Abstract
Using cDNA microarrays, we have conducted a systematic characterization of global gene expression in v-raf or v-raf/v-myc transformed rat liver epithelial (RLE) cell lines exhibiting both non-metastatic and metastatic phenotypes. Seven transformed cell lines were compared with the non-transformed parental RLE cell. The hierarchical clustering analysis of gene expression profiles revealed two groups reflecting the in vivo metastatic potential of the cells. Surprisingly, one non-metastatic cell line T1 was co-clustered with metastatic cell lines, suggesting that T1 underwent significant genetic changes. The T1 cell line was further compared against all the metastatic cell lines in order to reveal the critical genes required for metastatic conversion but not expressed in the T1. These data demonstrated that expression of genes involved in apoptosis and immune cell homing were altered in all metastatic cell lines. Survival of both intravasated cells in circulation systems and extravasated cells in a new tissue environment might be critical for the final step in the metastatic process. Our study provides gene expression signatures consistent with two critical events in the metastatic process, namely, the acquisition of early homing capacity and increased survival potential of the tumor cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The somatic mutation theory of carcinogenesis, currently the most widely adopted theory of tumor development, predicts that cancer arises from a step-wise accumulation of genetic changes that ultimately destroy the host (Hanahan and Weinberg 2000). The acquisition by the tumor of the capacity to invade and metastasize, the last step in the oncogenic process, is a common feature of all aggressive and lethal tumors. A hallmark of a metastatic tumor is the ability of the tumor cells to cross several biological barriers such as extracellular matrix (ECM), host connective tissue stroma, and junctions between vascular cells, as well as to survive and grow in tissue environments distant from the primary location. Modulation of a number of biological processes such as activation of extracelluar proteases, alteration of binding specificities of cadherins, CAM, and integrins are clearly important features of the metastatic phenotype (Liotta and Kohn 2001). However, re-examination of the data on the molecular mechanism(s) of tumor metastasis has formulated the notion that the metastatic process is driven by combined action of multiple genes involved in tumorigenesis (Savagner 2001). Moreover, it was suggested that the mutant alleles acquired early in tumor development might confer the proclivity to metastasize at later stages of tumorigenesis (Bernards and Weinberg 2002). The capacity of neoplastic cells to traverse biological barriers is frequently associated with a morphological transformation known as epithelial to mesenchymal transition (EMT), a phenomenon that is also observed during embryonic development (Savagner 2001). The structural characteristics of the mature epithelial phenotype include polarization of the cell surface into apical and basolateral domains, and formation of junctional complexes that mediate strong intercellular adhesions (Cereijido et al. 2000). The multiprotein complexes that comprise the tight junctions includes the transmembrane proteins occludin and claudins as well as a family of cytoplasmic proteins (ZO-1 and -3) that connect the junctions to the microfilaments and regulate its assembly (Tsukita et al. 2001). Similarly, the adherens junction in which E-cadherin forms homophilic contacts between adjacent cells and the catenins provide the connection between the cytoplasmic tail of E-cadherin and the sub-membranous cytoskeleton (Nagafuchi 2001). E-cadherin is a critical factor in the development of cell polarity and the establishment of solid adhesions, as well as being involved in growth control via contact inhibition and the maintenance of the overall differentiation state of the cells (Vleminckx and Kemler 1999). Although the mechanism(s) by which the highly structured epithelial cell transits into a mesenchymal phenotype during the EMT is not fully understood, a sustained cooperation between TGF-β1 signaling and oncogenic Ras has been identified as a critical event in driving both EMT and metastasis (Oft et al. 1996, 1998, 2002). In contrast, a number of agents including HGF, FGFs and TGF-β1 alone are capable of inducing an EMT phenotype that is fully reversible after withdrawal of anyone of these factors (Janda et al. 2002). Employing a combination of an in vitro/in vivo carcinogenesis model consisting of Ha-Ras transformed mammary epithelial cells possessing a competent TGF-β1 signaling system, Janda et al. (2002) were able to demonstrate with specific inhibitors and effector-specific Ras mutants that activation of Raf/MAPK is required for EMT. In addition, they showed that the activation of phosphatidylinositol 3-kinase (PI3K) induced scattering and protected against TGF-β1-induced apoptosis, and that activation of either the PI3K or the Raf/MAPK pathway is sufficient for tumorigenesis, whereas EMT in vivo and metastasis required a hyperactive Raf/MAPK pathway. These data clearly demonstrate the critical role of the Raf/MAPK pathway in the metastatic process.
