Abstract
The scalable XCAT voxelised phantom was used with the GATE Monte Carlo toolkit to investigate the effect of voxel size on dosimetry estimates of internally distributed radionuclide calculated using direct Monte Carlo simulation. A uniformly distributed Fluorine-18 source was simulated in the Kidneys of the XCAT phantom with the organ self dose (kidney ← kidney) and organ cross dose (liver ← kidney) being calculated for a number of organ and voxel sizes. Patient specific dose factors (DF) from a clinically acquired FDG PET/CT study have also been calculated for kidney self dose and liver ← kidney cross dose. Using the XCAT phantom it was found that significantly small voxel sizes are required to achieve accurate calculation of organ self dose. It has also been used to show that a voxel size of 2 mm or less is suitable for accurate calculations of organ cross dose. To compensate for insufficient voxel sampling a correction factor is proposed. This correction factor is applied to the patient specific dose factors calculated with the native voxel size of the PET/CT study.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The desire for patient specific radionuclide dosimetry estimates has become more practical with hybrid imaging technologies like PET/CT or SPECT/CT. In this case it is feasible that the complex geometries of human anatomy may be described as voxelised phantoms of specific patients. Many groups have investigated direct Monte Carlo simulation of patient images [1, 2] however little attention has been given to the effect of voxel size on the calculated mean absorbed dose per unit of cumulated activity, or organ dose factor (DF).
Traditionally the Monte Carlo calculation of DF values was performed by modeling mathematical anthropomorphic phantoms derived from simple geometric shapes. This approach addressed limitations in the computational methods available at the time. Although mathematical phantoms lack realism they do not suffer from insufficient regional sampling and are easily scaled. A good all round alternative to mathematical phantoms is the Extended Cardiac Torso (XCAT) [3]. XCAT provides a more realistic anthropomorphic voxelised phantom whilst maintaining scalable flexibility by using non-uniform rational b-splines (NURBS) surfaces [4].
It should however be noted that these model based phantoms represent reference individuals and may not necessarily be accurate to patient specific situations. In fact an insufficient representation of a real patient gives rise to large changes in the dose factor [2], where the magnitude of this change is dependent on the physical differences between the reference phantom compared to the patient. Scaling reference DF values to better represent specific patients is possible using the widely accepted OLINDA/EXM software [5]. This software scales the DF based on source organ mass, however it is still limited to using mathematical anthropomorphic phantoms. It is also understood that patient specific Monte Carlo based dosimetric methodologies can potentially incorporate minimal assumptions compared to scaling model based dosimetry methods. Therefore by far the most patient specific approach for dosimetry purposes is to use hybrid imaging technologies. For direct Monte Carlo calculation an anatomical image (e.g. CT or MRI) may be used to define organ geometries with the functional imaging modality (e.g. PET or SPECT) defining a heterogeneous source volume.
In this work we investigated the effects of insufficient voxel sampling on absorbed dose calculations using direct Monte Carlo simulation. Using Geant4 Application to Tomography Emission (GATE) [6] we aimed to find a suitable correction for DF values using the XCAT phantom. We then aimed to apply this correction to DF values that have been calculated using the native voxel size of a clinical FDG PET/CT scan.
Methods
Monte Carlo toolkit
This work used version 6.0p01 of the GATE toolkit. GATE was compiled with geant4 [7] version 4.9.2p02 and Class Libray for High Energy Physics (clhep) version 2.0.4.2. The low energy electromagnetic physics package based on the Livermore libraries has been used throughout this study. For all simulations a Fluorine-18 ion source has been used. The ion source type was defined as per the user manual [8] with ionic charge Q = 0 and excitation energy E = 0 keV. Voxelised source and attenuation media are passed to GATE in interfile format.
Calculations of absorbed Dose and its relationship to DF
Absorbed dose D (Gy) in any target organ may be calculated by equation 1, where N (Bq s) is the number of disintegrations that occur in the source organ and DF (Gy Bq−1 s−1) is the dose factor.
The dose factor is mathematically equivalent to the S-value described by the Medical Internal Radiation Dose (MIRD) committee [9]. The DF value is a function of the mass of the target region \(m_{k}\), the number of radiations \(n\) with energy \(E\) emitted per nuclear transition, E is the energy per radiation (MeV), for the \(i\)th radiation in the decay scheme of the radionuclide and the absorbed fraction \(\phi _{i}\). The absorbed fraction is defined as the ratio of the energy absorbed in the target region (\(k\)) by the energy emitted from the source organ (\(h\)). The specific absorbed fraction \(\varPhi _{i}\) (kg−1) may also be referred to and is the absorbed fraction over the mass of the target organ. DF may therefore be written as Eq. 2.
