oyster Crassostrea gigas exposed to prolonged thermal stress. FEBS J 274:6392–6402
Morga B, Arzul I, Faury N, Renault T (2010) Identification of genes from flat oyster Ostrea edulis as suitable housekeeping genes for quantitative real time PCR. Fish Shellfish Immunol 29:937–945
Niu D, Wang L, Bai Z, Xie S, Zhao H, Li J (2015) Identification and expression characterization of the myostatin (MSTN) gene and association analysis with growth traits in the razor clam Sinonovacula constricta. Gene 555:297–304
Niu D, Du Y, Wang Z, Xie S, Nguyen H, Dong Z, Shen H, Li J (2017) Construction of the First high-density genetic linkage map and analysis of quantitative trait loci for growth-related traits in Sinonovacula constricta. Mar Biotechnol 19:1–9
Peng C, Zhao X, Liu S, Shi W, Han Y, Guo C, Peng X, Chai X, Liu G (2017) Ocean acidification alters the burrowing behaviour, Ca2+/Mg2+-ATPase activity, metabolism, and gene expression of a bivalve species, Sinonovacula constricta. Mar Ecol Prog Ser 575:107–117
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Ran Z, Chen H, Ran Y, Yu S, Li S, Xu J, Liao K, Yu X, Zhong Y, Ye M (2017) Fatty acid and sterol changes in razor clam Sinonovacula constricta (Lamarck 1818) reared at different salinities. Aquaculture 473:493–500
Ren Q, Qi YL, Hui KM, Zhang Z, Zhang CY, Wang W (2012) Four invertebrate-type lysozyme genes from triangle-shell pearl mussel (Hyriopsis cumingii). Fish Shellfish Immunol 33:909–915
Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7:909–912
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Selvey S, Thompson EW, Matthaei K, Lea RA, Irving M, Griffiths LR (2001) β-Actin—an unsuitable internal control for RT-PCR. Mol Cell Probes 15:307–311
Shi J, Liu M, Shi J, Zheng G, Wang Y, Wang J, Chen Y, Lu C, Yin W (2012) Reference gene selection for qPCR in Ammopiptanthus mongolicus under abiotic stresses and expression analysis of seven ROS-scavenging enzyme genes. Plant Cell Rep 31:1245–1254
Song H, Dang X, He Y-q, Zhang T, Wang H-y (2017) Selection of housekeeping genes as internal controls for quantitative RT-PCR analysis of the veined rapa whelk (Rapana venosa). PeerJ 5:e3398
Stürzenbaum SR, Kille P (2001) Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Phy B 130:281–289
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295
Thompson FL, Iida T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol R 68:403–431
Thorrez L, Van Deun K, Tranchevent L-C, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F (2008) Using ribosomal protein genes as reference: a tale of caution. PLoS One 3:e1854
Valenzuela-Castillo A, Mendoza-Cano F, Enríquez-Espinosa T, Grijalva-Chon JM, Sánchez-Paz A (2017) Selection and validation of candidate reference genes for quantitative real-time PCR studies in the shrimp Penaeus vannamei under viral infection. Mol Cell Probes 33:42–50
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1
Volland M, Blasco J, Hampel M (2017) Validation of reference genes for RT-qPCR in marine bivalve ecotoxicology: systematic review and case study using copper treated primary Ruditapes philippinarum hemocytes. Aquatic Toxicol 185:86–94
Wang Z, Shao Y, Li C, Zhang W, Duan X, Zhao X, Qiu Q, Jin C (2016) RNA-seq analysis revealed ROS-mediated related genes involved in cadmium detoxification in the razor clam Sinonovacula constricta. Fish Shellfish Immunol 57:350–361
Zhang Y, He X, Yu Z (2011) Two homologues of inhibitor of NF-kappa B (IκB) are involved in the immune defense of the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol 30:1354–1361
Zhang Y, Chen D, Smith MA, Zhang B, Pan X (2012) Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS One 7:e31849
Zhao X, Duan X, Wang Z, Zhang W, Li Y, Jin C, Xiong J, Li C (2017) Comparative transcriptome analysis of Sinonovacula constricta in gills and hepatopancreas in response to Vibrio parahaemolyticus infection. Fish Shellfish Immunol 67:523–535
Zheng W-j, Sun L (2011) Evaluation of housekeeping genes as references for quantitative real time RT-PCR analysis of gene expression in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol 30:638–645
Acknowledgements
This work was financially supported by Zhejiang Major Program of Science and Technology (2016C02055-9, 2015C32004), Natural Science Foundation of Ningbo (2015C10009, 2015C10062), Start Research Fund projects for Xuelin Zhao, Ningbo University Fund (XYL17010) and the K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Xuelin Zhao declares that she does not have conflict of interest. Jianping Fu declares that he does not have conflict of interest. Liting Jiang declares that she does not have conflict of interest. Weiwei Zhang declares that she does not have conflict of interest. Yina Shao declares that hshe does not have conflict of interest. Chunhua Jin declares that he does not have conflict of interest. Jinbo Xiong declares that he does not have conflict of interest. Chenghua Li declares that he does not have conflict of interest.
Ethical approval
S. constricta is a commercially cultured animals, and all the experiments were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The study protocol was approved by the Experimental Animal Ethics Committee of Ningbo University, China.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
Melting curves for the eight candidate reference genes. (TIF 2487 KB)
Supplementary Table S1
Details of orthologues of clam Sinonovacula constricta blasting with housekeeping genes in Crassostrea gigas. (XLSX 11 KB)
Supplementary Table S2
Expression values (CT) of candidate reference genes in vibrio-infected samples. (XLSX 10 KB)
Supplementary Table S3
Expression values (CT) of candidate reference genes in five tissues. (XLSX 10 KB)
Rights and permissions
About this article
Cite this article
Zhao, X., Fu, J., Jiang, L. et al. Transcriptome-based identification of the optimal reference genes as internal controls for quantitative RT-PCR in razor clam (Sinonovacula constricta). Genes Genom 40, 603–613 (2018). https://doi.org/10.1007/s13258-018-0661-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0661-9