It has been demonstrated that rat liver epithelial (RLE) cells transduced with v-raf undergo EMT (Williams et al. 1992; Garfield et al. 1988). Six cloned cell lines (R3611-T lines) derived from a nude mouse xenograft of the v-raf transformed RLE cells showed different degrees of spontaneous metastatic potential when transplanted in nude mice (Bisgaard et al. 1997). All the R3611-T lines expressed high levels of both v-raf and c-raf as well as TGF-β1 and had competent TGF-β signaling (Hampton et al. 1990; Huggett et al. 1990). Furthermore, the metastatic conversion of the v-raf transformed RLE cells correlated with upregulation of urokinase-type plasminogen activator receptor (Bisgaard et al. 1997).
In this paper, we characterized gene expression profiles in the transformed RLE-derived cell lines and identified the genes that are highly correlated with invasive and metastatic potentials using two different cDNA microarrays.
Materials and methods
Cell culture
The RLE cell lines (T1, T2, T3, T4, T%, T7, and RJ2) used for study are described in the previous study (Bisgaard et al. 1997). The cell lines were grown in DMEM/F12 medium supplemented with glutamine (2 mM) and 10 % fetal bovine serum. To minimize the contribution of variations in culture condition or cell density to differential gene expression, the cells were harvested at about 80 % confluence level in plates.
Isolation of RNA and cDNA microarray
Total RNAs were isolated from the seven transformed RLE cell lines (T1, T2, T3, T4, T5, T7 and RJ2) using Trizol (Invitrogen) as described in the supplier’s instruction. Total RNA for reference RNA was also extracted from the parental RLE cell line. The RNAs (~5 μg) were electrophoresed in a 1.3 % agarose gel containing formaldehyde, and stained with ethidium bromide to determine the integrity of total RNAs. The customized 6.9 K rat cDNA microarrays and 9 K mouse cDNA microarrays were used for this study.
Microarray experiments
Labeling of total RNA for microarray analysis was performed using indirect methods as described below. Briefly, total RNA (20 μg) was reverse-transcribed for 1 h at 42 °C in a 50 μl reaction containing 2.5 μl of 20× dNTP mixture with 6 mM aminoallyl-dUTP (Sigma, St. Louis, MO), 10 μl of 5× first strand buffer, 5 μl of 0.1 M DDT, 1 μl of SUPERase In™ (10,000 unit, Ambion, Austin, TX), and 3 μl of Superscript II reverse transcriptase (200 μg/μl, Invitrogen). The synthesized cDNAs were precipitated with ethanol and resuspended in 10 μl of 0.1 M NaHCO3. For chemical coupling of aminoallyl groups in cDNAs with Cy-3 or Cy-5 dyes, monoreactive Cy-3 or Cy-5 dye (Amersham Pharmacia Biotech, Piscataway, CA) was dissolved in 0.1 M NaHCO3 and mixed with cDNAs. The mixture was incubated for 1 h at room temperature. Labeled cDNAs were precipitated with ethanol and purified using Nucleospin Extraction Kit (Clontech, Palo Alto, CA). The labeled cDNA probes were combined and concentrated to 11 μl by a Centricon-30 micro-concentrator (Millipore, Bedford, MA). The hybridization mix consisted of human COT-1 DNA (0.56 μg/μl, Boehringer Mannheim), poly-dA DNA (0.56 μg/μl, Pharmacia), yeast tRNA (0.22 μg/μl, Sigma), 5× SSC, 0.1 % SDS, and 25 % formamide in final volume of 28 μl. Before hybridization, the cDNA microarray slides were incubated with prehybridization buffer (5× SSC, 0.1 % SDS, and 1 % BSA) for 1 h at 42 °C and washed in water and isopropanol. Hybridization was carried out at 42 °C for overnight in a water