For non-penetrating radiation, self absorption is the dominate process and therefore the absorbed fraction has traditionally been set to 1.0 for organs that are much larger then the mean-free-path of the radiation [10]. This leads to the specific absorbed fraction being inversely proportional to the mass of the target organ. If the source and the target are the same organ then the reference DF may also be scaled by the organ mass for the non-penetrating component of the emission spectrum [11]. The MIRD perspective [12] scales the organ self dose by equation 3. Snyder [13] showed that if the absorbed fraction for non-penetrating radiation is set to 1.0 then organ mass has no effect on organ cross dose. This statement holds true if the source and target organs are sufficiently separated.
The procedure for calculating the absorbed fractions or DFs from a voxel phantom in GATE was the same regardless of complexity of the voxel data. Using the interfile method for dose calculation in GATE, the resulting output file was a dose histogram with the same dimensions as the entered interfile, with units of cGy. Multiplying the known mass of each voxel by the dose matrix results in the energy deposited in each voxel. A mask of the region of interest was then applied, where the sum represents the energy deposited in the region. Since the source energy and total energy emitted by the source was defined as values of the simulation the DF is easily calculated.
XCAT phantom simulations
Three male XCAT phantoms were created by using the dncat_bin program. The phantom height was kept constant at 175 cm, while the phantom weight was varied by scaling both organ masses and total body fat content. Thus creating three standardised phantoms with body mass index’s of 31.0 Kg m−2 (Large), 22.5 Kg m−2 (Medium) and 16.7 Kg m−2 (Small).
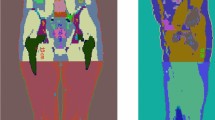
For each set of phantoms the voxel size was also varied between 0.4 mm and 16 mm voxels. Figure 1 shows the central coronal slice of the Large XCAT phantom with voxel sizes ranging from 1 to 8 mm.
For implementation into GATE a material lookup table was define to convert voxel values into tissue type. A uniformly distributed Fluorine-18 source was modeled in both kidneys. A total of \(10^{8}\) events was simulated, which is equivalent to N = 100 MBq · s.
Patient based simulation
A randomly selected routine clinically acquired \(^{18}\hbox {F-FDG}\) PET/CT scan was used as a basis for a retrospective calculation of the mean absorbed dose per unit of cumulated activity. The patient was a 75 year old male who was administered 365.75 MBq of \(^{18}\hbox {F-FDG}\) 70 minutes before the acquisition of the PET scan. The patient weighed 69 Kg and was 165 cm tall. Images were acquired using the Gemini PET/CT scanner (Philips Medical Systems). A total of seven emission bed positions were acquired with three minutes per bed position giving a total scan time of 21 min. PET images were then reconstructed with a 3D Row Action Maximum Likelihood Algorithm (3D-RAMLA [14]), the PET reconstructed voxel size was 4 mm × 4 mm × 4 mm. The helical CT attenuation scan had scan parameters of 60 mA, 30 s exposure time, pitch of 1.5 and 140 kVp. The reconstructed CT voxel size was 1.17 mm × 1.17 mm × 6.5 mm. The PET and CT images are inherently co-registered with identical patient positioning and coordinate system. Regular quality control of the Gemini PET/CT ensures co-registration between the PET and CT gantries. PET images were obtained in ugm (a vendor specific file format used by the Philips Allegro PET Scanner) format while CT images were obtained in dicom format.
Kidney and Liver regions were segmented manually from the CT image using an in-house software called Wasabi [15]. A voxel attenuation map was created by converting the patients CT from Hounsfield units to tissue type by use of a material lookup table.
The non-uniform activity distribution of the kidneys was found by applying the segmented kidney regions to the PET image which was normalised to the body weight Standardised Uptake Value \(\hbox {SUV}_{bw}\). The activity per gram at time \(t\) after injection was therefore found by scaling the PET image by equation 4, where \(A_{0}\) is the injected activity, \(M\) is the patient body weight and \(t_{1/2}\) is the radionuclide half-life.
Finally an activity distribution map in Bq per voxel was found by multiplying each voxel by the voxels density and volume (as defined from the attenuation map).
Results
For uniformly distributed activity in the kidneys the DF for self absorption are summarised in Table 1. It can be seen that as the phantom mass increased so did the absorbed fraction, where the kidney to kidney DF decreases as the phantom mass increases. A steady increase in the DF is also seen as the voxel size decreases.
