Abstract
Colletotrichum orbiculare causes anthracnose of Cucurbitaceae and is phylogenetically closely related to pathogens of several other herbaceous hosts belonging to the Asteraceae, Fabaceae and Malvaceae. Most of them are known for their hemibiotrophic infection strategy and as destructive pathogens either of field crops or weeds. In order to study the phylogenetic relationships of these fungi, a multilocus analysis (ITS, GAPDH, CHS-1, HIS3, ACT, TUB2, GS) of 42 strains of C. orbiculare and related species was conducted. The analysis resulted in nine clades that confirmed the four species previously known as belonging to this species complex, C. lindemuthianum, C. malvarum, C. orbiculare and C. trifolii, and recognised four new species from weeds, namely C. bidentis, C. sidae, C. spinosum and C. tebeestii. The name C. orbiculare itself is widely used in plant pathology and science, but is invalid according to current nomenclatural rules. Therefore we described a new species with the same epithet and a type specimen that agrees with our current understanding of this species, and is linked to a living culture. Following the recent epitypification of C. lindemuthianum, we chose appropriate specimens with associated strains to serve as epitypes of C. malvarum and C. trifolii, and selected an authentic specimen of C. trifolii as lectotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colletotrichum orbiculare is an important anthracnose pathogen of Cucurbitaceae, especially of cucumber (Cucumis sativus), melons (Cucumis melo), watermelon (Citrullus lanatus), pumpkin (Cucurbita pepo) and squash (Cucurbita maxima), but is reported on more than 40 plant host species worldwide (Farr and Rossman 2013). The pathogen causes lesions on seedlings, leaves, petioles, stems and fruits of cucurbits. On cucumber fruits, for example, circular sunken water-soaked lesions are formed that expand and turn black in moist weather, eventually becoming covered with pink spore masses. On leaves however, lesions are pale brown to reddish, and centres may crack and fall out (Sitterly and Keinath 1996).
Colletotrichum orbiculare is characterised by straight conidia with an obtuse apex and tapered towards the base, dark brown to black or greyish black cultures and a slower growth rate than C. gloeosporioides (Sutton 1992). Colletotrichum lagenarium is regarded as a synonym of C. orbiculare (von Arx 1957a) and sexual morphs linked to each of these names were described as Glomerella lagenaria by Stevens (1931) in the USA and by Watanabe and Tamura (1952) in Japan, and Glomerella cingulata var. orbiculare by Jenkins and Winstead (1962) in the USA.
Because of the morphological similarity of strains from beans and different Cucurbitaceae hosts, Halsted (1893a) considered C. lindemuthianum as a synonym of C. lagenarium. Other authors (Krüger 1913; Shear and Wood 1913) regarded C. lindemuthianum and C. lagenarium as separate species based on pathogenicity tests. Von Arx (1957a) regarded C. orbiculare as indistinguishable from C. gloeosporioides based on morphology, and considered C. orbiculare, C. lindemuthianum, C. mavarum and C. trifolii as divergent forms (‘abweichende formen’) of C. gloeosporioides that are specialised for particular host families. Von Arx and Müller (1954) considered G. lagenarium as well as G. lindemuthianum, the sexual morph of C. lindemuthianum, as synonyms of G. cingulata. However, several molecular studies based on DNA sequences of the nuclear ribosomal gene (Sherriff et al. 1994; Bailey et al. 1996; Sreenivasaprasad et al. 1996; Johnston and Jones 1997; Farr et al. 2006) have shown that C. orbiculare is distinct from other species including those in the C. gloeosporioides complex.
Sherriff et al. (1994) studied the large subunit of the nuclear ribosomal RNA gene (LSU) and the internal transcribed spacer 2 of the nuclear ribosomal RNA operon (ITS-2) of Colletotrichum isolates from several hosts. Because of the unifomity of the DNA sequence data they concluded C. orbiculare, C. lindemuthianum, C. malvarum and C. trifolii to be one species and suggested the name C. orbiculare in which host specific forms exist. However, based on sequence data of the glutamine synthase gene (GS) and a 200-bp intron of the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH), mitochondrial DNA restriction fragment length polymorphisms (mtDNA RFLP) and vegetative compatibility and pathogenicity tests of a large number of strains, Liu et al. (2007) recognised C. orbiculare as a species complex with C. lindemuthianum, C. malvarum, C. orbiculare and C. trifolii as distinct species. However, according to the phylogeny in Liu et al. (2007), C. orbiculare is paraphyletic. The C. orbiculare complex, represented by a few strains each, occupied a basal clade in previous Colletotrichum phylogenies based on sequence data of the 5.8S nuclear ribosomal gene with the two flanking internal transcribed spacers (ITS) and partial sequences of the beta-tubulin gene (TUB2) (Lubbe et al. 2004) and ITS/LSU sequence data (Farr et al. 2006). This was confirmed in recent multigene phylogenies generated by Cannon et al. (2012) and O’Connell et al. (2012).
The host plants of species of the C. orbiculare complex can be attacked by other Colletotrichum species as well. For example, some Colletotrichum strains from Cucurbitaceae hosts were included in previous multigene studies belonging to C. melonis (C. acutatum complex), C. karstii (C. boninense complex) and C. coccodes (Damm et al. 2012a, b; Liu et al. 2013). This can lead to misidentifications, because of the current host-dominated approach for identification of Colletotrichum species by some plant pathologists, and the overlapping morphological characters of many species that also vary depending on the literature used. For example, according to Baxter et al. (1983) the average conidial dimensions of C. orbiculare vary from 16.9 to 20.2 × 4.2–4.4 μm depending on the substrate, which is outside the size range of C. orbiculare conidia (10–15 × 4.5–6 μm) given by Sutton (1992), while according to von Arx (1957a) the conidia of C. orbiculare measure 11–19 × 4–6 μm. The uncertainty of the identification of species in the C. orbiculare complex was demonstrated by Cannon et al. (2012). In a phylogeny based on ITS sequences retrieved from GenBank, most of the sequences of C. lindemuthianum and C. orbiculare strains and three sequences of C. trifolii strains clustered in one clade. However a further three ITS sequences labelled as C. trifolii clustered with strains of C. destructivum and C. higginsianum, while one “C. lindemuthianum” strain clustered with C. boninense. No complete ITS sequence of C. malvarum is available in GenBank. Additionally, the phylogeny in the study of Bailey et al. (1996) indicates the existence of more than one species among strains associated with different Malvaceae hosts.
Because of their intracellular hemibiotrophic infection strategy and the ease with which they can be cultured and manipulated, Colletotrichum species have been used as model organisms in studies of host/parasite interactions. Species in the C. orbiculare complex, especially C. lindemuthianum and C. orbiculare, have been intensively studied in recent years to investigate infection structures and processes and their associated molecular and physiological traits (Perfect et al. 1999; Kubo and Takano 2013). This has included, for example, melanization and lipolysis in appressoria of C. orbiculare (Asakura et al. 2012; Lin et al. 2012). In order to study factors regulating the transition from biotrophy to necrotrophy, C. orbiculare strain MAFF 240422 (= CBS 514.97 = 104-T) has recently been subjected to whole genome-level analysis and compared with the genome of a C. gloeosporioides (s. lat.) strain (Gan et al. 2013). Resistance mechanisms in the Medicago-Colletotrichum pathosystem have been studied in order to find resistant lucerne cultivars, as well as factors regulating growth, development and infection of C. trifolii (Yang and Dickman 1999; Warwar et al. 2000; Chen et al. 2006; Mackie et al. 2007; Jaulneau et al. 2010). However, no sequence of the wild-type strain of C. trifolii race 1 (ATCC 66954) used in the study of Yang and Dickman (1999), and only a few sequences of the C. trifolii strains from other studies are available in GenBank; the identities of all these strains need confirmation.
Although strains identified as C. orbiculare, C. malvarum and C. trifolii have previously been intensively studied in various aspects, the application of these names is uncertain. Except for C. lindemuthianum, which was recently epitypified (Liu et al. 2013), no ex-type or authentic culture of species in the C. orbiculare complex is available and no multigene analysis has been applied to delineate the species in this complex. The aim of this study is therefore to revise the C. orbiculare species complex based on multigene and morphological analyses and to select lecto-and epitypes in order to stabilise phylogenetic application of these species names.
Materials and methods
Isolates
A total of 42 strains was studied, most of which were previously identified as C. orbiculare and C. lindemuthianum, as well as other related strains from the CBS and other culture collections. Type material (holotypes and epitypes) of the species studied is located in the Fungarium of the Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands, in the IMI and K(M) Fungaria, which are both based in the Royal Botanic Gardens, Kew, UK, the Herbário da Universidade Federal de Viçosa (VIC), Brazil and the US National Fungus Collections (BPI), Beltsville, Maryland, USA. All descriptions are based on ex-holotype or ex-epitype cultures as appropriate. Features of other strains or specimens are added if deviant. Subcultures of the holotypes and epitypes as well as all other isolates used for morphological and sequence analyses are maintained in the CBS culture collection or the Coleção Octávio Almeida Drummond (COAD), Universidade Federal de Viçosa, Brazil (Table 1).
Morphological analysis
To enhance sporulation, autoclaved filter paper and double-autoclaved stems of Anthriscus sylvestris were placed onto the surface of synthetic nutrient-poor agar medium (SNA, Nirenberg 1976). SNA and OA (oatmeal agar, Crous et al. 2009) cultures were incubated at 20 °C under near UV light with a 12 h photoperiod for 10 days. Measurements and photographs of characteristic structures were made according to Damm et al. (2007). Appressoria were observed on hyphae on the reverse side of SNA plates and on slide cultures. A slide culture was prepared by placing a 1 cm2 PCA (potato-carrot agar, Crous et al. 2009) block on a slide in a sterile petri dish containing sterile moist filter paper, inoculating its edges with conidia and covering it with a sterile cover slip. After 15 days the appressoria were studied on the underside of the cover slip and on the slide. Microscopic preparations were made in clear lactic acid, with 30 measurements per structure and observed with a Nikon SMZ1000 dissecting microscope (DM) or with a Nikon Eclipse 80i microscope using differential interference contrast (DIC) illumination.
Colony characters and pigment production on SNA and OA cultures incubated at 20 °C under near UV light with a 12 h photoperiod were noted after 10 days. Colony colours were rated according to Rayner (1970). Growth rates were measured after 7 and 10 d.
Phylogenetic analysis
Genomic DNA of the isolates was extracted using the method of Damm et al. (2008). The ITS, GAPDH, and partial sequences of the chitin synthase 1 (CHS-1), histone3 (HIS3), actin (ACT), TUB2 and GS genes were amplified and sequenced using the primer pairs ITS-1 F (Gardes and Bruns 1993) + ITS-4 (White et al. 1990), GDF1 + GDR1 (Guerber et al. 2003), CHS-354R + CHS-79 F (Carbone and Kohn 1999), CYLH3F + CYLH3R (Crous et al. 2004b), ACT-512 F + ACT-783R (Carbone and Kohn 1999), BT2Fd + BT4R (Woudenberg et al. 2009) or T1 (O’Donnell and Cigelnik 1997) + Bt-2b (Glass and Donaldson 1995) and GSF1 + GSR1 (Stephenson et al. 1997), respectively. The PCRs were performed in a 2720 Thermal Cycler (Applied Biosystems, Foster City, California) in a total volume of 12.5 μL. The GAPDH, CHS-1, HIS3, ACT and TUB2 PCR mixture contained 1 μL 20x diluted genomic DNA, 0.2 μM of each primer, 1x PCR buffer (Bioline, Luckenwalde, Germany), 2 mM MgCl2, 20 μM of each dNTP, 0.7 μL DMSO and 0.25 U Taq DNA polymerase (Bioline). Conditions for PCR of these genes constituted an initial denaturation step of 5 min at 94 °C, followed by 40 cycles of 30 s at 94 °C, 30 s at 52 °C and 30 s at 72 °C, and a final denaturation step of 7 min at 72 °C, while the ITS PCR was performed as described by Woudenberg et al. (2009). Some isolates occasionally gave two bands (GS), which were then amplified using a touchdown PCR program (Zhou et al. 2006). The DNA sequences generated with forward and reverse primers were used to obtain consensus sequences using Bionumerics v. 4.60 (Applied Maths, St-Marthens-Lathem, Belgium), and the alignment assembled and manually adjusted using Sequence Alignment Editor v. 2.0a11 (Rambaut 2002).
To determine whether the seven sequence datasets were congruent and combinable, tree topologies of 70 % reciprocal Neighbour-Joining bootstrap with Maximum Likelihood distances (10 000 replicates) with substitution models determined separately for each partition using MrModeltest v. 2.3 (Nylander 2004) were compared visually (Mason-Gamer and Kellogg 1996). A maximum parsimony analysis was performed on the multilocus alignment (ITS, GAPDH, CHS-1, HIS3, ACT, TUB2, GS) as well as for each gene separately with PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2000) using the heuristic search option with 100 random sequence additions and tree bisection and reconstruction (TBR) as the branch-swapping algorithm. Alignment gaps were treated as missing and all characters were unordered and of equal weight. The robustness of the trees obtained was evaluated by 100 000 bootstrap replications using the Fast-stepwise addition algorithm (Hillis and Bull 1993). Tree length, consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated for the resulting tree. A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v. 3.1.1 (Ronquist and Huelsenbeck 2003) for the combined sequence datasets. Models of nucleotide substitution for each gene determined by MrModeltest v. 2.3 were included for each gene partition. The analyses of two MCMC chains were run from random trees for 1000 000 generations and sampled every 100 generations. The first 25 % of trees were discarded as the burn-in phase of the analysis and posterior probabilities determined from the remaining trees. For additional comparison, a Neighbour-Joining analysis was performed on the multigene alignment using PAUP with 10 000 bootstrap replications. Sequences derived in this study have been lodged at GenBank, the alignment in TreeBASE (www.treebase.org/treebase-web/home.html) (S14347), and taxonomic novelties in MycoBank (Crous et al. 2004a).
Results
Phylogeny
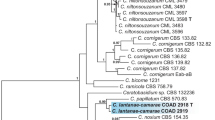
The seven sequence datasets did not show any conflicts in tree topology for the 70 % reciprocal bootstrap trees, which allowed us to combine them. In the multigene analyses (gene boundaries of ITS: 1–542, GAPDH: 553–815, CHS-1: 826–1123, HIS3: 1134–1532, ACT: 1543–1831, TUB2: 1842–2324; GS: 2335–3317) of 43 isolates of C. orbiculare and related Colletotrichum species and the outgroup (C. gloeosporioides strain CBS 112999), 3317 characters including the alignment gaps were processed, of which 240 characters were parsimony-informative, 604 parsimony-uninformative and 2473 constant. After a heuristic search using PAUP, 9366 most parsimonious trees were retained (tree length = 1004 steps, CI = 0.919, RI = 0.968, RC = 0.890, HI = 0.081) of which one is shown in Fig. 1. The topology of the 9366 trees was similar, which was verified for a large selection of trees. They differed only in the position of strains within the subclades. For Bayesian analysis, a GTR model was selected for ITS, a HKY + G model for GAPDH and ACT, a GTR + I model for CHS-1 and HIS3, a HKY model for TUB2 and a JC + I model for GS, and incorporated in the analysis.
One of 9366 most parsimonious trees obtained from a heuristic search of the combined ITS, GAPDH, CHS-1, HIS3, ACT, TUB2 and GS sequence alignment of the Colletotrichum orbiculare species complex. Bootstrap support values above 70 % (bold) and Bayesian posterior probability values above 0.95 are shown at the nodes. Colletotrichum gloeosporioides CBS 112999 is used as outgroup. Numbers of ex-holotype and ex-epitype strains are emphasised in bold. Strain numbers are followed by substrate (host genus) and country of origin, NL = Netherlands, unkn = unknown. Branches that are crossed by diagonal lines are shortened by 50 %
The analyses resulted in detection of 3 main clades and 9 subclades within the C. orbiculare species complex. The first main clade is formed by C. lindemuthianum strains and is well supported with a bootstrap support of 100 % and a Bayesian posterior probability value of 1.00. It consists of two subclades (bootstrap support/Bayesian posterior probability value 100/1.00 and 92/1.00) comprising about half of the strains each, both representing strains from Phaseolus vulgaris from Germany, USA and the Netherlands. The second main lineage is represented by a single strain of C. bidentis; the clustering with the third main clade is not supported. The third main clade (100/1.00) consists of six subclades: the clades representing C. trifolii and C. malvarum are well supported (100/1.00) and group with each other. A sister clade is formed by C. orbiculare containing the largest number of strains (100/1.00), two smaller subclades representing C. sidae (90/1.00) and C. spinosum (100/1.00) as well as a single-strain clade representing C. tebeestii. The NJ tree (not shown) confirmed the tree topology obtained with parsimony; the consensus tree obtained from Bayesian analyses only differs in the grouping of some of the subclades within the third main clade: C. sidae and C. tebestii cluster with each other (1.00) and C. spinosum forms a sister clade to both (1.00). The other Bayesian posterior probability values agreed with bootstrap supports (Fig. 1). The individual alignments and maximum parsimony analyses of the six single genes were compared with respect to their performance in species recognition. All clades are recognised with GS sequences, however some species differ only in one or few bp; other loci only recognised some of the species (see notes accompanying each species).