bath. Before scanning, slides were washed in 1× SSC with 0.1 % SDS for 2 min, then 1× SSC, 0.2× SSC, and 0.05× SSC, sequentially for 1 min. Hybridized arrays were scanned on a GenePix 4000A scanner (Axon Instrument, Union City, CA), and the resulting images were analyzed via GenePix Pro v3.0 (Axon Instrument) as described in the manufacturer’s manual. Each sample was examined at least twice by switching fluorescent dye Cy-5 and Cy-3. The variance in the duplicated fluorescence ratio measurements approached the minimum when the fluorescence signal was greater than approximately 0.4 % of the measurable total signal dynamic range above background in both channels of the hybridization. Average values of two experiments on each sample were obtained and values that were missing in two experiments were excluded from further analysis.
Data analysis
For cluster analysis, each Cy-3 or Cy-5 labeled experiments in 6.9 K rat cDNA microarrays and 9 K mouse cDNA microarrays were independently analyzed using BRB ArrayTool (http://linus.nci.nih.gov/BRB-ArrayTools.html). Before clustering, fluorescent intensity ratios of each spots were log2 transformed and each array is normalized separately by subtracting the median log-ratio from all log-ratios on that array. Hierarchical clustering algorithm was applied to cell lines using Pearson correlation coefficient as the measure of similarity and average linkage clustering. Examining cluster reproducibility was performed as described in user’s manual. Briefly, the reproducibility test is based on perturbing the normalized log-ratios by adding normally distributed random numbers with mean zero and variance equal to the estimated experimental noise variance. After the perturbation, the data was re-cluster to obtain clusters and comparison made between the original tree and the perturbed tree. The proportion of paired members remained in the same cluster after perturbation and re-clustering was computed (one represents a pair remain in the same cluster and zero represents members of a pair split to different cluster). The comparison was repeated 1,000 times and the proportion of remained pairs in the same cluster was averaged over the perturbed data sets resulting in robustness (R)-index. Discrepancy (D)-index was derived by determining a best match cluster in the perturbed data, defined as the one having the largest number of samples included in the original cluster. The number of discrepancies was computed as the number of samples in the best match cluster but not in the original cluster and the number of samples in the original cluster but not in the best match cluster. This number of discrepancies is computed for each original cluster and averaged over the repetitions. For analysis of average expression ratios from duplicated experiments from rat 6.9 K and mouse 9 K microarrays, genes with an expression ratio that has at least threefold ratio difference relative to the reference cell line in at least one cell lines were further selected. The hierarchical cluster analysis of combined rat and mouse data was performed as described. To select out the list of genes that discriminate observed clusters of cell lines, an F test was used to select genes which are differentially expressed between the two clusters at the given significance level (p value <0.01). Principal component analysis (PCA) was performed using GeneLinker™ Gold (Molecular Mining Corporation, Kingston, ON) as described in user’s manual.