Table 2 summarises the absorbed dose in the liver from F-18 uniformly distributed throughout the kidneys for the XCAT phantom. Again as the phantom mass increased the DF decreased and the absorbed fraction increases. The voxel size effects the DF differently for the kidney to liver situation compared to the kidney to kidney situation. A steady decrease in the DF was seen as the voxel size decreases.
Using the method described above, it was found that the kidney to kidney DF is 1.55 × 10−13 Gy Bq−1 s−1. The kidney to liver DF was also calculated as 2.43 × 10−15 Gy Bq−1 s−1. The absorbed fractions, kidney and liver masses and specific absorbed fractions are also presented in Table 3.
In “patient based simulation” section the absorbed dose for patient based simulation on a voxel-wise level is shown for the kidneys and liver in Fig. 2. The dose histogram is fused over the CT image to simplify organ location. The dose histogram is the result of 100 \(\hbox {MBq}\,\hbox {s}\) cumulated activity with a bio-distribution resulting from \(^{18}\hbox {F-FDG}\) at 70 min post injection. This differs from the XCAT phantom simulations which used a uniformly distributed source.
Discussion
Fitting data to different voxel sizes
It can be seen from Table 1 that a significantly small voxel size is required to achieve convergence for the organ self dose. Simulating small voxel sizes corrects this spatial sampling issue however this is limited to the computational and memory limitations of the computer hardware/software combination. These limitations are not only limited to the Monte Carlo process but also reduce the ability to accurately subsample patient data. In the kidney self-irradiation case the DFs only begin to converge as the voxel size decreases below the current limits of medical imaging equipment. A logical step to resolve this issue of inadequate voxel sampling is to consider fitting the DFs for the XCAT phantom.
The DF for uniform activity distributed throughout the kidneys for different voxel volumes are fitted to an equation of the form \(y=a-b\cdot x^{n}\) for the three XCAT phantoms. Where \(y\) is the DF in units of 10−13 Gy Bq−1 s−1 and × is the voxel volume in cm3. Table 4 summarises the fitting parameters for the three phantom sizes as well as the coefficient of determination r2. Figure 3a shows the fitted function for each phantom. Also shown for comparison is the kidney to kidney DF of the patient. As the voxel volume approaches zero (i.e. the geometric case) the DF becomes a × 10−13 Gy Bq−1 s−1, from the fitted parameters. The required correction is most significant for organ self dose.
Even though Table 1 shows that the kidney to liver DF converges for a voxel size 2 mm or less, values are also fitted to the equation of the form \(y=a-b\cdot x^{n}\). The fitting parameters for Fig. 3b are shown in Table 5, where a × 10−13 Gy Bq−1 s−1 is the kidney to liver DF.
From Fig. 3 it is noted that the DF increases with voxel size for organ self-dose but decreases with voxel size for organ cross-dose. Although it can not be conclusively ruled out from the results obtained, it is unlikely that this is a trend in all cases. Tables 1 and 2 show that a decrees in voxel size of the XCAT phantom corresponds to an increase in organ mass and absorbed fraction in all simulations performed, however this does not translate into the specific absorbed fraction and the DF. This is because the probability that the energy (assuming the same isotope) is deposited in the region is greatly effected by the regions geometry.
From the results of fitting the XCAT data it is suggested that as a rule of thumb a voxel size of 2 mm or less provides convergence for calculating organ cross doses, where for organ self dose a significantly smaller voxel size is required or must be corrected for.
Scaling patient DF for voxel volume
Using Fig. 3a the patient specific kidney to kidney DF can be scaled to account for voxel volume. The most obvious approach to achieve this scaling is to take the ratio of the DF reference phantom at a voxel volume of zero by the reference phantom DF at a voxel volume of x. Where x is the voxel volume of the patient dose histogram. Using the fitting parameters from the above section this can be written mathematically as Eq. 5.
Referring to Tables 1 and 2, it is decided to normalise the patient DF to the Large XCAT phantom as compared to the Medium phantom. This is because the patient and Large XCAT kidney masses are more closely matched. Even though the total body mass for the Medium phantom matches the patient mass the mass of the kidneys is approximately 17 g less then the patient kidney. Therefore the patient specific kidney to kidney DF becomes \(1.76 \times 10^{-13}\, Gy\, Bq^{-1}\, s^{-1}\) scaled up from \(1.55\times 10^{-13}\, Gy\, Bq^{-1}\, s^{-1}\). If normalisation was based on total body weight then the Medium phantom would have been used to obtain a DF of 1.82 \(\times 10^{-13}\, Gy\, Bq^{-1}\, s^{-1}\).