Taxonomy
Based on DNA sequence data and morphology, the 42 strains studied (Table 1) are assigned to eight species, including four species that have proved to be new to science. Additionally, C. orbiculare itself is described as a new species for technical nomenclatural reasons, as explained below. All species studied in culture are characterised below. Although there are two clades formed by strains from Phaseolus spp., here indicated as C. lindemuthianum 1 (including the ex-epitype strain) and C. lindemuthianum 2, they are not treated as separate species, because no difference in host preference, distribution or morphology was observed.
Colletotrichum bidentis Damm, Guatimosim & Vieira, sp. nov. Fig. 2
Colletotrichum bidentis (from ex-holotype strain COAD 1020). a–b. acervuli; c. setae; d–f. conidiophores; g. tip of a seta; h. seta; i–k. conidiophores; l–q. appressoria; r–s. conidia. a, c–f, r: from Anthriscus stem; b, g–k, s: from SNA, l–q from PCA slide culture. a–b: DM; c–s: DIC.—Scale bars: a = 100 μm; f = 10 μm; scale bar of a applies to a–b; scale bar of f applies to c–s
MycoBank MB 804691.
Etymology: The species epithet is derived from the host genus, Bidens.
Sexual morph not observed. Asexual morph on host plant. Lesions on stems starting as circular spots, cinnamon-brown, becoming discoid, orange, leading to blight, shrivelling and death of infected plants. Internal mycelium indistinct. External mycelium absent. Conidiomata acervular, solitary to crowded, subepidermal, olivaceous-brown, 25–125 μm wide. Conidiogenous cells terminal, hyaline, cylindrical, rarely ampuliform, with acute apices, (7–)11–20(−24) × 2–5 μm. Conidia aseptate, hyaline, smooth-walled, cylindrical, straight to slightly curved, ends rounded, strongly guttulate, (7–)9–18(−21) × (3–)4–5(−6) μm. Appressoria globose to ellipsoidal, sometimes irregular, dark brown to olivaceous brown, smooth-walled, wall one-layered, ca. 0.2 μm thick, (6–)7–10(−12) × (4–) 5–8(−9) μm.
Asexual morph on SNA. Vegetative hyphae 1.5–7 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata acervular, conidiophores and setae formed from a cushion of pale brown, angular cells 3.5–8 μm in diam. Setae medium to dark brown, basal cell usually pale brown, smooth-walled, 60–100 μm long, 1- to 3-septate, base cylindrical to ± inflated, 5.5–8.5 μm diam, tip round to slightly acute, sometimes ± hyaline. Conidiophores pale brown, smooth-walled to verruculose, septate, branched, to 60 μm long. Conidiogenous cells pale brown, smooth-walled to verruculose, cylindrical, sometimes surrounded by a gelatinous sheath, 13–28 × 3.5–7 μm, opening 0.5–1.5 μm diam, collarette 0.5 μm long, periclinal thickening not observed. Conidia hyaline, smooth-walled, aseptate, straight, sometimes slightly curved, cylindrical, with one end round and one end truncate, 12–15(−19.5) × (4–)4.5–5(−5.5) μm, mean ± SD = 13.6 ± 1.4 × 4.5 ± 0.3 μm, L/W ratio = 3.0.
Asexual morph on Anthriscus stem. Conidiomata acervular, conidiophores and setae formed from a cushion of pale brown, thick-walled, angular cells 3–8 μm in diam. Setae pale to medium brown, basal cell often paler, smooth-walled to verruculose, 29–60 μm long, 1- to 2-septate, base cylindrical to ± inflated, 4.5–7 μm diam, tip ± round to slightly acute, often hyaline. Conidiophores pale brown, smooth-walled to verruculose, septate, branched, to 60 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled to verruculose, cylindrical to ampulliform, sometimes surrounded by a gelatinous sheath, 8.5–31 × 3.5–5.5 μm, opening 0.5–1 μm diam, collarette 0.5 μm long, periclinal thickening sometimes observed. Conidia hyaline, smooth-walled, aseptate, straight, sometimes slightly curved, cylindrical to clavate, with one end round and one end truncate, (11–)11.5–14(−15.5) × (3.5–)4–4.5(−5) μm, mean ± SD = 12.7 ± 1.1 × 4.4 ± 0.3 μm, L/W ratio = 2.9.
Culture characteristics: Colonies on SNA flat to low convex, with entire margin, hyaline to buff, covered by thin, floccose, whitish, aerial mycelium, the Anthriscus stem, filter paper and medium partly covered by cinnamon to dark grey acervuli, the Anthriscus stem and filter paper partly pale olivaceous-grey to olivaceous-grey, reverse similar; growth 10–12 mm in 7 days (14.5–17 mm in 10 days). Colonies on OA flat with entire margin, buff, greyish sepia to olivaceous-grey in the centre, covered with ochraceous, dark grey to black acervuli, aerial mycelium lacking, reverse buff at the margin, gradually turning pale olivaceous-grey to iron-grey towards the centre, growth 11.5–13 mm in 7 days (19–21 mm in 10 days). Conidia in mass apricot to ochraceous.
Specimen examined: BRAZIL, Goiás, Jataí, road to Rio Verde, abandoned gas station, 17º50′30.6″S, 51º26′50.7″W, from anthracnose symptoms on stems of Bidens subalternans, 13 Feb. 2010, B.S. Vieira (VIC 31566 holotype, CBS H-21140 isotype, culture ex-holotype COAD 1020 = CPC 21930).
Notes: Bidens subalternans belongs to the B. pilosa complex (bur marigold, Asteraceae), and is often identified as B. pilosa because of similar morphology with this species (Magenta 1998; Grombone-Guaratini et al. 2006). It is distributed from Uruguay and central Argentina to northern and western Brazil (Sherff 1937; Cabrera 1974). Species of the B. pilosa complex infest at least 30 different crops in over 40 countries, and are known to significantly reduce crop yields (ISSG 2013).
One forma specialis of C. gloeosporioides and one Vermicularia species were previously described on Bidens. Colletotrichum gloeosporioides f. sp. pilosae [as ‘pilosa’] was described on leaves of Bidens pilosa in Varanasi, India (Singh 1974). The conidia are described as cylindrical to sybcylindrical with a size (10–21 × 3.3–4.4 μm) overlapping that of C. bidentis. However, the size range also suggests the conidia to be narrower and with a larger L/W ratio than C. bidentis. Since we do not have a living strain of C. gloeosporioides f. sp. pilosae, we are unable to confirm the identity of this fungus. Furthermore, formae speciales are host/pathogen combinations and do not have a taxonomic status, so cannot be considered to be formal synonyms. Vermicularia bidentis was described on Bidens tripartita in Belgium, and forms fusoid conidia with acute ends, measuring 12–16 × 3–4 μm (Verplancke and van den Broecke 1936). Although the conidial size also overlaps with C. bidentis, the shape suggests it to be a species of the C. acutatum species complex. Other reports of Colletotrichum on Bidens include C. dematium on Bidens pilosa in Cuba, Venezuela and the West Indies and Colletotrichum sp. on Bidens pilosa in Florida (Farr and Rossman 2013). However, there is no DNA sequence of a Colletotrichum species from Bidens in GenBank.
Colletotrichum bidentis is morphologically similar to C. lindemuthianum (see Liu et al. 2013), but unlike any of the other species in the C. orbiculare complex, conidia formed by C. bidentis are sometimes slightly curved, especially on SNA, and setae have a conspicuous white tip. However, the base of the setae of C. bidentis is wider (usually > 5 μm diam) than C. lindemuthianum and the basal cell is often pale brown. Additionally, C. bidentis is the slowest growing species in this complex on our diagnostic media.
Colletotrichum bidentis is separable from other species using all of the loci studied. The closest matches in a blastn search in GenBank with the ITS sequence of strain COAD 1020, with 99 % identity (4 bp differences), were C. orbiculare strains MAFF 306685 (AB269941) from Japan (Yoshida S, published 2007 in database only) and MX-2-153-Mexico (AY841133) from Annona cherimola (Villanueva-Arce et al. 2008). The closest match with the TUB2 sequence, with 97 % identity, was C. orbiculare (here: C. spinosum) strain STE-U 5296 (AY376589) from Xanthium spinosum in Argentina (Lubbe et al. 2004). There is no GS sequence closer than 91 % identical and no full-length GAPDH sequence with greater than 98 % homology available in GenBank.
Colletotrichum malvarum (A. Braun & Casp.) Southw., J. Mycol. 6(3): 116 (1891) Fig. 3
Colletotrichum malvarum (from ex-epitype strain CBS 521.97). a–b. acervuli; c. seta; d–f. conidiophores; g. tip of a seta; h. basis of a seta; i–l. conidiophores; m–r. appressoria; s–t. conidia. a, c–f, s: from Anthriscus stem; b, g–r, t: from SNA. a–b: DM; c–t: DIC.—Scale bars: a = 100 μm; f = 10 μm; scale bar of a applies to a–b; scale bar of f applies to c–t.
Basionym: Steirochaete malvarum A. Braun & Casp., Krankh. Pflanz.: 28 (1853)
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 2–6 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed (but see below). Conidiomata absent, conidiophores and setae formed directly on hyphae. Setae pale to medium brown, smooth-walled to verruculose, 65–130 μm long, 2–3-septate, base cylindrical, sometimes slightly inflated, 4–5.5(−6.5) μm diam, tip acute. Conidiophores pale brown, smooth-walled, septate, branched, to 50 μm long. Conidiogenous cells pale brown, smooth-walled, cylindrical, 9–22 × 3.5–4.5 μm, opening 1.5 μm diam, collarette 0.5 μm long, periclinal thickening sometimes visible. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to clavate, with one end round and the other truncate, (10.5–)11–12.5(−13) × 4–5(−5.5) μm, mean ± SD = 11.6 ± 0.7 × 4.5 ± 0.3 μm, L/W ratio = 2.6. Appressoria integrated in pale brown mycelium clusters, medium brown, smooth-walled, oblong in outline, often curved, with an entire margin, (4.5–)7.5–12.5(−16.5) × (3.5–)4.5–6(−7) μm, mean ± SD = 9.9 ± 2.5 × 5.3 ± 0.8 μm, L/W ratio = 1.9. Appressoria-like structures that possibly function as chlamydospores were found, measuring (5–)6–9.5(−11.5) × (4–)5.5–7.5(−8) μm, mean ± SD = 7.7 ± 1.7 × 6.4 ± 1.0 μm, L/W ratio = 1.2.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae densely arranged; no angular basal cells observed. Setae medium brown, smooth-walled to verruculose, 50–130 μm long, 1–2-septate, base cylindrical to conical, 3.5–5.5 μm diam, tip acute. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 20 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical, 15–22 × 3.5–4.5 μm, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening sometimes visible. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, sometimes clavate with one end round and the other truncate, (11–)11.5–13.5(−14) × 4.5–5(−5.5) μm, mean ± SD = 12.6 ± 0.9 × 4.8 ± 0.3 μm, L/W ratio = 2.6.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline, olivaceous-grey towards the centre, aerial mycelium lacking, reverse same colours; growth 15–16 mm in 7 days (20–23 mm in 10 days). Colonies on OA flat with entire margin, dark olivaceous, dark grey-olivaceous to olivaceous-black, aerial mycelium lacking, partly covered by rosy buff conidia oozing from black acervuli, reverse pale olivaceous-grey to olivaceous-grey; growth 11.5–14 mm in 7 days (16.5–18.5 mm in 10 days). Conidia in mass not visible.
Specimens examined: GERMANY, probably Berlin, from Malva sp., collection date and collector unknown (PC holotype, IMI 69872 slide from holotype). UK, from Lavatera trimestris cv. Mont Blanc, unknown collection date (deposited in CBS collection Feb. 1997 by J.A. Bailey), R. Maude (CBS H-20973 epitype, here designated, culture ex-epitype CBS 521.97 = LARS 720 = Lav-4; MBT175516). UNKNOWN COUNTRY, from unknown Malvaceae host, unknown collection date and collector (deposited in CBS collection Jul. 1924 by ABM Haye), culture CBS 123.24.
Notes: Steirochaete malvarum was described from cultivated Malva and Lavatera in Germany (Braun 1854). The fungus caused a disease of Malva verticillata, M. parviflora, M. mauritiana, M. bryoniifolia and Lavatera plebeja that starts with greenish black spots on leaves and stems and leads to wilt and plant death. The original publication states that the fungus forms stiff brown setae and aseptate, hyaline to slightly greenish, elliptical, ovoid to oblong conidia, and (probably inaccurately) that the spores are produced in weak chains of 3–4 from hyaline, filiform cells. Based on the drawing in the original description they measure ca 9–14 × 4.5–5 μm; Saccardo (1886) provided somewhat smaller measurements, 8–9 × 3–4 μm. The slide from the holotype in IMI is in poor condition, but measurements from the conidia are as follows: (10–)11.5–15(−16) × 4–5 μm, mean ± SD = 13.2 ± 1.8 × 4.6 ± 0.3 μm, L/W ratio = 2.9.
Southworth (1890) described a new species, C. althaeae that caused a destructive disease on hollyhock (Alcea rosea) in the USA and resembled C. lindemuthianum. Alcea and Althaea are actually different genera of Malvaceae that have frequently been confused. The following year, Southworth realised that a similar fungus had been previously described on Malva species. She regarded the species as synonyms and combined S. malvarum into Colletotrichum (Southworth 1891). She also reported on strains from prickly sida (Sida spinosa) from Kansas, USA that looked the same as those from hollyhock. Conidia from these strains did not produce a disease on Malva, but nevertheless she considered them to be identifiable as C. malvarum. These are likely to belong to C. sidae, described below.
Conidia of Southworth’s fungus from hollyhock were described as measuring 10–28 × 5 μm, which is larger than our measurements from the type of Steirochaete malvarum, and from the European strains we have assigned to this species. Bearing in mind the diversity of Colletotrichum species we have detected from the Malvaceae, it is quite possible that C. althaeae is actually not a synonym of S. malvarum, but the validity of the combination C. malvarum that Southworth (1891) made is not affected.
Grove (1937) listed some reports of C. malvarum in the UK: on a malvaceous plant from Perthshire (Cooke 1908), on Lavatera trimestris in Buckinghamshire and Hertfordshire (Smith 1909) and on stems of Malva sp. in Hampshire. The strain we have selected as epitype was isolated from Lavatera trimestris as well, although collected much more recently in the UK. The host genera Malva and Lavatera are closely related and both highly polyphyletic (García et al. 2009) and intergeneric hybrids have been reported (García et al. 2009; Hinsley 2013).
There are other species of Colletotrichum reported from hosts belonging to the Malvaceae. Colletotrichum magnusianum has larger conidia, which measure 16–20 × 4–5 μm and are cylindrical and slightly curved, based on the drawing in the original description. It was found on Malva neglecta close to Merano, Italy (Bresadola 1892). Allescher (1902) considered C. magnusianum to be different from C. malvarum. Magnus (1926) found a species of Colletotrichum on leaves of Malva neglecta in Austria and regarded it as C. magnusianum because of the shorter setae (30–40 μm) compared to those of C. malvarum (60–109 μm). We have not seen authentic material of Bresadola’s species, but it seems likely that it and C. malvarum are not conspecific.
Colletotrichum malvacearum was described on Hibiscus rosa-sinensis from India and has curved conidia (Pavgi and Singh 1965), while C. malvarum has straight conidia. The two species are clearly distinct.
Gloeosporium malvae was described by Sydow (1899), from leaves of Malva neglecta from Germany; the species has subcylindrical, straight to slightly curved conidia that are larger than those of C. malvarum or any of the other species on Malvaceae in this species complex, measuring 19–27 × 3–4 μm. It clearly belongs to a different species complex.
Mortensen (1991) reported on C. gloeosporioides strains isolated from severely diseased plants of Lavatera cvs. ‘Mont Blanc’ and ‘Silver Cup’ in Canada. These strains have a similar host range to C. gloeosporioides f. sp. malvae (= C. tebeestii, this study) from Malva pusilla (Mortensen 1988); however the disease symptoms were found to be different. The strains from Lavatera caused leaf and sometimes also stem lesions in the two Lavatera cultivars, several Malva spp., Althaea rosea and Carthamus tinctorius (Asteraceae), and to a lesser extent in other Malvaceae and non-malvaceous plants. The strains from Lavatera caused most severe disease on leaves, while C. gloeosporioides f. sp. malvae mostly attacks the stem. Mortensen (1991) also observed cultural differences: the Lavatera strains produced more aerial mycelium on PDA and showed less abundant sporulation compared with C. gloeosporioides f. sp. malvae. A culture was reportedly kept (DAOM 211155), but we have not been able to study it.