Results and discussion
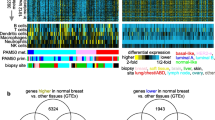
In order to identify the genes that are highly correlated with invasive and metastatic potentials, we characterized gene expression profiles in the transformed RLE-derived cell lines by using two different cDNA microarrays. Rat microarrays contained about 6,900 cDNA elements and mouse GEM2 cDNA microarrays contained about 9,000 cDNA elements on glass microscope slides. Total RNAs were purified from cell cultures when the cells were about 80 % confluent and used to prepare fluorescent cDNA probes with the Cy-3 or Cy-5 dyes by using an indirect labeling method. Using a matrix of Pearson correlation coefficients from the complete pair-wise comparison of all microarray experiments, the seven transformed RLE cell lines were displayed in hierarchical clustering dendrogram on the basis of similarity in gene expression pattern (Fig. 1a, b). Due to the intrinsic nature of the cluster algorithm that always detects clusters, even on random data, we used two independent approaches to test the reproducibility of cluster prediction in the transformed RLE cell lines. First, we replicated experiments using two different cDNA microarrays, 6.9 K rat and 9 K mouse microarrays. All experiments using rat or mouse microarrays were duplicated by labeling total RNAs from each cell line with the reciprocal fluorochrome. Four microarray experiments were performed with total RNA from the cell lines as experimental samples and the parental RLE cell line as a reference. All replicated microarray experiments were analyzed independently using BRB ArrayTools. All hierarchical cluster dendrograms from four experiments were consistent, and showed no discrepancy for the members of two clusters (Fig. 1a, b). As expected, all metastatic cell lines, RJ3611-T2lacZ (T2), RJ3611-T3lacZ (T3), RJ3611-T7lacZ (T7), and RJ2-14lacZ (RJ2), were clustered together and separated from non-metastatic cell lines, RJ3611-T4lacZ (T4) and RJ3611-T5lacZ (T5). Interestingly, non-metastatic cell line RJ3611-T1lacZ (T1) was co-clustered with metastatic cell lines. These results strongly suggest that the two major clusters of cell lines identified in this study are not due to artifacts from experimental errors or data processing.
Hierarchical cluster analysis of gene expression of transformed RLE cell lines. a Dendrogram representing similarities in gene expression patterns between cell lines in rat cDNA microarray experiments. All metastatic cell lines clustered together in large subgroup are highlighted in red. Duplicated experiments by using reciprocal fluorochrome were independently analyzed. Robustness index (R = 1.0) indicates perfect reproducibility of two clusters in both experiments. Discrepancy index (D = 0.0) indicates no discrepancy of membership in each cluster after perturbation of data. S.d. indicates standard deviation used for perturbation of data. Blue lines in dendrogram represent cut-off of clusters for reproducibility test. b Dendrogram representing similarities in gene expression patterns between cell lines in mouse cDNA microarray experiments. c Hierarchical clustering dendrogram of combined gene expression data. Data are presented in pseudo-colored matrix format; each column represents a cell line, and each row a cDNA. Red represents expression greater than RLE, green less than RLE, and gray missing or excluded data. Color intensity represents the magnitude of expression ratio as indicated. Genes with an expression ratio that has at least threefold ratio difference relative to RLE in at least 1 cell line were selected for hierarchical analysis. Red bar represents gene cluster enriched in genes involved in cell-to-cell interaction and adhesion. d Principal component analysis of cell lines indicated major cluster of five cell lines. Red- and blue-colored balls represent metastatic cell lines and non-metastatic cell lines respectively. Non-metastatic T1 cell line is labeled. (Color figure online)
To test the reproducibility of cluster prediction, we also evaluated cluster membership after introducing random perturbations to the data set. We perturbed data by adding normally distributed random numbers with mean equal to 0 and standard deviation equal to median variance of the log ratios of each gene across all the cell lines as indicated in Fig. 1a, b. An independent reproducibility test was also performed for each microarray experiment, and all the tests generated highly consistent indices. The R-index and D-index from the four experiments tested were 1 and 0 respectively, indicating that two major clusters of cell lines are not due to artifacts from statistical processing, and may reflect the biological characteristics among the cell lines. For further analysis, average expression ratios of each gene were calculated from duplicated experiments of each cell line, after inverting Cy-3/Cy-5 ratio values of a reciprocal experiment. Gene expression ratios that were missing in duplicated experiments were also eliminated. Hierarchical cluster analysis of combined gene expression ratios from rat and mouse microarray experiments is displayed in Fig. 1c. PCA of the combined data also supports our finding in the cluster analysis. Metastatic cell lines were grouped together and well separated from non-metastatic cell lines (Fig. 1d). Also, the non-metastatic T1 cell line was grouped with the metastatic cell lines as we observed in cluster analysis, suggesting that T1 had acquired a number of genetic alterations characteristic of the metastatic cell lines, but still lacked the critical traits needed for complete metastatic conversion.