From the results obtained the kidney to liver DF converges for larger voxel volumes compared to the kidney to kidney case. Therefore it is not necessary to scale the patient specific kidney to liver DF. It should however be noted that Eq. 5 still holds for this case as \(\frac{a}{a-b\cdot x^{n}}\) is equal to 1.
Agreement to MIRD and OLINDA\EXM
The most widely accepted values for DFs (or S-Values) for nuclear medicine applicable radionuclide are the original MIRD pamphlet 11 [16] and the internal dosimetry software MIRDOSE [17] which has been superseded by OLINDA\EXM [5]. For self dose in the kidneys pamphlet 11 and OLINDA\EXM quote a DF of 1.65 × 10−13 and 1.68 × 10−13 Gy Bq−1 s−1 respectively. Table 6 provides a summary of the calculated DFs for the kidney self dose.
When comparing the values in Table 6 the DFs for each of the three voxelised XCAT phantoms are notably higher compared to the geometrically defined MIRD 11 and OLINDA\EXM values. By using Eq. 5 to correct for insufficient voxel size it can be stated that any difference between the geometric (i.e. MIRD) and voxelised phantom results are due to differences in individual organ shape and size. The three XCAT phantoms have a significantly smaller kidney mass when compared to not only the geometric phantoms but also reference man, thus resulting in a higher DF. These differences again highlight the reliance on the phantom used to obtain the organ DFs.
As an additional attempt to compare the DFs for the model based phantoms it will be assumed that Eq. 3 is a reasonable correction for the kidney to kidney case. Table 6 shows results of scaling the kidney self dose DFs to an organ mass of 310 g i.e. reference man. Differences in the scaled DF can be explained by three key points.
First is differences in organ shape may be significant enough to effect the scaled DF, for example the kidneys in the mathematical phantom defined by two ellipsoids cut by a plane which are not as realistic as using the XCAT surface method. This effect on the DF is seen in the differences between the XCAT and OLINDA\EXM results.
The second explanation for differences in the DFs is from the differences in the Monte Carlo approaches used. In particular the methodologies for handling non-penetrating radiation in the MIRD pamphlets, which assumes that for self-absorption the absorbed fraction equals 1, and that when the source and target organ are separated by any distance the absorbed fraction equals 0. Where as GATE used a more physically rigorous model in the direct Monte Carlo simulation. This assumption is still useful in simple cases but it is important to note that the positrons of Fluorine-18 contribute most to organ self-absorption. As an approximation the OLINDA\EXM software can be used to estimate the contribution of the positrons to be 70 % of the total absorbed dose in the adult kidneys.
Finally when the scaled DFs for the three XCAT phantoms are compared to each other the differences are not due to differences in methodology, or organ shape. Some small differences may have been introduced by the scaling performed to create the data sets, however this is considered minimal since NURBS surfaces have been used. This comparison leads to the conclusion that organ mass scaling is only a crude approximation. From the results obtained it is suggested that adapting phantom results to individual patients through mass scaling is not necessarily appropriate. The scaled patient specific kidney to kidney DF (shown in Table 6) adds additional evidence for this.
The patient specific kidney to kidney DF is calculated by direct Monte Carlo simulation which takes into account not only the patient’s organ size and shape but also its non-uniform activity distribution. As a direct comparison, the patient kidneys have a mass (0.269 kg) between the Medium (0.252 kg) and Large (0.283 kg) XCAT phantoms. However the calculated patient specific DF of 1.78 × 10−13 Gy Bq−1 s−1 is lower then the Medium (2.12 × 10−13 Gy Bq−1 s−1) and Large (1.86 × 10−13 Gy Bq−1 s−1) XCAT phantoms. If only the organ mass is considered then the patient specific DF should have fallen between the Medium and Large XCAT values, however this is not the case. It is therefore unreasonable to assume organ masses alone will be able to correct for differences in organ geometry or source distribution. This highlights that considerable care is required to adapt phantom results to individual patients.
In the kidney to liver DF case, pamphlet 11 and OLINDA\EXM quote DFs of 2.18 × 10−15 and 2.06 × 10−15 Gy Bq−1 s−1 respectively. Table 7 shows a summary of the kidney to liver DFs for each phantom model. Again when comparing the source to target DFs for all phantoms and reference values the DFs are significantly different. With a range between 2.01 × 10−15 and 3.20 × 10−15 Gy Bq−1 s−1. For the XCAT phantoms the possible error in kidney self-dose for a voxel size of 2 mm is in the order of 12–15 %. This demonstrates yet another possible source of error when taking into account patient specific radionuclide dosimetry and again suggests that considerable care is required when adapting phantom results to individual patients.