Colletotrichum malvarum was reported as an anthracnose pathogen of Althaea officinalis in greenhouses in Italy, producing dark colonies with whitish aerial mycelium and cylindrical conidia measuring 14–25 × 3–6 μm on PDA (Tosi et al. 2004). The large size of the conidia suggests a possible relationship with C. althaeae or C. magnusianum. An anthracnose disease of the same host in Switzerland was attributed to C. orbiculare “f. sp. from A. officinalis” (Michel 2005). The isolate formed smaller conidia measuring 10–13 × 3–4 μm, and setae which were 62–75 μm long, was seed transmitted and caused anthracnose of A. officinalis, A. rosea and Malva alcea. Other plants were unaffected, including Lavatera trimestris, M. crispa, M. moschata, M. sylvestris, Fragaria × ananassa and Hypericum perforatum. Further studies are needed, but these strains may well belong to C. malvarum s. str.
There are two reports on C. malvarum on Malva verticillata in Korea (Cho and Shin 2004; Kim et al. 2008) and one on Malva sylvestris in China (Tai 1979). Since Malva verticillata was listed as one of the host species of C. malvarum in the original description of the species, it is possible that those reports refer to C. malvarum s. str. However strains from those collections need to be re-identified using DNA sequence data.
Detailed molecular phylogenetic analysis of strains assigned to C. malvarum has not previously been carried out. Based on morphological similarity, affinities to the lectin Bauhinia purpurea agglutinin (BPA) and the monoclonal anibody UB 20 and ITS-2/D2 rDNA data, Bailey et al. (1996) treated isolates from Lavatera trimestris, Malva pusilla and Sida spinosa as one species belonging to the C. orbiculare species aggregate. We have reassigned some of these strains as C. malvarum, C. tebeestii and C. sidae. The four strains from Lavatera trimestris included in the ITS-2/D2 phylogeny of Bailey et al. (1996) formed a uniform clade, but two strains from Sida spinosa apparently had identical ITS-2/D2 sequences. It is now widely accepted that such sequences are too conservative in Colletotrichum to separate taxa robustly at species level (Cannon et al. 2012). Hyde et al. (2009) speculated that C. malvarum might subsequently be shown not only to be distinct from C. orbiculare, but to represent a species complex in its own right. That view has been substantiated in the current study.
Colletotrichum malvarum is one of the slowest growing species in the C. orbiculare complex; all other species, exept for C. bidentis, reached >20 mm in 10 days on SNA. Colletotrichum malvarum is closely related to C. trifolii; the ITS and CHS-1 sequences are identical. However, the two species can be separated with all other genes studied, and most robustly with GS sequences. Sequences currently lodged in GenBank as C. malvarum (Z18981, DQ792859, DQ792860, DQ792892, DQ792893) belong to C. sidae and C. spinosum according to our study; sequences of Colletotrichum from Malva in GenBank are apparently all from strains from Malva pusilla in Canada, described as C. tebeestii below. The only sequences of C. malvarum s. str. in GenBank are very short ITS-2 and LSU (D2) sequences of C. “orbiculare” generated in the study of Bailey et al. (1996).
Colletotrichum orbiculare Damm, P.F. Cannon & Crous, sp. nov. Fig. 4
Colletotrichum orbiculare (a–g, j–r from ex-holotype strain CBS 570.97, h, i from strain CBS 133196). a–b. acervuli; c. tip of a seta; d. basis of a seta; e–f. conidiophores; g. seta; h–j. conidiophores; k–p. appressoria; q–r. conidia. a, c–f, q: from Anthriscus stem; b, g–p, r: from SNA. a–b: DM; c–r: DIC.—Scale bars: a = 100 μm; e = 10 μm; scale bar of a applies to a–b; scale bar of e applies to c–r.
≠ Cytospora orbicularis Berk. [as ‘Cytispora orbicularis’], Annls Nat. Hist. 1: 207 (1838)
-
≡ Gloeosporium orbiculare (Berk.) Berk., Some Notes upon the Cryptogamic Portion of the Plants Collected in Portugal 1842–50 by Dr Friedr. Welwitsch. The Fungi: 7 (1853)
-
≡ Myxosporium orbiculare (Berk.) Berk. Outlines of British Fungology: 325 (1860)
-
≡ Colletotrichum orbiculare (Berk.) Arx [as ‘(Berk. & Mont.) Arx’], Verh. K. Akad. Wet. Amsterdam, tweede sect. 51(3): 112 (1957), nom. inval. (Art. 34.2)
-
≡ Sirogloea orbicularis (Berk.) Arx, Verh. K. Akad. Wet. Amsterdam, tweede sect. 51(3): 113 (1957), nom. inval. (Art. 34.2)
MycoBank MB 804706
Etymology: The species epithet is based on the widely used but invalid name by which this species has become known.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 2–5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata absent, conidiophores and setae formed directly from hyphae. Setae pale to medium brown, smooth-walled, 30–140 μm long, 1–5-septate, base cylindrical, 3–6.5 μm diam, the tip ± acute or ± rounded, sometimes ending in a conidiogenous opening. Conidiophores hyaline, smooth-walled, fast disintegrating. Conidiogenous cells hyaline, smooth-walled, fast disintegrating, collarette not observed, periclinal thickening not observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to clavate, with one end round and the other truncate, (9–)10.5–12.5(−14) × 4–4.5(−5) μm, mean ± SD = 11.6 ± 1.1 × 4.4 ± 0.2 μm, L/W ratio = 2.7, conidia of strain CBS 514.97 shorter and broader, measuring (9.5–)10–11.5(−13) × (4–)4.5–5.5(−6.5) μm, mean ± SD = 10.8 ± 0.9 × 5.2 ± 0.6 μm, L/W ratio = 2.1. Appressoria (none observed after 10 days, n = 11 after 4 weeks) single, pale to medium brown, smooth-walled, ovate or clavate in outline, with an entire to undulate margin, (5.5–)6–8(−8.5) × (4–)4.5–6(−6.5) μm, mean ± SD = 7.2 ± 1.0 × 5.1 ± 0.8 μm, L/W ratio = 1.4.
Asexual morph on Anthriscus stem. Conidiomata densely arranged with setae enclosing conidiophore clusters; no angular basal cells observed. Setae pale to medium brown, smooth-walled to verruculose, 60–130 μm long, 2–4(−6)-septate, base cylindrical to conical, 3–7.5 μm diam, the tip ± rounded. Conidiophores hyaline, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical, 13–24 × 3–4 μm, opening 1–1.5 μm diam, collarette not observed, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, sometimes clavate with one end round and one end truncate, (11–)12–14.5(−16.5) × 4–4.5(−5) μm, mean ± SD = 13.3 ± 1.3 × 4.4 ± 0.3 μm, L/W ratio = 3.0.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to honey, olivaceous-grey in the centre due to formation of acervuli, aerial mycelium lacking, reverse same colours; growth 16–19.5 mm in 7 days (23–26 mm in 10 days). Colonies on OA flat with entire margin; buff, with olivaceous to olivaceous-grey sectors covered with very short whitish aerial mycelium, reverse buff, with pale olivaceous-grey to olivaceous-grey sectors, growth 17.5–18 mm in 7 days (26.5–27.5 mm in 10 days). Conidia in mass not visible.
Specimens examined: ITALY, Orto Botanico di Parma, on skin of Lagenaria fruit, Nov. 1867, G. Passerini (Erbar. Crittogam. Ital., ser II, no. 148; K isotype of Fusarium lagenaria); Orto Botanico e orti privati dei dintorni di Pavia, summer 1889 (Briosi G and Cavara F, I Funghi Parassiti delle Piante Coltivate ed Utila no. 99; K isotype of C. oligochaetum). JAPAN, Kyoto, on leaves of Cucumis melo, Aug. 2011, Dongliang Jiang, culture CBS 133196 = KTU-H5. PORTUGAL, vicinity of Lisbon, from fruit lesions on skin of Cucurbita lagenaria (possibly Lagenaria siceraria), 1842–1850, F. Welwitsch (K, Cryptotheca Lusitana no. 25, ex. Herb. Berkeley). UK, King’s Cliffe, on small orange gourds (Cucurbita pepo?), collection date unknown, M.J. Berkeley; K, holotype of Cytospora orbicularis, CBS H-4203, isotype. UNKNOWN COUNTRY IN EUROPE (probably UK), from Cucumis sativus, collection date unknown (deposited in CBS collection Mar. 1997 by J.A. Bailey), G.A. Carter (CBS H-20976 holotype of C . orbiculare, culture ex-holotype CBS 570.97 = LARS 73). UNKNOWN COUNTRY, from Cucumis sativus, unknown collection date and collector (deposited in CBS collection Aug. 1924 by W.F. Bewley), culture CBS 122.24.
Notes: Cytospora orbicularis was published by Berkeley (1838) with type material collected in King’s Cliffe, UK on small orange gourds (Cucurbita pepo?) that belongs to a fungus unrelated to that described here. It forms small straight, ellipsoidal conidia (CBS H-4203 measures (4–)5–6(−7) × 2–3(−5) μm, mean ± SD = 5.6 ± 0.7 × 2.5 ± 0.7 μm, L/W ratio = 2.2), that are discharged in tendrils from acervular conidiomata, and is not a Colletotrichum species.
A collection from Portugal (listed above) distributed by Welwitsch as part of his exsiccata set Cryptotheca Lusitana, six specimens of which are preserved at Kew, was examined by Berkeley as well. Berkeley (1853) stated that he received the sample from Montagne, who considered it to belong to a new taxon with the provisional name Gloeosporium welwitschii. Although that name was not taken up and has apparently never been validly published, Berkeley considered Welwitsch’s specimens as a species of Gloeosporium as well. However he incorrectly considered it as a synonym of his previously published species Cytospora orbicularis, and cited this species as the basionym of the new combination Gloeosporium orbiculare, which is therefore not a Colletotrichum either. A few years later Berkeley combined Cytospora orbicularis in Myxosporium as M. orbiculare, referring to the minute conidia in tendrils. He did however not mention the name Gloeosporium orbiculare in his “Outlines of British Fungology” (Berkeley 1860), and subsequent authors assumed that the judgement made by Berkeley (1853) was correct, and therefore listed Gloeosporium orbiculare with Cytospora orbicularis as basionym (e.g. Allescher 1902; Grove 1937; Vassiljevski and Karakulin 1950).
The discordance between the two collections discussed in the previous paragraphs was noted by von Arx (1957a, b). His solution was to treat the name Gloeosporium orbiculare as introduced by Berkeley (1853) as a new species (attributed inaccurately to Berkeley and Montagne) rather than a new combination based on Cytospora orbicularis as made clear by Berkeley. His new combination into Colletotrichum was based on this notional taxon. The type of Cytospora orbicularis was considered by von Arx to be a species of Sirogloea, an obscure coelomycete genus described by Petrak (1923); neither of the two species assigned to this genus appears to have been studied subsequently. Von Arx’s (1957a, b) interpretation of C. orbiculare in the sense of Berkeley (1853) has been followed by the vast majority of Colletotrichum workers, whether they are taxonomists or pathologists. Regrettably, however, von Arx’s actions to preserve the name C. orbiculare in its modern interpretation do not conform to modern nomenclatural rules. To treat Gloeosporium orbiculare as a new species was an invalid action, because Berkeley (1853) clearly cites Cytospora orbicularis as the basionym of Gloeosporium orbiculare. Consequently, the combinations Colletotrichum orbiculare (Berk.) Arx and Sirogloea orbicularis (Berk.) Arx are both invalid (Art. 34.2) as they are alternative names based on the same type.
It would be highly desirable to retain the name for this economically important cucurbit pathogen, as accepted here. This is possible under the current nomenclatural rules: since the name C. orbiculare (Berk.) Arx is invalid and therefore does not exist in nomenclatural terms, the epithet is available again. This allows us to describe the fungus as a new species with the same epithet, based on a specimen that conforms to our present understanding of this species and linked to a living culture. The alternative would be to re-adopt the name C. lagenaria (based on Fusarium lagenaria Pass.), which admittedly was used by some authors for the fungus in question in the earlier part of the 20th century. This would however cause widespread confusion in the applied mycological community. Additionally, we are not certain if the two species are indeed synonyms (see below). Since the invalid name C. orbiculare (Berk.) Arx agrees with the fungus we describe here as new, we omit the authorities in the following discussion on the species and elswhere in this study. Apparently all authors who took up the name Gloeosporium orbiculare from Berkeley (1853) used it incorrectly for a species of the genus Colletotrichum as we understand it today; we assume it might in most cases be the same fungus newly described here as C. orbiculare.
According to von Arx (1957a), C. orbiculare forms leaf spots and fruit anthracnose on several Cucurbitaceae hosts. The fungus usually forms dark mycelium in culture with little aerial mycelium; a typical form on melon with paler aerial mycelium was also isolated. Two typical C. orbiculare isolates from cucumbers did not cause a disease on inoculated bean plants, while cucumber seedlings and apple fruits were infected and an isolate from melon with paler aerial mycelium caused fruit rot on apples as well.
Colletotrichum orbiculare as described here might have a number of earlier names, none of which is confirmed here (see below). In order to avoid further name changes and confusion of this important plant pathogen, we expect the name C. orbiculare described here to be included in the list of Colletotrichum names presented for nomenclatural protection at the 10th International Mycological Congress 2014.
Fusarium lagenaria was described by Passerini (1868) from Cucumis melo and Colocynthis (= Citrullus) sp. in Italy. Roumeguère (1880a) described a collection from France initially identified as Gloeosporium reticulatum (Mont.) Roum. (syn. Fusarium reticulatum Mont.) but corrected the identification shortly afterwards (Roumeguère 1880b) to Fusarium lagenaria. In the same publication, Roumeguère transferred the fungus to Gloeosporium as G. lagenaria (Pass.) Sacc & Roum. Shortly afterwards, Halsted (1893a) combined the name into Colletotrichum, based on studies of strains from different hosts in the USA. The holotype specimen of Fusarium lagenaria could not be located (e-mail from Antonio De Natale: herbarium PORU and POR are closed due to renovation of the building), but an isotype is included in the Kew collection. Its similarity with C. orbiculare (at least in terms of morphological characters) can be broadly confirmed, though the collection has conidia that are very variable in size and shape, with a substantial proportion of conidia that are clavate rather than cylindrical. Conidial measurements from the isotype measure (12.2–)13.5–16.7(−17.5) × (4.2–)4.5–5.7 μm, mean ± SD = 15.0 ± 1.1 × 4.9 ± 0.4 μm, L/W ratio = 3.0. Wollenweber and Hochapfel (1949) studied the holotype specimen of F. lagenaria; the conidia measured 15–18 × 4.5–6 μm, mean = 16.5 × 5.3 μm. The conidial length/width ratio of the holo- and isotypes of C. lagenaria is thus somewhat larger than that calculated for the type of C. orbiculare as described here. Several authors listed Gloeosporium orbiculare and C. lagenaria (or Gloeosporium lagenaria) as two separate anthracnose pathogens of Curcurbitaceae (Grove 1937; Vassiljevski and Karakulin 1950; Wollenweber and Hochapfel 1949). Wollenweber and Hochapfel (1949) keyed the two species out according to their average conidial size (based on living strains on different media) with 14 × 5 μm as Gloeosporium orbiculare and 17 × 5.3 μm as Gloeosporium lagenaria. However, both dimensions are somewhat larger than our measurements of the type of the newly described C. orbiculare and that of Fusarium lagenaria, respectively. In the absence of authentic cultures it is not possible to confirm the synonymy of Fusarium lagenaria with C. orbiculare using current species recognition criteria.
Gloeosporium cucurbitarum was described by Berkeley and Broome (1882) from the skin of gourds (Lagenaria or Cucurbita) in Brisbane, Australia. They stated that clavate, short-stipitate conidia 0.0004 to 0.0009 in. (ca. 10–23 μm) in length are formed in small cirri that arise from depressed patches on the host cuticle. Conidia and patches are bright orange coloured. Two syntypes were assigned, both collected by F.M. Bailey, both of which are present in Kew. Bailey 393 is fragmentary and rather decomposed, but two specimens of Bailey 371 show a Colletotrichum-like acervular fungus lacking setae. Conidia from Bailey 371 are variable in size and shape, measuring (11–)12.5–18 × (3.5–)4–5.5 μm, mean ± SD = 14.7 ± 1.7 × 4.6 ± 0.4 μm, L/W ratio = 3.2. Their dimensions and shape are therefore very similar to those of C. lagenaria. Von Arx (1957a) listed C. lagenaria and Gloeosporium cucurbitarum as synonyms of C. orbiculare, but their precise taxonomic position is difficult to ascertain without molecular data.
Colletotrichum oligochaetum was described by Cavara (1889) on leaves and stems of Lagenaria vulgaris from Italy. Conidia were considered to be aseptate, hyaline, cylindrical or ovoid, often constricted in the centre and with one or both sides truncate, measuring 13–15 × 4–5 μm. Setae were olivaceous, 1–2-septate, with an inflated base and obtuse tip, measuring 60–70 × 5–7 μm. Examination of an isotype of C. oligochaetum from Kew confirms these observations, and it is possible that Cavara’s species represents an earlier name of C. orbiculare in the restricted sense of the species described here. Conidial measurements are as follows: (10.5–)11–15(−17) × 4–5 μm, mean ± SD = 13.1 ± 1.5 × 4.3 ± 0.3 μm, L/W ratio = 3.1. Cavara’s species name has not been used in recent years, and sequence data from type material are not available. In the interests of nomenclatural stability, it would not be appropriate to adopt it as the correct name for C. orbiculare.