Several independent gene expression clusters were identified in the analysis of the expression patterns. The largest distinctive gene expression cluster within the cluster diagram was enriched for genes involved in cell–cell interaction and adhesion (Fig. 1c). Expression of these genes varied widely among the cell lines but was well correlated with the metastatic potentials of the cells.
We applied the F test for gene selection to create a more limited set of genes for future exploration. At the given significance level (p value <0.01), genes that discriminate the metastatic cell lines including T1 from non-metastatic cell lines was created. Applying the F test on gene expression data produced 148 cDNAs from rat and mouse microarray experiments (Fig. 2). More than 90 % of genes in the list displayed lower expression in the metastatic cell lines than the non-metastatic cell lines, indicating that loss of function was a more critical determinant for metastatic conversion than gain of function. The majority of genes in the list were, not surprisingly, directly and indirectly involved in cell-to-cell interactions and cell adhesion. ECM proteins, such as Col1a1, Col1a2, Col2a1, Col3a1, Col4a1, Col4a5, Col5a1, Col5a2, Col9a1, Col12a1, Col14a1, and Fn1, were less expressed in metastatic cell lines when compared with non-metastatic cell lines. In addition to ECM proteins, cytoskeletal proteins and their binding proteins such as, Krtl-16, Krtl-17, Arpc1b, Plec1, Tmsb4X, Tmsb10, and Cfl2 were much less expressed in metastatic cell lines. Likewise, expression of Pg25, Fhl2, Fhl3, Gja1, Emb, Ncam1, Sparcl1, Sparc, and Cdh11, genes involved in cell adhesion, was down-regulated in metastatic cell lines. Importantly, expression of thrombospondin-1, -2, and -3, an inhibitor of angiogenesis, was down-regulated in all the metastatic cell lines and up-regulated in non-metastatic cell lines relative to the parental RLE cells (used as a reference). These results indicate that disruption of (ECM and loss of cell-to-cell interaction appear to be important events for metastatic conversion of transformed RLE cell lines.
Discriminative gene expression between two clusters. F test was performed to select genes which are differentially expressed between the two clusters discovered in hierarchical cluster analysis at the given significance level (p value <0.01) by using BRB ArrayTools. After F test, genes with an expression ratio that has at least twofold ratio difference relative to RLE in at least two cell lines were further selected. Data are presented in pseudo-colored matrix format as described in Fig. 1. Name of genes, clone ID, and functional description of each gene are also provided in each column. Genes involved in ECM and cytoskeletal remodeling and cell-to-cell interaction are highlighted in blue. (Color figure online)
Another F test (p value <0.01) was applied to select genes that could discriminate between metastatic and non-metastatic cell lines. The list of selected genes proved less instructive than previous F test results (Fig. 3). Among 36 selected genes, only a few (i.e. Ctsd) were known to associated with metastasis. This is probably due to the fact that the gene expression profile of the non-metastatic T1 cell line is more similar to metastatic cell lines than to the T4 and T5 cell lines. Interestingly, the expression of Eps8 was enhanced only in the metastatic cell lines. Eps8 is a substrate of EGFR and activates small GTPase Rac that promotes anchorage-independent growth in fibroblasts (Ruggieri et al. 2001). This may account for anchorage-independent growth of v-raf transformed metastatic cell lines in the presence of EGF. Also, reduced expression of Elm1 in metastatic cells is particularly interesting, since its expression was very low in our metastatic cell lines and over-expression of Elm1 in metastatic melanoma cells reduced the frequency in lung metastasis (Hashimoto et al. 1998).