Variations in human anatomy
The dose factors calculated in this work have solely concentrated on the kidneys and liver of the XCAT phantom and an individual patient. Both the XCAT phantom and the patient have organs defined that are individual to either the standardised phantom or individual patient. It is however important to recognise that the significant variability between individual patient organ shapes and sizes have not been taken into account. This variability between patients is particularly true for the liver that may vary significantly in not only mass but also shape and will undoubtedly lead to a change in the dose factors calculated.
Conclusions
This work has shown the effects of voxel size on absorbed dose calculations and proposed a simple correction factor for insufficient voxel sampling. Using the XCAT phantom it was found that significantly small voxel sizes are required to achieve convergence for organ self dose calculations. It has also been shown that for organ cross dose convergence may be achieved at a voxel size of 2 mm or less.
Future work will extend the fitting parameters to other organs and radioactive sources.
References
Denis-Bacelar AM, Romanchikova M, Chittenden S, Saran FH, Mandeville H, Du Y, Flux GD (2013) Patient-specific dosimetry for intracavitary 32p-chromic phosphate colloid therapy of cystic brain tumours. Eur J Nucl Med Mol Imag 40:1532–1541
Zankl M, Petoussi-Henss N, Fill U, Regulla D (2003) The application of voxel phantoms to the internal dosimetry of radionuclides. Radiat Protect Dosim 105(1–4):539
Segars W, Tsui B (2009) Mcat to xcat: the evolution of 4-d computerized phantoms for imaging research. Proc IEEE 97(12):1954–1968
Piegl L (1991) On nurbs: a survey. Comput Graph Appl 11(1):55–71
Stabin M, Sparks R, Crowe E (2005) Olinda/exm: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med 46(6):1023
Jan S, Santin G, Strul D, Staelens S, Assié K, Autret D, Avner S, Barbier R, Bardiès M, Bloomfield PM et al (2004) Gate: a simulation toolkit for pet and spect. Phys Med Biol 49:4543–4561
Agostinelli S, Allison J, Amako K, Apostolakis J, Araujo H, Arce P, Asai M, Axen D, Banerjee S, Barrand G et al (2003) Geant4-a simulation toolkit. Nucl Instrum Method Phys Res Sect A 506(3):250–303
Jan S, Santin G, Strul D, Staelens S, Assié K, Autret D, Avner S, Barbier R, Bardiès M, Bloomfield PM, et al. (2007) GATE users guide. http://www.opengatecollaboration.org
Loevinger R, Budinger T, Watson E (1988) Mird primer for absorbed dose calculations. The Society of Nuclear Medicine, New York
Stabin M, Siegel J, Sparks R, Eckerman K, Breitz H (2001) Contribution to red marrow absorbed dose from total body activity: a correction to the Mird method. J Nucl Med 42(3):492
Williams L, Liu A, Yamauchi D, Lopatin G, Raubitschek A, Wong J (2002) The two types of correction of absorbed dose estimates for internal emitters. Cancer 94(S4):1231–1234
Howell R, Wessels B, Loevinger R, Watson E, Bolch W, Brill A, Charkes N, Fisher D, Hays M, Robertson J et al (1999) The Mird perspective 1999. Medical internal radiation dose committee. J Nucl Med 40(1):3S–10S
Cloutier RJ, Edwards CL, Snyder WS (1970) Medical radionuclides: radiation dose and effects. J Radiat Biol 18(4):399
Browne J, De Pierro A (1996) A row-action alternative to the em algorithm for maximizing likelihood in emission tomography. IEEE Trans Med Imag 15(5):687–699
Saunder T (2006) Wasabi, medical image quantitative analysis and visualisation package. http://wasabi.petnm.unimelb.edu.au/index.html
Snyder W, Ford M, Warner G, Watson S (1975) S”: absorbed dose per unit cumulated activity for selected radionuclides and organs. Mird pamphlet no. 11. Society of Nuclear Medicine, New York
Stabin M (1996) Mirdose: personal computer software for internal dose assessment in nuclear medicine. J Nucl Med 37(3):538
Acknowledgments
We would like to acknowledge the assistance of staff from the Victorian Partnership for Advanced Computing. We would also like to acknowledge the assistance of Dr. Rick Franich, Dr. Jianfeng He, Mr. Tim Saunder, Mr. Gareth Jones and Dr. Sylvia J Gong.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hickson, K.J., O’Keefe, G.J. Effect of voxel size when calculating patient specific radionuclide dosimetry estimates using direct Monte Carlo simulation. Australas Phys Eng Sci Med 37, 495–503 (2014). https://doi.org/10.1007/s13246-014-0277-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-014-0277-6