Gloeosporium lagenaria var. citrulli was described by Potebnia (1907) on the epicarp of Citrullus vulgaris in South Russia where the taxon was noted to be very common and destructive. Conidia of this species were observed to be hyaline and 14 × 5 μm in size, with conidiophores measuring 20–30 × 5 μm. No setae were observed, in contrast to other strains from Russia that were identified as C. oligochaetum. Bearing in mind that many strains of C. orbiculare have poorly developed setae, we think it is possible that Potebnia’s variety belongs to that species. However, we have not seen material of this taxon to confirm this. Nomenclatural priority is rank-limited, so Potebnia’s name would not threaten uptake of C. orbiculare as a new species.
Colletotrichum gloeosporioides f. sp. cucurbitae was first introduced for isolates from cucumber that were used in a pathogenicity trial (Menten et al. 1980) and later reported from Citrullus lanatus, Cyclanthera pedata and Solanum muricatum (sweet pepino) (Cardoso et al. 2001). Formae speciales are host/pathogen combinations and do not belong in the taxonomic hierarchy of the pathogen, so cannot be considered to be formal synonyms. We have not been able to assess the phylogenetic position of these strains, but it is likely that the fungi concerned belong to C. orbiculare.
Ellis and Everhart (1889) observed a fungus on banana rind in the USA that they could not differentiate from Gloeosporium lagenaria on Cucurbitaceae species and therefore called it Gloeosporium lagenaria var. musarum. However, a large number of strains from Musa species has been studied belonging to different species in several Colletotrichum species complexes, including C. paxtonii in the C. acutatum complex (Damm et al. 2012a), C. karstii in the C. boninense complex (Damm et al. 2012b) and C. musae in the C. gloeosporioides complex (Weir et al. 2012); but none of them belongs to the C. orbiculare complex (Damm, unpublished data). Conidia of C. karstii that occurs on banana in Central and South America have a broadly similar shape and similar dimensions (Yang et al. 2011; Damm et al. 2012b) as those of the isotype of Fusarium lagenaria, that could not be confirmed to belong to the C. orbiculare complex in our study (see above). It is therefore possible that Gloeosporium lagenaria var. musarum is a synonym of C. karstii. No material has been seen and no morphological details were provided by Ellis and Everhart (1889), but we think that it is more likely that their fungus belongs to a different species complex.
Glomerella magna was described from diseased Citrullus lanatus (watermelon) in the USA (Jenkins and Winstead 1962, 1964). It has larger conidia than C. orbiculare, measuring 24–40 × 4–6 μm. Based on mtDNA RFLP, DNA fingerprinting and RAPD PCR by Correll et al. (1993), Glomerella magna isolates formed a haplotype different from that of C. orbiculare. This agrees with the study of Liu et al. (2007) and preliminary results based on sequence data (U. Damm, unpublished data), indicating that Glomerella magna belongs to a different species complex. Wasilwa et al. (1993) found that strains of this species were less virulent compared with those of C. orbiculare.
We have not seen isolates producing sexual structures that belong to C. orbiculare s. str., but sexual taxa linked by their authors to C. orbiculare and its relatives have been observed. Glomerella lagenaria was described by Stevens (1931) from ultraviolet-irradiated corn meal agar cultures of strains identified as C. lagenaria from Georgia and Illinois, USA. One of these was isolated by Stevens himself from melons (host species not mentioned); the other was isolated by Dr B.B. Higgins in Georgia. Perithecia were described as globose, dark, up to 110 μm diam, with numerous asci containing hyaline, aseptate, immature ascospores. The name G. lagenaria was introduced as a new combination based on Fusarium lagenaria Pass. Some years later, Watanabe and Tamura (1952) observed asci and ascospores in potato agar cultures of fungi identified as C. lagenaria from cucumber in Japan, and also described the sexual stage as Glomerella lagenaria. A further study noted a sexual morph referred to as Glomerella cingulata var. orbiculare (Jenkins and Winstead 1961). This developed from an isolate identified as belonging to C. orbiculare race 1 (see Goode 1958), from edible gourd (Lagenaria leucantha var. longissima) in North Carolina, USA. The isolate on its own produced few ascospores, but abundant ascospore production was observed when the strain was mated with certain other isolates assigned to C. orbiculare, and the ascospores themselves when cultured produced fertile perithecia. Jenkins and Winstead (1961) regarded their isolate as different from Glomerella lagenaria as described by Watanabe and Tamura (1952). The name Ga. cingulata var. orbiculare is invalid as the abstract published by the authors was not intended formally to introduce the name, and no Latin description was provided (Weir et al. 2012).
In the study of Correll et al. (1993) apparently authentic isolates of the study of Jenkins and Winstead (1961) from cucuzzi gourd (Lagenaria leucantha var. longissima) and two further isolates from honeydew fruits (Cucumis melo) formed a mtDNA RFLP haplotype different from those of C. orbiculare (s. str.) and Glomerella magna, suggesting these isolates to belong to a different species, maybe even a different species complex. One of these isolates was observed to be homothallic, producing the sexual morph in culture (J. C. Correll, unpublished data). In inoculation experiments by Wasilwa et al. (1993), in contrast to isolates of C. orbiculare (s. str.), isolates of this species were avirulent or only weakly virulent to cucurbits. This species could well belong to the C. boninense species complex. Damm et al. (2012b) recently identified two strains from Citrullus lanatus and Cucumis melo as C. karstii, a species of the C. boninense complex that produces a sexual morph in culture without crossing, suggesting it to be homothallic. Its conidia are slightly larger but have a similar shape to those of C. orbiculare, and the size of asci and ascospores of C. karstii is similar to that of the sexual stage described by Jenkins and Winstead (1962). However, the strains from the study of Correll et al. (1993) need to be re-identified using sequence data. None of the teleomorphic strains referred to in the papers of Stevens (1931) and Watanabe and Tamura (1952) have been re-examined, and we are not confident that any of them represent a real sexual stage of C. orbiculare either.
There are further species of Colletotrichum associated with diseases of cucurbits belonging to other Colletotrichum species complexes (Cannon et al. 2012). One Colletotrichum strain from Cucurbita pepo in the CBS collection belongs to C. coccodes (Liu et al. 2013) and one strain from Cucumis melo to the recently described species C. melonis belonging to the C. acutatum species complex (Damm et al. 2012a). Colletotrichum coccodes has longer and narrower conidia than C. orbiculare, while conidia of C. melonis have at least one acute end and a larger L/W ratio (≥ 3.5) than C. orbiculare (≤ 3.0). There are uncharacterised specimens from the C. gloeosporioides complex in the IMI dried collection associated with several species of cucurbits.
Among the many historical taxa assigned to Colletotrichum and Vermicularia (a generic synonym; Cannon et al. 2012) that have not been assessed using molecular methods, there are some associated with cucurbits that do not appear to belong to the C. orbiculare species complex. Vermicularia cucurbitae (Cooke 1878) was described as forming globose “perithecia” with linear, tiny (15 × 3 μm) conidia and hyaline, lanceolate, acuminate, triseptate conidia measuring 50 × 5 μm in fruits of Cucurbitaceae from Aiken, South Carolina, USA. It can therefore be discounted as a potential earlier name of C. orbiculare, and is probably not a species of Colletotrichum. Another species on skin of rotten Cucurbita lagenaria in Germany and Italy, Vermicularia wallrothii (Saccardo 1884) produces curved conidia that are longer than those of C. orbiculare, measuring 25–28 × 3–3.5 μm. It belongs to a different species complex.
Colletotrichum orbiculare is closely related to C. sidae, C. spinosum and C. tebeestii; their CHS-1 sequences are identical. Additionally, the GAPDH and TUB2 sequences are identical with those of C. sidae, and ITS sequences are not always different from C. spinosum. However, the species can be separated from the other species with its unique GS, ACT and HIS3 sequences, and GS sequences provide the most effective differential data separating these taxa.
Comparison of sequences retrieved from NIAS Genbank (http://www.gene.affrc.go.jp) showed that strains MAFF 306685, MAFF 306518, MAFF 306681 and MAFF 306589 from Citrullus vulgaris, Cucumis melo, Cucumis sativus and Gerbera, respectively, in Japan have identical ITS, GAPDH, ACT and TUB2 sequences with strain CBS 570.97. In blastn searches, the ITS sequence of CBS 570.97 is identical to those of strain 11–045 (JX997422) from Citrullus lanatus, four strains (11–100, 11–169, 11–209, 10–151) from Cucumis sativus (JX997424-JX997427) and strain 11–061 (JX997423) from an unidentified cucurbitaceous rootstock (all from an unpublished study from Korea by Han KS, Lee SC, Lee JS, Soh JW and Park MJ), further to strains MAFF 306518 (AB042308) and MAFF 306589 (AB042309) (Moriwaki et al. 2002) and to previously published or generated ITS sequences of two strains that are included in this study, 104-T (= CBS 514.97, JQ005778, O’Connell et al. 2012) and BBA 71046 (= CBS 570.97, Nirenberg et al. 2002). With 99 % similarity (one bp difference), follow Glomerella cingulata var. orbiculare strain ATCC 42085 (KC146360) from cucumber in the USA (unpublished study by Gujjari P, Suh S-O and Zhou J), C. orbiculare strains MAFF 306685 (AB269941) from watermelon in Japan and MAFF 306681 (AB269939) from cucumber in Japan (both published in database only by Yoshida S, 2007) and C. orbiculare strain MX-2-153-Mexico (AY841133) from Annona cherimola in Mexico (Villanueva-Arce et al. 2008). The closest match with the TUB2 sequence of strain CBS 570.97, with 100 % identity, is C. orbiculare strain 104-T (= CBS 514.97, JQ005862, O’Connell et al. 2012), followed by C. spinosum (as C. orbiculare) strain STE-U 5296 (= CBS 113171, AY376589, Lubbe et al. 2004); both strains are included in this study. The GAPDH sequence of strain CBS 570.97 was 100 % identical to that of C. orbiculare s. str. strains DAR61396 (DQ792862) and MH2 (DQ792863) and 99 % identical (1 bp difference) to that of C. orbiculare s. str. strains CP6 (DQ792867), JC1 (DQ792866) and RL1 (DQ792868) as well as two strains from Sida (here C. sidae) 3-7-11 (DQ792869), 4-3-12 (DQ792870), all generated by Liu et al. (2007). In a blastn search with the GS sequence of strain CBS 570.97, all GS sequences of C. orbiculare (s. str.) strains from the study of Liu et al. (2007) were 99 % identical (1–4 bp differences): strains AK9 (DQ792884), JC1 (DQ792889), DAR61396 (DQ792885), JX13 (DQ792888), RL1 (DQ792891), JX10 (DQ792887), MH2 (DQ792886), CP6 (DQ792890). GS sequences of the two C. orbiculare strains from Xanthium (here treated as C. spinosum) were only 93 % identical.
Colletotrichum orbiculare has been used extensively in recent years to investigate host-pathogen interactions (Asakura et al. 2009, 2012; Tanaka et al. 2009; Fujihara et al. 2010; Kubo and Takano 2013), and strain MAFF 240422 (= CBS 514.97 = 104-T, included in this study) has been subjected to whole genome-level analysis (Gan et al. 2013).
Colletotrichum sidae Damm & P.F. Cannon, sp. nov. Fig. 5
Colletotrichum sidae (from ex-holotype strain CBS 504.97). a–b. acervuli; c. seta; d–f. conidiophores; g. seta; h–j. conidiophores; k–n. appressoria-like structures; o–p. conidia. a, c–f, o: from Anthriscus stem; b, g–n, p: from SNA. a–b: DM; c–p: DIC.—Scale bars: a = 100 μm; f = 10 μm; scale bar of a applies to a–b; scale bar of f applies to c–p.
MycoBank MB 804692
Etymology: The species epithet is derived from the host genus name.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–8.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed (but see below). Conidiomata absent, conidiophores and setae formed directly on hyphae. Setae pale to medium brown, smooth-walled to finely verruculose, 40–70 μm long, 1–2-septate, base cylindrical, 3.5–5 μm diam, tip ± acute to ± rounded. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 35 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical to ampulliform, 12–18.5 × 3.5–4.5 μm, polyphialides observed, opening 0.5–1.5 μm diam, collarette 0.5–1 μm long, periclinal thickening not observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, with one end round and the other truncate, (9–)10–12(−14) × (4–)4.5–5.5 μm, mean ± SD = 11.1 ± 1.0 × 4.9 ± 0.4 μm, L/W ratio = 2.3. Appressoria not observed on the undersurface of the medium, but in old cultures appressoria-like structures that possibly function as chlamydospores were observed within the medium. These are single, medium brown, smooth-walled, subglobose, ovate to broadly elliptical in outline, with an entire or undulate margin, (3–)4–6(−7.5) × (2.5–)3.5–5(−6) μm, mean ± SD = 5.2 ± 1.0 × 4.3 ± 0.7 μm, L/W ratio = 1.2.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells, 3–8 μm diam. Setae pale to medium brown, verruculose, 30–75 μm long, 1–2(−3)-septate, base cylindrical, sometimes inflated, 4–8.5 μm diam, tip ± acute to ± rounded. Conidiophores pale brown, smooth-walled, septate, branched, to 50 μm long. Conidiogenous cells pale brown, smooth-walled, cylindrical, 7–18 × 3.5–6 μm, opening 1 μm diam, collarette not observed, periclinal thickening rarely visible. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, with one end round and the other truncate, (12–)12.5–14(−15.5) × 4.5–5(−5.5) μm, mean ± SD = 13.3 ± ?>0.9 × 4.8 ± 0.2 μm, L/W ratio = 2.8.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline; agar medium, filter paper and Anthriscus stem partly covered with sparse grey to black acervuli, Anthriscus stem partly covered with low white aerial mycelium, reverse same colours; growth 18.5–20 mm in 7 days (26.5–29 mm in 10 days). Colonies on OA flat with entire margin; dark olivaceous to olivaceous-black, with a buff margin, partly covered with very short aerial mycelium, reverse olivaceous-grey, growth 19.5–21 mm in 7 days (29.5–30.5 mm in 10 days). Conidia in mass whitish to rosy-buff.
Specimens examined: USA, Arkansas, from Sida spinosa, 1 May 1988 (deposited in CBS collection Feb. 1997 by J.A. Bailey), D.O. TeBeest (CBS H-20975 holotype, culture ex-holotype CBS 504.97 = ATCC 58399 = NRRL 8096 = LARS 76); Arkansas, Stuttgart, from Sida spinosa, unknown collection date (deposited in CBS collection Mar. 1997 by J.A. Bailey), D.O. TeBeest (culture CBS 574.97 = ATCC 96725 = 3-1-1 = LARS 625 = Cm-9); Arkansas, from Sida spinosa, unknown collection date (deposited in CBS collection Feb. 1997 by J.A. Bailey), D.O. TeBeest (culture CBS 518.97 = LARS 629 = Cm-4).
Notes: Prickly sida (Sida spinosa) is an arable weed in the USA, especially in cotton and soybean fields, where it reduces the yield severely. It is especially difficult to control in cotton, because both plants belong to the Malvaceae (Templeton 1974). A fungus naturally occuring on this weed in Arkansas, USA, identified as C. malvarum was tested as a mycoherbicide for biological control of prickly sida (Templeton 1974; Kirkpatrick et al. 1982). This fungus first causes stem spots and later a devasting seedling blight of young weed seedlings. According to GenBank accession Z18981 (28S ribosomal RNA), LARS 076 is the same strain as C. malvarum ATCC 58399 (= NRRL 8096) that was patented for biological control of prickly sida by Templeton (1976). Strains tested by Kirkpatrick et al. (1982) were only pathogenic to hollyhock (Althaea rosea) and prickly sida, both Malvaceae. No infection occurred in any other of the 38 plants tested, including soybeans, cotton, wheat and tomatoes.
Another C. malvarum strain from Sida spinosa, ATCC 96725, was incompatible when mated with C. gloeosporioides f. sp. aeschynomene strain ATCC 96723 (= Clar-5a) from Aeschynomene viginica (Cisar et al. 1994), which belongs to the C. gloeosporioides species complex and has recently been described as C. aeschynomenes by Weir et al. (2012).
Bailey et al. (1996) recognised this fungus as belonging to the C. orbiculare species complex, but the five strains from Sida spinosa included in their ITS-2/D2 phylogeny did not group; some of them had the same sequences as those from Lavatera trimestris, which is regarded as C. malvarum s. str. in this paper. The sequence is too conserved to be diagnostic at this level. Liu et al. (2007) showed the isolates from Sida spinosa differed from C. orbiculare and several other closely related species based on RFLPs and sequences from the GS and GAPDH genes.