Discriminative gene expression between metastatic and non-metastatic cell lines. F test was performed to select genes that are differentially expressed between the metastatic and non-metastatic cell lines at the given significance level (p value < 0.01). After F test, genes with an expression ratio that has at least twofold ratio difference relative to RLE in at least two cell lines were further selected. Data are presented in pseudo-colored matrix format as described in Fig. 1. Name of genes and clone ID of each gene are also provided in each column. (Color figure online)
To define the critical genetic elements required for complete metastatic conversion but are missing in the T1 cell line, we performed another microarray experiment comparing the metastatic cell lines with T1 cells using three different cDNA microarrays. In addition to 6.9 K rat and 9 K mouse cDNA microarrays used in previous experiments, a larger microarray containing mouse 36 K cDNAs was used for the gene expression profile study of the metastatic cell lines using T1 as a common reference. All experiments were duplicated and average expression ratios of each gene were calculated from a subset of well-measured spots as described in previous experiments. In order to identify the differentially expressed genes in all four metastatic cell lines when compared with the T1 cell line, an expression ratio of at least 1.7-fold difference relative to T1 in all four cell lines were selected for further analysis. To define the genes that are most likely to be critical in metastatic conversion, each gene was assigned to four non-mutually exclusive critical processes after invasion and intravasation in the metastatic cascade, namely, extravasation, survival/apoptosis, proliferation, and angiogenesis (Table 1). 15 of the 43 known genes could be assigned to the categories. The most frequently assigned category among the metastatic cell lines was survival/apoptosis, suggesting that survival in circulation and/or new environments might be the most critical factor for the final step in the metastatic cascade.
We determined the Pearson correlation coefficient of each gene to define those displaying strong positive and/or negative correlation with the metastatic potentials of the cell lines. Among selected genes whose expression was negatively correlated with the incidence of metastasis (Fig. 4), the expression of Alrp/Carp, an inhibitor of cell proliferation (Kanai et al. 2001), had the highest correlation (r = −0.96). Notably, three members (Elm1, Ctfg and Cyr61) of the CCN family known to mediate cell adhesion to ECM (Lau and Lam 1999) were grouped together as negatively correlated genes. Moreover, metastasis suppressor Elm1 that was less expressed in metastatic cell lines when compared with non-metastatic cell lines (Fig. 3) had the second highest negative correlation (r = −0.81) with metastatic potential of cell lines. These data suggest that an inverse correlation between the expression of the CCN family genes and the incidence of metastasis in metastatic cell lines may not be coincidental. Among positively correlated genes, Mmp16 displayed the strongest correlation. Mmp16 is a member of membrane-type matrix metalloproteinases known to be involved in tumor invasion (Sato et al. 1997). It is noteworthy that the well characterized metastasis gene osteopontin (OPN) was selected both as the commonly over-expressed gene in all metastatic cell lines (Table 1) and positively correlated with metastasis (Fig. 4). Galectin-3 is another over-expressed gene in the metastatic cell lines. Similar to OPN, galectin-3 also plays multiple roles in metastasis (Table 1). Galectin-3 inhibits nitrogen free radical-mediated apoptosis (Song et al. 2002; Moon et al. 2001), one of the major death pathways activated during ischemia–reperfusion injury that often is caused by the arrest of tumor cells within the microcirculation due to its size restriction (Weiss et al. 1989; Edmiston et al. 1998). Like OPN, Galectin-3 participates in immune cell extravasation (Sato et al. 2002), indicating that galectin-3 play a significant role in extravasation of tumor cells. In addition, galectin-3 is also involved in angiogenesis (Nangia-Makker et al. 2000). Over-expression of versatile proteins, such as OPN and galectin-3, might provide a great advantage for cells to complete the metastatic process.