Colletotrichum sidae is only known from Sidae spinosa from the USA. Few other species are known on Sida spp. (Farr and Rossman 2013). For example, C. capsici was reported on Sida acuta in India and Sida spinosa in the USA (Sarbhoy and Agarwal 1990; McLean and Roy 1991); C. capsici has strongly curved conidia and was recently revealed to be a synonym of C. truncatum (Damm et al. 2009). Thaung (2008) lists C. gloeosporioides on Sida sp. in Myanmar; this is the only report of this species from Sida. Because of the wide concept of C. gloeosporioides in the past (Von Arx 1957a), is possible that this report actually also represents C. sidae.
Colletotrichum sidae is closely related to C. orbiculare, C. spinosum and C. tebeestii; their CHS-1 sequences are identical. Additionally, the HIS3 and ACT sequences of C. sidae are the same as those of C. tebestii, while the GAPDH and TUB2 sequences are identical with those of C. orbiculare. Therefore, blastn searches with the TUB2 and GAPDH sequences of strain CBS 504.97 resulted in the same closest matches as with that of the ex-epitype strain of C. orbiculare. However, the species can be separated with GS and ITS sequences. Closest match in a blastn search with the GS sequence of strain CBS 504.97 was with 100 % identity that of strain 4-3-12 (DQ792893), followed by that of strain 3-7-11 with 99 % (6 bp differences, DQ792893), both from Sida spinosa from the study of Liu et al. (2007). The closest matches with the ITS sequence of strain CBS 504.97 with 99 % identity (1 bp difference) are C. orbiculare MAFF 306685 (AB269941) from Japan (Yoshida S, published 2007 in database only) and C. orbiculare MX-2-153-Mexico (AY841133) from Annona cherimola (Villanueva-Arce et al. 2008). All other sequences of Colletotrichum strains from Sida that could be located in GenBank are short ITS-2/LSU (D2) sequences from the study of Bailey et al. (1996).
Colletotrichum spinosum Damm & P.F. Cannon, sp. nov.Fig. 6
Colletotrichum spinosum (from ex-holotype strain CBS 515.97). a–b. acervuli; c. seta; d–f. conidiophores; g. seta; h–i. conidiophores; j–o. appressoria; p–q. conidia. a, c–f, p: from Anthriscus stem; b, g–o, q: from SNA. a–b: DM; c–q: DIC.—Scale bars: a = 100 μm; f = 10 μm; scale bar of a applies to a–b; scale bar of f applies to c–q.
MycoBank MB 804693
Etymology: A reflection both of the setose nature of the conidiomata, and of the name of its host.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–8 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata absent, conidiophores and setae formed directly from hyphae. Setae pale brown, smooth-walled, 25–80 μm long, 0–3-septate, base cylindrical or inflated, 3.5–6 μm diam, tip ± rounded or ± acute. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical to doliiform, sometimes with a mucous coating, 8–20 × 3.5–5 μm, opening 1.5–2 μm diam, collarette not observed, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to clavate, with one end round and the other truncate, (9.5–)12.5–14.5(−23) × (4–)4.5–5 μm, mean ± SD = 13.5 ± 1.2 × 4.6 ± 0.3 μm, L/W ratio = 3.0. Appressoria (after 4 weeks) formed singly, pale to medium brown, smooth-walled, subglobose, ovate or oblong in outline, with an entire to undulate margin, (5.5–)6.5–10.5(−13) × (4–)5–6.5(−7) μm, mean ± SD = 8.5 ± 1.9 × 5.6 ± 0.9 μm, L/W ratio = 1.5.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae densely arranged; no angular basal cells observed. Setae medium brown, smooth-walled to verruculose, 40–95 μm long, 0–2(−3)-septate, base cylindrical to ± inflated, 4–9 μm diam, tip ± acute to ± rounded. Conidiophores pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells pale brown, smooth-walled, cylindrical, 9–24 × 3–5.5 μm, sometimes with a mucous coating, opening 1–2 μm diam, collarette not observed, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, sometimes clavate with one end round and the other truncate, (12–)12.5–14.5(−16) × 4.5–5(−5.5) μm, mean ± SD = 13.5 ± 1.0 × 4.9 ± 0.3 μm, L/W ratio = 2.8.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale cinnamon; the agar medium, filter paper and Anthriscus stem partly covered with saffron, apricot to dark grey acervuli, aerial mycelium lacking, reverse same colours; growth 18.5–19.5 mm in 7 days (28.5 mm in 10 days). Colonies on OA flat with entire margin; surface buff, partly olivaceous-grey to iron-black, margin buff, covered with black acervuli with saffron to apricot conidial masses oozing out, reverse olivaceous-grey with a pale purplish grey margin, growth 19.5–20.5 mm in 7 days (28–29.5 mm in 10 days). Conidia in mass saffron to apricot.
Specimens examined: AUSTRALIA, New South Wales, Coolah, from stem lesion of Xanthium spinosum, 20 Mar. 1983 (deposited in CBS collection Feb. 1997 by J.A. Bailey), B. Auld (CBS H-20977 holotype, culture ex-holotype CBS 515.97 = LARS 465 = DAR 48942). ARGENTINA, Chepes, from Xanthium spinosum, collection date unknown (deposited in IMI collection 1995 by C. Ellison and H.C. Evans and in CBS collection 2002 by P.W. Crous), culture CBS 113171 = IMI 368075.
Notes: Xanthium spinosum (bathurst burr, spiny cocklebur, Asteraceae) is a widespread noxious weed of pastures and crops (e.g. cotton, soybean, lucerne) in Australia, where it was introduced during the white settlement period (Auld and Medd 1987). It is native to Argentina (Wapshere et al. 1995). Colletotrichum spinosum was first reported in Queensland, Australia as Colletotrichum sp. (Veitch 1942) and in 1941 in New South Wales (Walker 1962), where it caused severe damage (50–80 % mortality) in the late 1940s (Butler 1951). The species was originally identified as C. xanthii (Butler 1951) and later considered to belong to C. orbiculare (Simmonds 1965, 1966; Walker et al. 1991).
Colletotrichum spinosum (as C. orbiculare) was found to be a common pathogen of Xanthium spinosum in eastern Australia, causing seedling blight and stem anthracnose (Walker et al. 1991). It was tested as a mycoherbicide against this weed (Auld et al. 1988, 1990; Auld and Say 1999). Both isolates included in our study were used in an evaluation of strains from Argentina and Australia for biological control of the weed in Australia (Auld and Say 1999). The occurrence of strains from both Australia and Argentina suggests that this fungus was introduced to Australia with its host plant. Conidia from the type strain of C. spinosum measured 11–15(−18) × (4–)4.5–5 μm on Xanthium host tissue, and 11–13 × 4.5–5(−5.5) μm on water agar and carnation leaf and stem pieces, with only a few appressoria being produced on glass slides or cover slips (Walker et al. 1991).
In pathogenicity tests (Walker et al. 1991), isolate DAR 48942 (= CBS 515.97) was found to be highly virulent to its host plants (plants killed), and caused leaf and stem lesions on some other Xanthium spp. as well as some other Asteraceae, especially Carthamus tinctorius cv. Gila (not on two other cultivars tested). In addition, it was found to infect Citrullus lanatus var. lanatus (Cucurbitaceae), Eucalyptus cinerea (Myrtaceae) and two Acacia spp. (Mimosaceae), while other Cucurbitaceae showed no reaction. Additionally, it caused fruit rots of melon, cucumber, apple, pear and tomato.
Another species from Xanthium, C. xanthii, was described on stems of Xanthium canadense in the USA by Halsted (1893b). Walker et al. (1991) studied the type material of C. xanthii and found acute conidia, and concluded this fungus to be a synonym of C. acutatum. Even if it is not identical with C. acutatum s. str. as defined by Damm et al. (2012a), C. xanthii is clearly a different fungus to C. spinosum and almost certainly belongs to the C. acutatum species complex. Other Colletotrichum species reported on Xanthium spp. are C. dematium, C. coccodes and C. truncatum (Alcorn 1976; Roy 1982; Hartman et al. 1986; Walker et al. 1991), all belonging to different species complexes (Cannon et al. 2012). One strain from Xanthium sp. (CBS 125346 = DAOM 212643) was confirmed as being C. dematium s. str. (Damm et al. 2009).
Colletotrichum spinosum is closely related to C. orbiculare, C. sidae and C. tebestii; the four species have identical CHS-1 sequences. In a blastn search on GenBank, the ITS sequence of strain CBS 515.97 was 100 % identical with that of C. orbiculare strain MAFF 306685 from watermelon in Japan (AB269941, Yoshida S, published 2007 in database only) and C. orbiculare MX-2-153-Mexico (AY841133) from Annona cherimola (Villanueva-Arce et al. 2008). In our study, one of the C. orbiculare strains from Japan (CBS 133197) also had the same ITS sequence, while all others differed. This means that ITS sequences are not reliable for distinguishing C. spinosum from other species.
While there is only one bp difference in the ACT sequences of C. spinosum and C. orbiculare, C. spinosum can be separated effectively based on GAPDH, HIS3, TUB2 and GS sequences. Some strains from previous studies were confirmed as being C. spinosum by performing blastn searches in GenBank with TUB2, GAPDH and GS sequences of strain CBS 515.97: the TUB2 sequence of the strain was identical with that of strain STE-U 5296 (= CBS 113171, AY376589) from Xanthium, another strain of C. spinosum included in this paper previously treated as C. orbiculare by Lubbe et al. (2004), while GS (DQ792882, DQ792882) and GAPDH (DQ792859, DQ792860) sequences of strains LW1 and LW6 from Xanthium from the study of Liu et al. (2007) matched with those of strain CBS 515.97.
Colletotrichum tebeestii Damm & P.F. Cannon, sp. nov. Fig. 7
Colletotrichum tebeestii (from ex-holotype strain CBS 522.97). a–b. acervuli; c. seta; d–f. conidiophores; g. tip of a seta; h. basis of a seta; i–k. conidiophores; l–q. appressoria-like structures; r–s. conidia. a, c–f, r: from Anthriscus stem; b, g–q, s: from SNA. a–b: DM; c–s: DIC.—Scale bars: a = 100 μm; f = 10 μm; scale bar of a applies to a–b; scale bar of f applies to c–s.
MycoBank MB 804694
Etymology: Named in honour of David TeBeest, a prominent researcher on pathology and management of Colletotrichum diseases and the development of mycoherbicides.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–7 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed (but see below). Conidiomata, conidiophores and setae formed directly on hyphae or on pale brown angular cells, 3–7 μm diam. Setae medium brown, verruculose, 35–80 μm long, 1–3-septate, base cylindrical, sometimes slightly inflated, 3.5–6 μm diam, tip ± acute. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 60 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical to obclavate, sometimes extending to form new conidiogenous loci, 11–18 × 3.5–5.5 μm, opening 1–1.5(−2) μm diam, collarette not observed, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to clavate, with one end round and the other truncate, (7.5–)11–14(−15.5) × 4–4.5(−5) μm, mean ± SD = 12.5 ± 1.3 × 4.4 ± 0.3 μm, L/W ratio = 2.8. Appressoria not formed after 10 days on the underside of the medium, but in old cultures appressorium-like structures are formed within the medium that could function as chlamydospores; these are single or in small dense clusters, pale to medium brown, smooth-walled, subglobose, elliptical or clavate in outline, with an entire to undulate margin, (4–)5–7.5(−10) × (3.5–)4–6(−8) μm, mean ± SD = 6.3 ± 1.4 × 4.9 ± 1.0 μm, L/W ratio = 1.3.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells, 3.5–8 μm diam. Setae medium brown, smooth-walled, upper part verruculose, 25–100 μm long, 1–3-septate, base cylindrical, sometimes slightly inflated, 4.5–8 μm diam, tip acute. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical to subulate, sometimes extending to form new conidiogenous loci, 9–18 × 3–5 μm, opening 1–1.5 μm diam, collarette 0.5–1 μm long, but often not visible, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to clavate, with one end round and the other truncate, (8.5–)11.5–13.5(−14.5) × 4–4.5(−5) μm, mean ± SD = 12.5 ± 1.2 × 4.5 ± 0.3 μm, L/W ratio = 2.8.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale cinnamon, filter paper sometimes partly grey-olivaceous; the agar medium, Anthriscus stem and filter paper partly covered with sparse low white to grey aerial mycelium and salmon to black acervuli, reverse filter paper sometimes partly grey-olivaceous; growth 15–19 mm in 7 days (21.5–25.5 mm in 10 days). Colonies on OA flat with entire margin, dark grey-olivaceous to olivaceous-black with a buff margin, aerial mycelium lacking, partly covered by salmon conidia oozing from black acervuli, reverse pale olivaceous-grey to olivaceous-grey, growth 18–20.5 mm in 7 days (26–29 mm in 10 days). Conidia in mass salmon.
Specimen examined: CANADA, Saskatchewan, Raymore, from Malva pusilla, unknown collection date (probably 1982, deposited in CBS collection Feb 1997 by J.A. Bailey), D.O. TeBeest (CBS H-20974 holotype, culture ex-holotype CBS 522.97 = LARS 733 = No. 83–43).
Notes: Mortensen (1988) tested the pathogenicity of seven strains identified as C. gloeosporioides f. sp. malvae from different locations in Saskatchewan, and one from Manitoba, Canada. An isolate selected from these strains was developed as a mycoherbicide against the annual weeds round-leaved mallow (Malva pusilla) and velvetleaf (Abutilon theophrasti) occuring in lentil and wheat fields in Canada (Mortensen 1988; Makowski and Mortensen 1992). It was registered under the name BioMal® by Philom Bios, Saskatoon, Canada, but to our knowledge is not commercially available. The fungus attacks leaves, stems and petioles of Malva pusilla. The control of round-leaved mallow in strawberry plots by C. gloeosporioides f. sp. malvae was found to increase the strawberry yield. Strawberry plants themselves were apparently infected, but the fungus does not cause disease symptoms (Mortensen and Makowski 1995). In other field crops such as wheat, lentil and sugar beet, the fungus causes only latent infections of non-target species, except for safflower (Makowski and Mortensen 1998). Bearing in mind the apparently strong host specificity of species in the C. orbiculare clade, the identity of fungi isolated from such non-target sources should be reconfirmed. Goodwin (2001) studied the hemibiotrophic interaction of strains of this biological control agent with its host. Host and pathogen genes expressed during infection were studied by Dean et al. (2003) and Goodwin and Chen (2002a, b).
The strain we have studied (CBS 522.97 = LARS 733 = No. 83–43, from Raymore, Saskatchewan, Canada) is not the same strain that was patented and registered as BioMal (Mortensen et al. 1994; Mortensen 1987); the latter was designated as ATCC 20767 (= Regina 83-43-1), from Regina, also in Saskatchewan. Bailey et al. (1996) list a strain from the USA (94116) as the BioMal fungus; we are unsure how this discrepancy arose. All seven strains from Malva pusilla included in the ITS-2/D2 phylogeny of Bailey et al. (1996) formed a uniform clade, one of which is included in our study; we assume they all belong to the species described here as C. tebeestii.
Because of the similar morphology and the limited sequence variation (ITS-2/D2) of the strains from Malva pusilla compared to strains from other Malvaceae genera, Medicago and Cucumis sativus, Bailey et al. (1996) recognised this fungus as being one of the forms of the “C. orbiculare aggregate species”. Consequently, all of their sequences are lodged in GenBank as C. orbiculare. However, the multigene phylogeny in our study revealed this fungus to form a separate clade. Colletotrichum tebeestii is indeed closely related to C. orbiculare, C. sidae and C. spinosum; their CHS-1 sequences are identical. Additionally, ACT and HIS3 sequences are identical to those of C. sidae, while there is only 1 bp difference in the ITS, TUB2 and GS sequences between C. tebeestii and one or two of the other three species. Colletotrichum tebeestii can best be differentiated from other species by its unique GAPDH sequence.
Closest match in a blastn search with the ITS sequence of strain CBS 522.97 was (with 100 % identity) C. “gloeosporioides” strain DAOM 212647 (EU400138, Chen et al. 2012). Closest matches of the TUB2 sequence of strain CBS 522.97 (with 99 % identity) were C. spinosum (as C. orbiculare) strain STE-U 5296 (= CBS 113171, AY376589, Lubbe et al. 2004) and C. orbiculare strain 104-T (= CBS 514.97, JQ005862, O’Connell et al. 2012); both strains are included in this paper. The GS sequence is 99 % (1 or 8 bp difference(s), respectively) identical with C. sidae (as C. malvarum) strains 3-7-11 and 4-3-12 (DQ792892, DQ792893) and C. spinosum (as C. orbiculare) strains LW1 and LW6 (DQ792882, DQ792882) from the study of Liu et al. (2007). The latter two strains (from Xanthium) lodged as C. “malvarum” also had the most similar GAPDH sequences, but these were only 93 % identical with that of the ex-type strain of C. tebeestii.
Colletotrichum trifolii Bain, in Bain & Essary, J. Mycol. 12(5): 193 (1906). Fig. 8
Colletotrichum trifolii (a–h. from lectotype BPI 399624; i–ad. from ex-epitype strain CBS 158.83). a–b. acervuli on host tissue; c–e. conidiophores; f. conidia; g–h. setae; i–j. acervuli; k. tip of a seta; l. basis of a seta; m–p. conidiophores; q. tip of a seta; r. basis of a seta; s–v. conidiophores; w–ab. appressoria; ac–ad. conidia. a–h: from host tissue; i, k–p, ac: from Anthriscus stem; j, q–ab, ad: from SNA. a–b, i–j: DM; c–h, k–ad: DIC.—Scale bars: a = 1 mm; b = 100 μm; f = 10 μm; i = 100 μm; p = 10 μm; scale bar of f applies to c–h; scale bar of i applies to i–j; scale bar of p applies to k–ad.
Sexual morph not observed. Asexual morph on stem of Trifolium pratense (BPI 399624). Conidiomata, conidiophores and setae formed on pale brown angular cells 4–8.5 μm diam. Setae medium to dark brown, smooth-walled to verruculose, flexuous, 41–76 μm long, 1–2-septate, base cylindrical to conical, 4.5–6.5 μm diam, tip acute to ± rounded. Conidiophores pale brown, smooth-walled, septate, branched, to 30 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical, 7.5–22 × 3–6 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening visible. Conidia hyaline, smooth-walled, aseptate, straight, ellipsoidal to cylindrical, sometimes clavate, with both ends rounded or one end round and the other truncate, (10–)10.5–13(−16) × (4.5–)5–6(−6.5) μm, mean ± SD = 11.7 ± 1.4 × 5.4 ± 0.5 μm, L/W ratio = 2.2.
Asexual morph on SNA. Vegetative hyphae 1–7 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata absent, conidiophores and setae formed directly on pale brown, verruculose hyphae. Setae medium to dark brown, smooth-walled to verruculose, 35–110 μm long, 1–3-septate, base cylindrical, 2.5–6.5 μm diam, tip acute, sometimes ± rounded. Conidiophores pale brown, smooth-walled to verruculose, septate, branched, up to 40 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled to verruculose, cylindrical, 8–31 × 4–4.5 μm, opening 1.5–2.5 μm diam, collarette 1–1.5 μm long (rarely observed), periclinal thickening sometimes visible. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, sometimes slightly constricted in the middle, with one end round and one end truncate, (8.5–)9.5–12.5(−16) × 4–5 μm, mean ± SD = 11.1 ± 1.5 × 4.5 ± 0.3 μm, L/W ratio = 2.5, conidia of strain CBS 128554 shorter and broader, measuring (7–)8–10(−12) × (5–)5–6(−6.5) μm, mean ± SD = 8.9 ± 1.2 × 5.5 ± 0.4 μm, L/W ratio = 1.6. Appressoria (not formed after 10 day, n = 22 after 4 weeks) single or in small dense clusters, pale to medium brown, smooth-walled, subglobose, elliptical or clavate in outline, with an entire to undulate margin, (4.5–)5–9.5(−14) × (4–)4.5–6(−6.5) μm, mean ± SD = 7.4 ± 2.3 × 5.3 ± 0.7 μm, L/W ratio = 1.4.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, verruculose hyphae; no angular basal cells observed. Setae medium to dark brown, smooth-walled to verruculose, bent outwards (giving the appearance of a sea anemone), 60–120 μm long, 2–3-septate, base cylindrical-conical, sometimes with a short branch, 3.5–6.5(−9.5) μm diam, tip acute, sometimes ± rounded. Conidiophores rarely observed, pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells rarely observed, very inconspicuous; pale brown, smooth-walled, cylindrical, 10–26 × 4–5 μm, opening 1–2 μm diam, collarette 1 μm long, periclinal thickening sometimes visible. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, sometimes clavate with one end round and the other truncate, (9–)10.5–12.5(−14.5) × (3.5–)4–4.5(−5) μm, mean ± SD = 11.5 ± 1.2 × 4.2 ± 0.3 μm, L/W ratio = 2.7. Conidia of strain CBS 128554 shorter and broader, measuring (6.5–)8–10.5(−12) × (4.5–)5–6(−6.5) μm, mean ± SD = 9.4 ± 1.4 × 5.5 ± 0.4 μm, L/W ratio = 1.7.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale honey; agar medium, Anthriscus stem and filter paper partly covered with grey to black acervuli, aerial mycelium lacking, reverse filter paper partly pale olivaceous-grey; 16.5–18 mm in 7 days (24–25.5 mm in 10 days). Colonies on OA flat with entire margin; olivaceous-black with a buff margin, aerial mycelium lacking, partly covered by rosy-buff acervuli, reverse olivaceous-grey to iron-grey, 18–19.5 mm in 7 days (25.5–28 mm in 10 days). Conidia in mass rosy-buff.
Specimens examined: USA, Tennessee, Knoxville, from Trifolium pratense, 1 Aug 1905, S.H. Essary, S.W. Bain, (BPI 399624 lectotype, here designated; MBT175536); from Trifolium sp., collection date and collector unknown (deposited in CBS collection Jan. 1983 by Z Klocokar-Smit, No. 255) (CBS H-20978 epitype, here designated, culture ex-epitype CBS 158.83 = BBA 70709; MBT175537); Wisconsin, from stem lesion of Medicago sativa, collection date and collector unknown (deposited in CBS collection 2010 by PR Johnston: C1210-3, who received it from SL Nygaard: N85ANW), CBS 128554 = ICMP 12934. UNKNOWN COUNTRY, unknown host, (deposited in CBS collection 1983) culture CBS 425.83.
Notes: Bain and Essary described a new anthracnose disease on clover in the USA that caused 25–75 % yield loss in Tennessee and was also prevalent in Ohio and West Virginia (US Department of Agriculture 1905: 609). The species was described as C. trifolii, and observed on stems, petioles and rarely leaves of red clover (Trifolium pratense) and alfalfa (Medicago sativa) and additionally reported from Kentucky and Arkansas (Bain and Essary 1906). The conidia were reported as measuring 11–13 × 3–4 μm and straight with rounded ends. Conidia measured in this study from the lectotype BPI 399624 were found to be (10–)10.5–13(−16) × (4.5–)5–6(−6.5) μm in size, agreeing in length. Conidia from both the lectotype and the living cultures (CBS 158.83, ex-epitype, and CBS 128554) are therefore broader than indicated by Bain and Essary (1906).
Bain and Essary (1906) did not explicitly cite type material, but list the two host species and the states of the USA where the species was collected or reported from: “Tennessee, Kentucky, Arkansas; Virginia (J. M. Westgate); West Virginia, Ohio (Yearbook U. S. Department of Agriculture, 1905, p. 609)”. There are several specimens identified as this species in the USDA collection (BPI) that were collected at locations listed in the prologue of the species prior to its publication and can reasonably be assumed to be specimens on which the name C. trifolii was based. One of these was collected from Trifolium pratense (BPI 399624), collected in Aug. 1905 from Tennessee, USA by the authors of the species. Three further specimens (BPI 399604, BPI 399605 and BPI 399606) from Medicago sativa in Virginia and collected in Jul. 1905 may also be authentic material, one of which lists as collector “Westgate?”. An earlier collection on M. sativa from Mississippi from Nov. 1902 (BPI 399607) was not cited by Bain and Essary, and cannot be considered part of the type material. BPI 399624 is an obvious choice for the lectotype, as the only available material from Trifolium and the only collection definitely linked to the authors of the species. Moreover, this collection is from Knoxville; Bain and Essary were apparently working at the University of Tennessee in Knoxville at that time, because this institution is listed at the end of their publication, just after the description of the species. The dried material is in very good condition, but we nevertheless designate an epitype based on a sequenced culture also to fix application of the name on a molecular basis.
Several other Colletotrichum and Gloeosporium species have been described from Trifolium and Medicago. O’Gara found a Colletotrichum species on red clover and alsike clover (T. hybridum) in Utah, USA that attacks leaves and petioles of these hosts and named it C. destructivum (O’Gara 1915). It forms conidia that are longer than those of C. trifolii, measuring 14–22 × 3.5–5 μm, and are straight to slightly curved, while conidia of C. trifolii are straight and measure (10–)10.5–13(−16) × (4.5–)5–6(−6.5) on the host (BPI 399624); conidia of C. trifolii formed on SNA or Anthriscus stem had similar dimensions or were even shorter. Preliminary studies indicate that strains matching the conidial features of C. destructivum (U. Damm, unpublished data) occupy a distinct clade within Colletotrichum indicated as destructivum clade in Cannon et al. (2012) and O’Connell et al. (2012). An extensive study on C. destructivum and related species is in progress (U. Damm, unpublished data).
Gloeosporium trifolii was described by Peck (1879, publ. 1883) from red clover in Albany, NY, USA. Conidial measurements were given as 0.006–0.009 × 0.00016–0.00025 in. (= 15–23 × 4–6.3 μm). Based on its similar conidia size, it seems possible that this taxon might provide an earlier name for C. destructivum, although von Arx (1957a) suggested that Peck’s fungus was synonymous with Stagonospora meliloti (Lasch) Petrak. Gloeosporium medicaginis was described on leaves, petioles and stipules of Medicago sativa from Kansas, USA by Ellis and Kellerman (1887), with conidia broadly similar to those of Gloeosporium trifolii. Von Arx (1957a) also considered this fungus to be a synonym of S. meliloti. However, material of these species has not been examined, and living cultures derived from their types are not available.
More recently described taxa associated with Trifolium and Medicago include Gloeosporium trifoliorum Rothers, a species described from living leaves of Trifolium in Russia that forms short-cylindrical conidia, measuring 4–8(−9) × 1.5–2(−2.5) μm (Vassiljevski and Karakulin 1950). Von Arx (1957a) suggested that this species might belong in Sporonema. Colletotrichum medicaginis-denticulatae was described on leaves of Medicago denticulata from Taiwan (Sawada 1933). Its affinities are currently unknown. Colletotrichum medicaginis was described on stems of Medicago sativa in Uttar Pradesh, India (Pavgi and Singh 1964). Conidia were described as crescent-shaped and pointed at both ends; this suggests a relationship with the C. truncatum or C. dematium clades (Cannon et al. 2012). Damm et al. (2009) reported strains from M. sativa assigned to C. spinaciae (dematium clade) and C. truncatum s. str.
Colletotrichum trifolii has more conspicuous setae than the other species in this species complex that are darker and often knobby. The species is closely related to C. malvarum; the ITS and CHS-1 sequences of the two species are identical. However, they can be separated with all other loci studied, best with GS sequences. The ITS sequence of strain CBS 158.83 is 100 % identical with C. trifolii strains DAOM 225587 (EU400136, Chen et al. 2012) and BBA 70709 (= CBS 158.83, AJ301941, Nirenberg et al. 2002) and C. “gloeosporioides” strain DAOM 212639 (EU400137, Chen et al. 2012), while strains UQ349, CBS 149.34 and UM71, the ITS sequence (AF451909, AJ301942, HQ148103) of which lodged in GenBank as C. trifolii by Ford et al. (2004), Nirenberg et al. (2002) and Rosa et al. (2012), do not belong to the C. orbiculare species complex (not shown). Closest matches in blastn searches with the TUB2 sequence of strain CBS 158.83 were with 99 % identity C. orbiculare strain 104-T (= CBS 514.97, JQ005862, O’Connell et al. 2012) and with 98 % identity C. spinosum (as C. orbiculare) strain STE-U 5296 (= CBS 113171, AY376589, Lubbe et al. 2004). The GS sequences of strain CBS 158.83 is 100 % identical with that of C. trifolii strain 14-2-63 (DQ792877) and 99 % identical with C. trifolii strains ON7 (DQ792876) and ON12 (DQ792875), all from Medicago sativa (Liu et al. 2007). The GAPDH sequences of the latter two strains (DQ792852, DQ792853) are identical with that of CBS 158.83. Additionally, strain MAFF 510847 from Medicago sativa from Japan has identical ITS, GAPDH, ACT and TUB2 sequences as CBS 158.83 (sequences retrieved from NIAS Genbank (http://www.gene.affrc.go.jp); the ITS sequence (AB087223) was also included in Moriwaki et al. (2003).
Discussion
The species of the C. orbiculare species complex appear to be restricted to specific herbaceous host species/genera in four plant families, Asteraceae, Cucurbitaceae, Fabaceae and Malvaceae. Colletotrichum lindemuthianum is restricted to Phaseolus vulgaris and P. coccineus (Fabaceae), while C. orbiculare attacks species of several genera in the Cucurbitaceae, and possibly some plants in other families (e.g. Gerbera - Asteraceae). Colletotrichum trifolii causes anthracnose on Trifolium and Medicago, closely related genera belonging to the Fabaceae, while C. malvarum, C. tebeestii and C. sidae seem to be restricted to only one or few host species of the Malvaceae. Colletotrichum spinosum and C. bidentis are so far only known from Xanthium spinosum and Bidens subalternans, respectively, both belonging to the Asteraceae. However, the number of strains included in our study is comparatively low; future collections are needed to confirm or extend the known host spectra of these species. Three of the species of the C. orbiculare species complex, C. lindemuthianum, C. orbiculare and C. trifolii are serious pathogens of agricultural crops, while four are pathogens of important weeds, namely: C. bidentis, C. sidae, C. spinosum and C. tebeestii.
Some collections and pathogenicity tests in previous studies suggest a wider host range and cross-pathogenicity of some of the species in the C. orbiculare complex. For example Halsted (1893a) observed that isolates from bean and watermelon could both infect citron fruits. Simmonds (1965) reported C. orbiculare from cucurbits, safflower, bathurst burr and celery based on cultural and conidial characters. However isolations from celery and safflower were not pathogenic to watermelon. Walker et al. (1991) mention inoculation tests (J. Walker, unpublished data) in which a C. orbiculare isolate from safflower caused stem lesions and shoot blight on seedlings of bathurst burr. Walker et al. (1991) showed that strains from cucurbits could infect a wide range of plants, and morphologically similar strains from Xanthium could infect cucurbits. Shen et al. (2001) demonstrated that a strain of C. orbiculare from Malva pusilla (here C. tebeestii) could also infect Nicotiana species (Solanaceae). According to von Arx (1957a) typical C. orbiculare strains from cucumber caused a disease on cucumber seedlings and a root rot on apples, but no disease on young bean plants.
Inoculation experiments by von Arx and van der Velden (1961) demonstrate the influence of the inoculation technique on the presumed host spectrum: without wounding, three C. orbiculare strains caused leaf spots or necroses on all tested Cucurbitaceae, while all other host plants (Nicotiana spp., Solanum lycopersicum, Phaseolus vulgaris, Lathyrus sp.) remained healthy. None of these hosts showed disease symptoms when inoculated with one of two Glomerella cingulata strains. However, after wound inoculation, all five strains caused fruit rot of cucumber, melon, apple, tomatoes and oranges and developed conidia on the fruits. The inoculation method and host organ tested might explain some of the controversial results of previous studies. However, even if a species has the potential to cause an infection on a specific host or host organ in an experiment, it might usually not come in contact with it in nature. Additionally, the identity of many of the C. orbiculare (or C. lagenarium) isolates used in previous studies (e.g. Halsted 1893a; von Arx 1957a; von Arx and van der Velden 1961; Simmonds 1965; Walker et al. 1991) has not been confirmed by molecular methods.
The infection strategy of these fungi is highly relevant to pathogenicity, and also to host specificity. Biotrophic fungi are usually highly host specific, but do not (or do not quickly) kill the host, while necrotrophic fungi usually have a broad host range but are vulnerable to non-specific host defence mechanisms. Hemibiotrophic fungi combine both strategies: they have an initial biotrophic phase (which predicates host specificity) followed by a virulent necrotrophic phase leading to quick death of the target plants. This makes them especially destructive to their hosts, and especially qualified as biological control agents, if they are highly host-specific (Goodwin 2001).
The infection strategy of most of the species in the C. orbiculare species complex, specifically of C. lindemuthianum, C. orbiculare, C. sidae (as forms of C. orbiculare) and C. tebeestii (as C. gloeosporioides f. sp. malvae), has been well studied and characterised as hemibiotrophic (Dargent and Touzé 1974; Stumm and Gessler 1984; O’Connell et al. 1985; Xuei et al. 1988; ; Bailey et al. 1996; Morin et al. 1996; Wei et al. 1997; Goodwin 2001). In the study of Bailey et al. (1996), strain LARS 629 (= CBS 518.97, included in our study as C. sidae) formed large globular intracellular infection vesicles and primary hyphae inside epidermal cells of Sida spinosa leaves, which they considered as typical for all forms of the “C. orbiculare aggregate species”. Pain et al. (1992) raised antibodies against C. lindemuthianum germlings that showed cross-reaction to C. lindemuthianum from beans and strains here accepted as C. sidae, C. orbiculare, C. spinosum and C. trifolii, but not to other Colletotrichum species, but also to a C. “lindemuthianum” strain from Vigna that does not belong in the C. orbiculare complex (U. Damm, unpublished data). All the species that showed a reaction with the antibodies formed intracellular infection vesicles after penetrating epidermal cells of their hosts (R. J. O’Connell, unpublished data, see Pain et al. 1992) that is typical for hemibiotropic pathogens. Therefore, in all known species in the C. orbiculare complex (except for the recently collected C. bidentis), typical structures of hemibiotropic infection strategy have been observed. The reasons why Colletotrichum species from the C. orbiculare complex that establish hemibiotrophic relationships are highly host specific are probably to be found in the close cytoplasmatic interactions between plant and pathogen that is developed during the initial, biotrophic phase of the infection (Bailey et al. 1992). In contrast, the intramural pathogen C. capsici grows only in cell walls and has a wide host range (Roberts and Snow 1990).