Genes displaying correlated expression with metastatic potential of cell lines. Average expression ratios of each gene were calculated from the results of all three microarray experiments using 6.9 K rat and 9 and 36 K mouse cDNA microarrays. Gene expression ratios from rat and mouse microarray experiments were combined. Before analysis, genes with low variation across all of the arrays were excluded by using p values (>0.01) as described in methods and materials. Total 266 cDNAs were selected. In each gene, Pearson correlation coefficient between gene expression and the incidence of metastasis in cell lines was determined and genes with significant correlation (>0.5 or <−0.5) were further selected. a Expression patterns of genes negatively correlated with the incidence of metastasis. b Expression patterns of genes positively correlated with the incidence of metastasis. Thin colored lines represent expression pattern of each genes. Thick blue line represents the incidence of metastasis in lung. Genbank accession number, Pearson correlation coefficient, and name of genes are provided in each column. (Color figure online)
The current multi-step tumor model has been fundamental for the characterization of oncogenes and tumor-suppressor genes (Hanahan and Weinberg 2000). However, the conversion from benign to malignant tumors with metastatic potential requires additional changes, not adequately account for by classical oncogenes and tumor-suppressor genes. In particular, current tumor paradigms have so far failed to identify metastasis-associated genes as a unique group of tumor genes. The formation of clinical metastasis depends on the completion of every step in a process often referred to as the metastatic cascade (Fidler 2002). Due to the complexity of the metastatic process, it may be useful, as illustrated in the current work, to apply systematic analysis of global gene expression to a simplified version of metastatic conversion in order to better understand this complex phenomenon.
In conclusion, adopting transformed non-metastatic and metastatic RLE cell lines as a model system and global gene expression analysis, we have discovered gene expression signatures characteristic for distinct steps in metastatic conversion (Fig. 5). Our results suggested that increased proclivity for escaping from primary tumor sites is the first critical step in the metastatic process. However, genes conferring survival both in the circulation and at distant metastatic sites may constitute the ultimate determinants for establishing the metastatic phenotype. To this end expression of multi-functional proteins, such as OPN and galectin-3, may be an integral part of empowering the tumor cell to complete the metastatic process.
Metastatic pathway of transformed RLE cells. Green and red arrows represent under- or over-expression of genes in metastatic cell lines when compared with non-metastatic cell lines. Double arrows represent more altered expression of genes in highly metastatic cell lines than less metastatic cell lines. (Color figure online)
References
Bernards R, Weinberg RA (2002) A progression puzzle. Nature 418:823
Bisgaard HC, Mackay AR, Gomez DE, Ton PT, Thorgeirsson SS, Thorgeirsson UP (1997) Spontaneous metastasis of rat liver epithelial cells transformed with v-raf and v-raf/v-myc: association with different phenotypic properties. Invasion Metastasis 17:240–250
Cereijido M, Shoshani L, Contreras RG (2000) Molecular physiology and pathophysiology of tight junctions. I. Biogenesis of tight junctions and epithelial polarity. Am J Physiol Gastrointest Liver Physiol 279:G477–G482
Edmiston KH, Shoji Y, Mizoi T, Ford R, Nachman A, Jessup JM (1998) Role of nitric oxide and superoxide anion in elimination of low metastatic human colorectal carcinomas by unstimulated hepatic sinusoidal endothelial cells. Cancer Res 58:1524–1531
Fidler IJ (2002) Critical determinants of metastasis. Semin Cancer Biol 12:89–96
Garfield S, Huber BE, Nagy P, Cordingley MG, Thorgeirsson SS (1988) Neoplastic transformation and lineage switching of rat liver epithelial cells by retrovirus-associated oncogenes. Mol Carcinog 1:189–195