The formation of two clades within C. lindemuthianum strains was noticed by Cannon et al. (2012) based on ITS data retrieved from GenBank. They suggested that C. lindemuthianum could represent more than one taxon. The phylogenies presented by Balardin et al. (1999) indicate an even higher variability within the ITS region of C. lindemuthianum strains from different countries in the Americas. However, the sequences generated in that study were not deposited in GenBank and therefore cannot easily be examined. Manual comparison of the printed sequences indicates that Balardin et al.’s strain ARG81 has an ITS sequence that differs by only 1 bp from that of the epitype of C. lindemuthianum (Liu et al. 2013), but their most divergent strain MX457 differs by 13 of 546 bp. Analysis of the sequence differences as given in Balardin et al. (1999) indicates that most of the discrepancies occur as variations in the number of bases in strings of identical bases (e.g. AAA versus AAAA, or AAA- versus AAAA), suggesting that many of them may be due to mis-reads. The genetic diversity that they observed may therefore be artefactual.
The recent study of Liu et al. (2013) typifying C. lindemuthianum based on a multigene phylogeny does not support different taxa for the bean pathogen, although the same two groups as observed by Cannon et al. (2012) were formed by ITS data only (three bp differences, not shown). However, these differences overlapped with the results of the other four loci included that were either uniform for all strains or grouped differently. The strains and sequences included in the study of Liu et al. (2013) are the same as in our study. However, in our study GS sequences were generated additionally that support the grouping found with ITS data, displaying 20 bp differences and a 4-bp indel. No consistency of the two clusters (indicated as C. lindemuthianum 1 and 2 in Fig. 1) with host preference, distribution or morphology was found in our study. Both clades contained strains from Phaseolus vulgaris in Europe (especially Germany and Netherlands), North (USA) and Central/South America. Based on comparison with ITS or GS sequences generated in previous studies (data not shown), all four strains from the study of Liu et al. (2007), most of the strains from the study of Chen et al. (2012) and one strain from the study of Moriwaki et al. (2003) belong to C. lindemuthianum 2, while one strain from Chen et al. (2012) belongs to C. lindemuthianum 1. The strains included in Chen et al. (2012) were assigned to different races. However, due to the small number of strains and races included, no general connection can be made between the two groups and previously described races. Future research will show if physiology, pathogenicity or other characteristics support describing C. lindemuthianum 2 as a separate species.
Although the species of the C. orbiculare complex occupy a basal clade in the generic phylogeny derived of known Colletotrichum spp. (Cannon et al. 2012), a separation of these taxa from Colletotrichum at generic level is not suggested. A preliminary LSU analysis of representatives of the known families of the Glomerellales (Réblová et al. 2011) and all previously well studied Colletotrichum species complexes (Cannon et al. 2012) suggests the genus Colletotrichum to be monophyletic (not shown). Presently however, not all Colletotrichum species and species complexes are sufficiently known from DNA sequence data; some of them might have an intermediate position between C. orbiculare and other species complexes. Moreover, the species of the C. orbiculare complex agree with the typical morphological features of the genus Colletotrichum as currently accepted; they all form large aseptate, hyaline (in mass bright-coloured) conidia with enteroblastic, non-catenate succession from phalidic conidiogenous cells on densely arranged conidiophores, often together with stiff brown, septate, setae; in nature usually in acervuli erumpent from the epidermis of plant cells, as well as brown appressoria formed in nature after the germination of conidia on the plant surface, facilitating host infection. The species of the C. orbiculare complex further share many other morphological characters with certain species and species complexes that are not monophyletic within Colletotrichum, for example the shape of conidia and appressoria and the formation of vesicles and broad primary hyphae inside the host tissue, characteristic for the biotrophic stage of the hemibiotropic infection process. Last but not least, these species cause the same kind of symptoms on their host plants, generally referred to as anthracnose.
References
Alcorn JL (1976) Host index of plant diseases in Queensland. Supplement 1, Queensland Department of Primary Industries, Indoorpilly
Allescher A (1902) Fungi Imperfecti. In: Rabenhorst’s Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz. 2d edn,.1(7):385–704
Asakura M, Yoshino K, Hill AM, Kubo Y, Sakai Y, Takano Y (2012) Primary and secondary metabolism regulates lipolysis in appressoria of Colletotrichum orbiculare. Fungal Gen Biol 49:967–975
Asakura M, Ninomiya S, Sugimoto M, Oku M, Yamashita SI, Okuno T, Sakai Y, Takano Y (2009) Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 21:1291–1304
Auld BA, McRae CF, Say MM (1988) Possible control of Xanthium spinosum by a fungus. Agr Ecosyst Environ 21:219–223
Auld BA, Medd RW (1987) Weeds. An illustrated botanical guide to the weeds of Australia. Inkata Press, Melbourne
Auld BA, Say MM (1999) Comparison of isolates of Colletotrichum orbiculare from Argentina and Australia as potential bioherbicides for Xanthium spinosum in Australia. Agr Ecosyst Environ 72:53–58
Auld BA, Say MM, Ridings HI, Andrews J (1990) Field applications of Colletotrichum orbiculare to control Xanthium spinosum. Agr Ecosyst Environ 32:315–323
Bailey JA, Nash C, Morgan LW, O’Connell RJ, TeBeest DO (1996) Molecular taxonomy of Colletotrichum species causing anthracnose on the Malvaceae. Phytopathology 86:1076–1083
Bailey JA, O’Connell RJ, Pring RJ, Nash C (1992) Infection strategies of Colletotrichum species. In: Bailey JA, Jeger MJ (eds) Colletotrichum: Biology and control. CAB International, Wallingford, pp 88–120
Bain SM, Essary SH (1906) A new anthracnose of alfalfa and red clover. J Mycol 12:192–193
Balardin RS, Smith JJ, Kelly JD (1999) Ribosomal DNA polymorphism in Colletotrichum lindemuthianum. Mycol Res 103:841–848
Baxter AP, van der Westhuizen GCA, Eicker A (1983) Morphology and taxonomy of South African isolates of Colletotrichum. S Afr J Bot 2:259–289
Berkeley MJ (1838) Notices of British fungi [59–107]. Annals and Magazine of Natural History 1:198–208
Berkeley MJ (1853) Some notes upon the cryptogamic portion of the plants collected in portugal 1842–50 by Dr Friedr. Welwitsch. The Fungi, London
Berkeley MJ (1860) Outlines of British fungology. UK, London
Berkeley MJ, Broome CE (1882) List of fungi from Brisbane. Queensland: With descriptions of new species.-Part II. Transactions of the Linnean Society of London 2:53–73
Braun A (1854) Über einige neue oder weniger bekannte Krankheiten der Pflanzen, welche durch Pilze erzeugt werden. Verlag der Nicolai’schen Buchhandlung, Berlin
Bresadola G (1892) Fungi Tridentini novi, vel nondum delineati, descripti, et iconibus illustrati. Lith. Typ. J. Zippel, Tridenti
Butler FC (1951) Anthracnose and seedling blight of bathurst burr caused by Colletotrichum xanthii Halst. Aust J Agr Res 2:401–410
Cabrera AL (1974) Flora ilustrada de Entre-Rios (Argentina). Colección Científica del INTA, Buenos Aires 6:106–554
Cannon PF, Damm U, Johnston PR, Weir BS (2012) Colletotrichum – current status and future directions. Stud Mycol 73:181–213
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Cardoso MO, Boher B, de Avila AC, Assis LAG (2001) Doenças das Cucurbitáceas no Estado do Amazonas. Embrapa Amazônia Ocidental. Circular Técnica 9:2–14
Cavara F (1889) Matériaux de mycologie Lombarde. Revue mycologique 11(44):173–192
Chen C, Ha Y, Min J, Memmott SD, Dickman MB (2006) Cdc42 is required for proper growth and development in the fungal pathogen Colletotrichum trifolii. Eukaryot Cell 5:155–166
Chen Y-Y, Conner RL, Gillard CL, McLaren DL, Boland GJ, Balasubramanian PM, Stasolla C, Zhou QX, Hwang SF, Chang K-F, Babcock C (2012) A quantitative real-time PCR assay for detection of Colletotrichum lindemuthianum in navy bean seeds. Plant Patholog (early view). doi:10.1111/j.1365-3059.2012.02692
Cho WD, Shin HD (2004) List of plant diseases in Korea, 4th edn. Korean Society of Plant Pathology, Korea
Cisar CR, Spiegel FW, TeBeest DO, Trout C (1994) Evidence for mating between isolates of Colletotrichum gloeosporioides with different host specificities. Curr Genet 25:330–335
Cooke MC (1878) North American fungi. Hedwigia 17:37–40
Cooke MC (1908) Fungus notes for 1907. T Brit Mycol Soc 3:34–46
Correll JC, Rhoads DD, Guerber JC (1993) Examination of mitochondrial DNA restriction fragment length polymorphisms, DNA fingerprints, and randomly amplified polymorphic DNA of Colletotrichum orbiculare. Phytopathology 83:1199–1204
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004a) MycoBank: An online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22
Crous PW, Groenewald JZ, Risede JM, Hywel-Jones NL (2004b) Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Stud Mycol 50:415–430
Crous PW, Verkleij GJM, Groenewald JZ, Samson RA (2009) Fungal biodiversity. CBS laboratory manual series 1. Centraalbureau voor Schimmelcultures, Utrecht
Damm U, Cannon PF, Woudenberg JHC, Crous PW (2012a) The Colletotrichum acutatum species complex. Stud Mycol 73:37–113
Damm U, Cannon PF, Woudenberg JHC, Johnston PR, Weir B, Tan YP, Shivas RG, Crous PW (2012b) The Colletotrichum boninense species complex. Stud Mycol 73:1–36
Damm U, Crous PW, Fourie PH (2007) Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora spp. nov. Mycologia 99:664–680
Damm U, Mostert L, Crous PW, Fourie PH (2008) Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia 20:87–102
Damm U, Woudenberg JHC, Cannon PF, Crous PW (2009) Colletotrichum species with curved conidia from herbaceous hosts. Fungal Divers 39:45–87
Dargent R, Touzé A (1974) Etude cinétique, en microscopie électronique, des interactions entre Colletotrichum lagenarium et les cellules foliaires de Cucumis melo. Can J Bot 52:1319–1323
Dean JD, Goodwin PH, Hsiang T (2003) Colletotrichum gloeosporioides infection induces differential expression of glutathione S-transferase genes in Malva pusilla. Funct Plant Biol 30:821–828
Ellis JB, Everhart BM (1889) New and rare species of North American fungi (Sphaeropsideae). J Mycol 5:145–157
Ellis JB, Kellerman WA (1887) New Kansas fungi. J Mycol 3(9):102–105
Farr DF, Aime MC, Rossman AY, Palm ME (2006) Species of Colletotrichum on Agavaceae. Mycol Res 110:1395–1408
Farr DF, Rossman AY (2013) Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. Retrieved April 5, 2013, from http://nt.ars-grin.gov/fungaldatabases/
Ford R, Banniza S, Photita W, Taylor PWJ (2004) Morphological and molecular discrimination of Colletotrichum truncatum causing anthracnose on lentil in Canada. Australas Plant Path 33:559–569
Fujihara N, Sakaguchi A, Tanaka S, Fujii S, Tsuji G, Shiraishi T, O’Connell R, Kubo Y (2010) Peroxisome biogenesis factor PEX13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare. Mol Plant Microbe In 23:436–445
Gan P, Ikeda K, Irieda H, Narusaka M, O’Connell RJ, NarusakaY TY, Kubo Y, Shirasu K (2013) Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol 197:1236–1249
García PE, Schönswetter P, Aguilar JF, Feliner GN, Schneeweiss GM (2009) Five molecular markers reveal extensive morphological homoplasy and reticulate evolution in the Malva alliance (Malvaceae). Mol Phylogenet Evol 50:226–239
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Glass NL, Donaldson G (1995) Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microb 61:1323–1330
Goode MJ (1958) Physiological specialization in Colletotrichum lagenarium. Phytopathology 48:79–83
Goodwin PH (2001) A molecular weed-mycoherbicide interaction: Colletotrichum gloeosporioides f. sp. malvae and round-leaved mallow, Malva pusilla. Can J Plant Pathol 23:28–35
Goodwin PH, Chen GY (2002a) High expression of a sucrose non-fermenting (SNF1)-related protein kinase from Colletotrichum gloeosporoides f. sp malvae is associated with penetration of Malva pusilla. FEMS Microbiol Lett 215:169–174
Goodwin PH, Chen GYJ (2002b) Expression of a glycogen synthase protein kinase homolog from Colletotrichum gloeosporioides f.sp malvae during infection of Malva pusilla. Can J Microbiol 48:1035–1039
Grombone-Guaratini MT, Mansanares ME, Semir J, Solferini VN (2006) Chromosomal studies of three species of Bidens (L.) (Asteraceae). Caryologia 59:14–18
Grove WB (1937) British stem and leaf fungi (Coelomycetes). University Press, Cambridge
Guerber JC, Liu B, Correll JC, Johnston PR (2003) Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95:872–895
Halsted BD (1893a) Identity of anthracnose of the bean and watermelon. B Torrey Bot Club 20:246–250
Halsted BD (1893b) Some new weed fungi. B Torrey Bot Club 20:250–252
Hartman GL, Manandhar JB, Sinclair JB (1986) Incidence of Colletotrichum spp. on soybeans and weeds in Illinois and pathogenicity of Colletotrichum truncatum. Plant Dis 70:780–782
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192
Hinsley SR (2013) Malvaceae Info: Intergeneric hybrids in Malvaceae. Retrieved May 22, 2013, from www.malvaceae.info/Genera/Hybrids/Hybrids.php
Hyde KD, Cai L, Cannon PF, Crouch JA, Crous PW, Damm U, Goodwin PH, Chen H, Johnston PR, Jones EBG, Liu ZY, McKenzie EHC, Moriwaki J, Noireung P, Pennycook SR, Pfenning LH, Prihastuti H, Sato T, Shivas RG, Tan YP, Taylor PWJ, Weir BS, Yang YL, Zhang JZ (2009) Colletotrichum – names in current use. Fungal Divers 39:147–182
ISSG (2013) Global Invasive Species Database: Bidens pilosa (herb). Retrieved May 22, 2013, from: www.issg.org/database/species/impact_info.asp?si=1431&fr=1&sts=&lang=EN
Jaulneau V, Cazaux M, Hoi JWS, Fournier S, Esquerré-Tugayé M-T, Jaquet C, Dumas B (2010) Host and nonhost resistance in Medicago-Colletotrichum interactions. Mol Plant Microbe In 23:1107–1117
Jenkins SF, Winstead NN (1961) Observations on the sexual stage of Colletotrichum orbiculare. Science 133:581–582
Jenkins SF, Winstead NN (1962) Morphology, taxonomy and sexuality of the ascogenous stages of two Colletotrichum spp. that attack cucurbits. Phytopathology 52:15
Jenkins SF, Winstead NN (1964) Glomerella magna, cause of a new anthracnose of cucurbits. Phytopathology 54:452–454
Johnston PR, Jones D (1997) Relationships among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia 89:420–430
Kim WG, Hong SK, Kim JH (2008) Occurrence of anthracnose on Chinese mallow caused by Colletotrichum malvarum. Mycobiology 36:139–141
Kirkpatrick TL, Templeton GE, TeBeest DO (1982) Potential of Colletotrichum malvarum for biological control of prickly sida. Plant Dis 66:323–325
Krüger F (1913) Beiträge zur Kenntnis einiger Gloeosporien. Arbeiten aus der Kaiserlichen Biologischen Anstalt für Land- und Forstwirtschaft 9:233–323
Kubo Y, Takano Y (2013) Dynamics infection-related morphogenesis and pathogenesis in Colletotrichum orbiculare. J Gen Plant Pathol. doi:10.1007/s10327-013-0451-9
Lin SY, Okuda A, Ikeda K, Okuno T, Takano Y (2012) LAC2 Encoding a secreted laccase is involved in appressorial melanization and conidial pigmentation in Colletotrichum orbiculare. Mol Plant Microbe In 25:1552–1561
Liu B, Wasilwa LA, Morelock TE, O’Neill NR, Correll JC (2007) Comparison of Colletotrichum orbiculare and several allied Colletotrichum spp. for mtDNA RFLPs, intron RFLP and sequence variation, vegetative compatibility and host specificity. Phytopathology 97:1305–1314
Liu F, Cai L, Crous PW, Damm U (2013) Circumscription of the anthracnose pathogens Colletotrichum lindemuthianum and C. nigrum. Mycologia. doi:10.3852/12-315
Lubbe CM, Denman S, Cannon PF, Groenewald JZ, Lamprecht SC, Crous PW (2004) Characterization of Colletotrichum species associated with diseases of Proteaceae. Mycologia 96:1268–1279
Mackie J, Musial JM, Armour DJ, Phan HT, Ellwood SE, Aitken KS, Irwin JA (2007) Identification of QTL for reaction to three races of Colletotrichum trifolii and further analysis of inheritance of resistance in autotetraploid lucerne. Theor Appl Genet 114:1417–1426
Magenta MAG (1998) As subtribos Ambrosiinae, Galinsoginae e Coreopsidinae (Heliantheae-Asteraceae) no Estado de São Paulo. USP, São Paulo
Magnus P (1926) Colletotrichum magnusianum. Berichte des Naturwissenschaftlichen-Medizinischen Vereins in Innsbruck 40:280–281
Makowski RMD, Mortensen K (1998) Latent infections and penetration of the bioherbicide agent Colletotrichum gloeosporioides f. sp. malvae in non-target field crops under controlled environmental conditions. Mycol Res 102:1545–1552
Makowski RMD, Mortensen K (1992) The first mycoherbicide in Canada: Colletotrichum gloeosporioides f. sp. malvae for round-leafed mallow control. In: Richardson RG (ed) Proceedings of the first international weed control Congress. Monash University, Melbourne, 1992, Commonwealth Agricultural Bureau, Wallingford, UK, pp 298–300
Mason-Gamer RJ, Kellogg EA (1996) Chloroplast DNA analysis of the monogenomic Triticeae: phylogenetic implications and genome-specific markers. In: Jauhar PP (ed) Methods of genome analysis in plants. CRC Press, Boca Raton, pp 301–325
McLean KS, Roy KW (1991) Weeds as a cource of Colletotrichum capsici causing anthracnose on tomato fruit and cotton seedlings. Can J Plant Pathol 13:131–134
Menten JOM, Kimati H, Costa CP (1980) [1979] Reaction of cucumber Cucumis sativus L. populations to Colletotrichum gloeosporioides f.sp. cucurbitae nov. comb. Summa Phytopathologica 5:127–133
Michel VV (2005) First report of anthracnose caused by Colletotrichum orbiculare f. sp. from A. officinalis of marsh mallow (Althaea officinalis) in Switzerland. Plant Dis 89:687
Morin L, Derby JAL, Kokko EG (1996) Infection process of Colletotrichum gloeosporioides f sp malvae on Malvaceae weeds. Mycol Res 100:165–172
Moriwaki J, Sato T, Tsukiboshi T (2003) Morphological and molecular characterisation of Colletotrichum boninense sp. nov. from Japan. Mycoscience 44:47–53
Moriwaki J, Tsukiboshi T, Sato T (2002) Grouping of Colletotrichum species in Japan based on rDNA sequences. J Gen Plant Pathol 68:307–320
Mortensen K (1987) Control of round-leaved mallow and velvetleaf weeds with C. gloeosporioides. EP0218386 dated Apr 15 1994
Mortensen K (1988) The Potential of an endemic fungus, Collectotrichum gloeosporioides, for biological control of round-leaved mallow (Malva pusilla) and velvetleaf (Abutilon theophrasti). Weed Sci 36:473–478
Mortensen K (1991) Colletotrichum gloeosporioides causing anthracnose of Lavatera sp. Can Plant Dis Surv 71:155–159
Mortensen K, Makowski RMD (1995) Tolerance of strawberries to Colletotrichum gloeosporioides f. sp. malvae, a mycoherbicide for control of round-leaved mallow (Malva pusilla). Weed Sci 43:429–433
Mortensen K, Makowski RMD, Cunningham JE, Carmichael RD (1994) Method and compositions for controlling round-leaved mallow using Colletotrichum gloeosporioides f. sp. malvae, ATCC 20767. US Patent US5296369 dated Mar 22 1994
Nirenberg HI (1976) Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169:1–117
Nirenberg HI, Feiler U, Hagedorn G (2002) Description of Colletotrichum lupini comb. nov. in modern terms. Mycologia 94:307–320
Nylander JAA (2004) MrModeltest v.2. Program distributed by the author. Evol Biol Centre, Uppsala University, Uppsala
O’Connell RJ, Bailey JA, Richmond DV (1985) Cytology and physiology of infection of Phaseolus vulgaris by Colletotrichum lindemuthianum. Physiol Plant Pathol 27:75–98
O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J et al (2012) Life-style transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44:1060–1065
O’Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116
O’Gara PJ (1915) New species of Colletotrichum and Phoma. Mycologia 7:38–41
Pain NA, O’Connell RJ, Bailey JA, Green JR (1992) Monoclonal antibodies which show restricted binding to four Colletotrichum species: C. lindemuthianum, C. malvarum, C. orbiculare and C. trifolii. Physiol Mol Plant P 41:111–126
Passerini G (1868) Erbario Crittogamico Italiano. Ser. II, Fasc. III no. 148. In: Notaris G de, Baglietto F (1868) Erbario Crittogamico Italiano. Ser. II, Fasc. III (nos 101–150)
Pavgi MS, Singh UP (1964) Parasitic fungi from North India – III. Mycopathol Mycol Appl 24:355–361
Pavgi MS, Singh UP (1965) Parasitic fungi from North India – IV. Mycopathol Mycol Appl 27:81–88
Peck CH (1879) publ.1883) Report of the Botanist no. 33. Assembly Document, New York State Legislature no 120(8):11–49
Perfect SE, Hughes HB, O’Connell RJ, Green JR (1999) Colletotrichum: a model genus for studies on pathology and fungal–plant interactions. Fungal Genet Biol 27:186–198
Petrak F (1923) Mykologische Notizen. VI. Annales Mycologici 21:182–335
Potebnia A (1907) Mycologische Studien. Annales Mycologici 5:1–28
Rambaut A (2002) Sequence alignment editor. Version 2.0. University of Oxford, Oxford
Rayner RW (1970) A mycological colour chart. Commonwealth Mycological Institute, Kew
Réblová M, Gams W, Seifert KA (2011) Moniliochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Stud Mycol 68:163–191
Roberts RG, Snow JP (1990) Morphological and pathological studies of Colletotrichum capsici and Colletotrichum indicum. Mycologia 82:82–90
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rosa LH, Tabanca N, Techen N, Wedge DE, Pan Z, Bernier UR, Becnel JJ, Agramonte NM, Walker LA, Moraes RM (2012) Diversity and biological activities of endophytic fungi associated with micropropagated medicinal plant Echinacea purpurea (L.) Moench. Am J Plant Sci 3:1105–1114
Roumeguère C (1880a) Nouvelle apparition en France du Gloeosporium (Fusarium) reticulatum Mt., destructeur des melons. Revue Mycologique 2:169–172
Roumeguère C (1880b) Fungi Gallici Exsiccati. Cent. X. Revue Mycologique 2:200–202
Roy KW (1982) Seedling diseases caused in soybean by species of Colletotrichum and Glomerella. Phytopathology 72:1093–1096
Saccardo PA (1884) Sylloge Fungorum vol. 3: i-ii, 1–860. Saccardo, Padua, Italy
Saccardo PA (1886) Sylloge Fungorum, vol 4: i-v, 1–807. Saccardo, Padua, Italy
Sarbhoy AK, Agarwal DK (1990) Descriptions of tropical plant pathogenic fungi. Set 1. Malhotra Publ. House, New Delhi
Sawada K (1933) Descriptive catalogue of the Formosan fungi. Part VI. Report of the Department of Agriculture, Goverment Research Institute of Formosa 61:1–99
Shear C, Wood A (1913) Studies of fungous parasites belonging to the genus Glomerella. U.S. Department of Agriculture (USDA) 252:1–110
Shen S, Goodwin PH, Hsiang T (2001) Infection of Nicotiana species by the anthracnose fungus Colletotrichum orbiculare. Eur J Plant Pathol 107:767–773
Sherff EE (1937) The genus Bidens. Field Museum of Natural History Botanica Seria 16:16–484
Sherriff C, Whelan MJ, Arnold GM, Lafay J-F, Brygoo Y, Bailey JA (1994) Ribosomal DNA sequence analysis reveals new species groupings in the genus Colletotrichum. Exp Mycol 18:121–138
Simmonds JH (1965) A study of the species of Colletotrichum causing ripe fruit rots in Queensland. Queensland Journal of Agricultural and Animal Sciences 22:437–459
Simmonds JH (1966) Host index of plant diseases in Queensland. Queensland Department of Primary Industries. Brisbane, Australia
Singh UP (1974) A new forma specialis of Colletotrichum gloeosporioides Penz. from India. Beihefte zur Nova Hedwigia 47:451–452
Sitterly WR, Keinath AP (1996) Anthracnose. In: Zitter TA, Hopkins DL, Thomas CE (eds) Compendium of cucurbit diseases. APS Press, pp 24–25
Smith AL (1909) New or rare microfungi. T Brit Mycol Soc 3:111–124
Southworth EA (1890) A new hollyhock disease. J Mycol 6(2):45–50
Southworth EA (1891) Additional observations on anthracnose of the hollyhock. J Mycol 6(3):115–116
Sreenivasaprasad S, Mills PR, Meehan BM, Brown AE (1996) Phylogeny and systematics of 18 Colletotrichum species based on ribosomal DNA spacer sequences. Genome 39:499–512
Stephenson SA, Green JR, Manners JM, Maclean DJ (1997) Cloning and characterisation of glutamine synthetase from Colletotrichum gloeosporioides and demonstration of elevated expression during pathogenesis on Stylosanthes guianensis. Curr Genet 31:447–454
Stevens FL (1931) The ascigerous stage of Colletotrichum lagenarium induced by ultra-violet irradiation. Mycologia 23(2):134–139
Stumm D, Gessler C (1984) A technique for continuous observation of the infection process in cucumber anthracnose. Phytopathol Z 111:312–316
Sutton BC (1992) The genus Glomerella and its anamorph Colletotrichum. In: Bailey JA, Jeger MJ (eds) Colletotrichum: Biology, pathology and control. CAB International, Wallingford, pp 1–26
Swofford DL (2000) PAUP* 4.0: Phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland
Sydow P (1899) Beiträge zur Kenntnis der Pilzflora der Mark Brandenburg. II. Hedwigia Beiblatt 38:134–140
Tai FL (1979) Sylloge fungorum sinicorum. Sci. Press, Acad. Sin, Peking
Tanaka S, Ishihama N, Yoshioka H, Huser A, O’Connell R, Tsuji G, Tsuge S, Kubo Y (2009) The Colletotrichum orbiculare ssd1 mutant enhances Nicotiana benthamiana basal resistance by activating a mitogen-activated protein kinase pathway. Plant Cell 21:2517–2526
Templeton GE (1974) Endemic fungus disease for control of prickly sida in cotton and soybeans. Arkansas Farm Res 23(4):12
Templeton GE (1976) C. malvarum spore concentrate, formulation, and agricultural process. US Patent 3,999,973 dated Dec 28 1976
Thaung MM (2008) Biodiversity survey of coelomycetes in Burma. Australas Mycol 27:74–110
Tosi L, Buonaurio R, Cappellii (2004) Occurrence of anthracnose caused by Colletotrichum malvarum on Althaea officinalis in Italy. Plant Dis 88:425
US Department of Agriculture (1905) Yearbook of the United States Department of Agriculture 1905, USA
Vassiljevski NI, Karakulin BP (1950) Fungi imperfecti parasitici: Pars II. Melanconiales. Academiae Scientiarum URSS, Moscow and Leningrad, Russia
Veitch R (1942) Report of the Director of Plant Industry (Research). Report of the Department of Agriculture Queensland 1941–1942:5–8
Verplancke G, van den Broecke R (1936) Contribution a la flore mycologique Belge. Bull Soc Roy Bot Belgique 69(II):69–95
Villanueva-Arce R, Yáñez-Morales M, Hernández-Anguiano AM (2008) Especies de Colletotrichum en Chirimoya (Annona cherimola Mill.). Agrociencia 42:689–701
von Arx JA (1957a) Die Arten der Gattung Colletotrichum Cda. Phytopathol Z 29:413–4684
von Arx JA (1957b) Revision der zu Gloeosporium gestellten Pilze. Verhandelingen der Koninklijke Nederlandse Akademie van Wetenschappen, Afd. Natuurkunde, Tweede reeks 51:1–153
von Arx JA, Müller E (1954) Die Gattungen der amerosporen Pyrenomyceten. Beitr zur Kryptogamenfl der Schweiz 11(1):1–434
von Arx JA, van der Velden FJJA (1961) Das Colletotrichum der Gurkengewächse. Phytopathol Z 41:228–235
Walker J (1962) Notes on plant parasitic fungi. Proceedings of the Linnean Society of New South Wales 87:162–176
Walker J, Nikandrow A, Millar GD (1991) Species of Colletotrichum on Xanthium (Asteraceae) with comments on some taxonomic and nomenclatural problems in Colletotrichum. Mycol Res 95:1175–1193
Wapshere AJ, Erb H, Bunster L (1995) Implications of a primary survey on the biological control prospects for the pasture and rangleland weed bathurst burr, Xanthium spinosum in Australia. In: Delfosse ES, Scott RR (eds) Proceedings of the Eighth International Symposium of the Biological Contol of Weeds, Lincoln, New Zealand, 1992, DSIR/Csiro: Melbourne, Australia, pp 355–359
Warwar V, Oved S, Dickman MB (2000) Antisense expression of the calmodulin gene from Colletotrichum trifolii impairs prepenetration development. FEMS Microbiol Lett 191:213–219
Wasilwa LA, Correll JC, Morelock TE, McNew RE (1993) Reexamination of races of the cucurbit anthracnose pathogen Colletotrichum orbiculare. Phytopathology 83:1190–1198
Watanabe T, Tamura M (1952) Studies on the perfect stage of the causal fungus of the anthracnose of cucumber. Ann Phytopathological Soc Japan 16:137–140
Wei YD, Byer KN, Goodwin PH (1997) Hemibiotrophic infection of round-leaved mallow by Colletotrichum gloeosporioides f. sp. malvae in relation to leaf senescence and reducing reagents. Mycol Res 101:357–364
Weir B, Damm U, Johnston PR (2012) The Colletotrichum gloeosporioides species complex. Stud Mycol 73:115–180
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: White TJ, Sninsky JJ, Gelfand DH, Innin MA (eds) PCR protocols: A guide to methods and applications. Academic, San Diego, pp 315–322
Wollenweber HW, Hochapfel H (1949) Beiträge zur Kenntnis parasitärer und saprophytischer Pilze. VI. Vermicularia, Colletotrichum, Gloeosporium, Glomerella und ihre Beziehung zur Fruchtfäule. Z Parasitenkd 14:181–268
Woudenberg JHC, Aveskamp MM, de Gruyter J, Spiers AG, Crous PW (2009) Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 22:56–62
Xuei XL, Järlfors U, Kuć J (1988) Ultrastructural changes associated with induced systemic resistance of cucumber to disease: Host response and development of Colletotrichum lagenarium in systemically protected leaves. Can J Bot 66:1028–1038
Yang Z, Dickman MB (1999) Colletotrichum trifolii mutants disrupted in the catalytic subunit of cAMP-dependent protein kinase are nonpathogenic. Mol Plant Microbe In 12:430–439
Yang YL, Cai L, Yu ZN, Liu ZY, Hyde KD (2011) Colletotrichum species on Orchidaceae in southwest China. Cryptogamie Mycol 32:229–253
Zhou H, Brockington M, Jungbluth H, Monk D, Stanier P, Sewry CA, Moore GE, Muntoni F (2006) Epigenetic allele silencing unveils recessive RYR1 mutations in core myopathies. Am J Hum Genet 79:859–868
Acknowledgements
We thank the curators and staff of the CBS culture collections as well as Dr Richard J. O’Connell (BIOGER-CPP, INRA-AgroParisTech, Thiverval-Grignon, France), Prof. dr Yasuyuki Kubo (Kyoto Prefectural University, Kyoto, Japan), Dr Peter Johnston and Dr Bevan Weir (Landcare Research, Auckland, New Zealand), Prof. dr Lisa Vaillancourt (Department of Plant Pathology, University of Kentucky, USA), Yonghong Ge and Prof. dr David I. Guest (Faculty of Agriculture, Food and Natural Resources, The University of Sydney, Australia) for kindly supplying isolates for this study. We kindly thank the curators of the fungaria at the Royal Botanic Gardens in Kew (IMI and K(M)), UK and at the US National Fungus Collections (BPI), Beltsville, Maryland, USA for providing access to historical type specimens. Dr Paul Kirk (Royal Botanic Gardens, Kew, UK) is thanked for advice on nomenclatural matters. This research was supported by the Dutch Ministry of Agriculture, Nature and Food Quality through an endowment of the FES programme “Versterking infrastructuur Plantgezondheid”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Damm, U., Cannon, P.F., Liu, F. et al. The Colletotrichum orbiculare species complex: Important pathogens of field crops and weeds. Fungal Diversity 61, 29–59 (2013). https://doi.org/10.1007/s13225-013-0255-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-013-0255-4