Hampton LL, Worland PJ, Yu B, Thorgeirsson SS, Huggett AC (1990) Expression of growth-related genes during tumor progression in v-raf-transformed rat liver epithelial cells. Cancer Res 50:7460–7467
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Hashimoto Y, Shindo-Okada N, Tani M, Nagamachi Y, Takeuchi K, Shiroishi T, Toma H, Yokota J (1998) Expression of the Elm1 gene, a novel gene of the CCN (connective tissue growth factor, Cyr61/Cef10, and neuroblastoma overexpressed gene) family, suppresses In vivo tumor growth and metastasis of K-1735 murine melanoma cells. J Exp Med 187:289–296
Huggett AC, Hampton LL, Ford CP, Wirth PJ, Thorgeirsson SS (1990) Altered responsiveness of rat liver epithelial cells to transforming growth factor beta 1 following their transformation with v-raf. Cancer Res 50:7468–7475
Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S (2002) Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol 156:299–313
Kanai H, Tanaka T, Aihara Y, Takeda S, Kawabata M, Miyazono K, Nagai R, Kurabayashi M (2001) Transforming growth factor-beta/Smads signaling induces transcription of the cell type-restricted ankyrin repeat protein CARP gene through CAGA motif in vascular smooth muscle cells. Circ Res 88:30–36
Lau LF, Lam SC (1999) The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res 248:44–57
Liotta LA, Kohn EC (2001) The microenvironment of the tumour-host interface. Nature 411:375–379
Moon BK, Lee YJ, Battle P, Jessup JM, Raz A, Kim HR (2001) Galectin-3 protects human breast carcinoma cells against nitric oxide-induced apoptosis: implication of galectin-3 function during metastasis. Am J Pathol 159:1055–1060
Nagafuchi A (2001) Molecular architecture of adherens junctions. Curr Opin Cell Biol 13:600–603
Nangia-Makker P, Honjo Y, Sarvis R, Akahani S, Hogan V, Pienta KJ, Raz A (2000) Galectin-3 induces endothelial cell morphogenesis and angiogenesis. Am J Pathol 156:899–909
Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E (1996) TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev 10:2462–2477
Oft M, Heider KH, Beug H (1998) TGF-beta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol 8:1243–1252
Oft M, Akhurst RJ, Balmain A (2002) Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol 4:487–494
Ruggieri R, Chuang YY, Symons M (2001) The small GTPase Rac suppresses apoptosis caused by serum deprivation in fibroblasts. Mol Med 7:293–300
Sato H, Okada Y, Seiki M (1997) Membrane-type matrix metalloproteinases (MT-MMPs) in cell invasion. Thromb Haemost 78:497–500
Sato S, Ouellet N, Pelletier I, Simard M, Rancourt A, Bergeron MG (2002) Role of galectin-3 as an adhesion molecule for neutrophil extravasation during streptococcal pneumonia. J Immunol 168:1813–1822
Savagner P (2001) Leaving the neighborhood: molecular mechanisms involved during epithelial–mesenchymal transition. BioEssays 23:912–923
Song YK, Billiar TR, Lee YJ (2002) Role of galectin-3 in breast cancer metastasis: involvement of nitric oxide. Am J Pathol 160:1069–1075
Tsukita S, Furuse M, Itoh M (2001) Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2:285–293
Vleminckx K, Kemler R (1999) Cadherins and tissue formation: integrating adhesion and signaling. BioEssays 21:211–220
Weiss L, Orr FW, Honn KV (1989) Interactions between cancer cells and the microvasculature: a rate-regulator for metastasis. Clin Exp Metastasis 7:127–167
Williams AO, Huggett AC, Thorgeirsson SS (1992) Pathology of spontaneous and oncogene transformed rat liver epithelial cells and derived tumours in nude mice. Int J Exp Pathol 73:99–114
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (2012-0000481) and the Ministry of Education, Science and Technology (2012R1A1A4A01010039). Microarray study was performed by the Shared Research Equipment Assistance Program by Korea Basic Science Institute, MEST.
Source(s) of funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (2012-0000481) and the Ministry of Education, Science and Technology (2012R1A1A4A01010039).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Heo, J., Lee, Js. & Leem, SH. Distinct gene expression signatures during development of distant metastasis. Genes Genom 35, 511–522 (2013). https://doi.org/10.1007/s13258-013-0097-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-013-0097-1