Abstract
Classical (one-variable-at-a-time) and statistical methods (Plackett-Burman and Central composite design) were used to optimise growth medium for the production of cholesterol oxidase (COX) from Rhodococcus sp. NCIM 2891. COX activities from the classically and statistically optimised media were 0.75 and 3.25 U/ml, respectively. The statistically optimised medium had 4.33- and 9.7-fold higher enzymatic activity than the classically optimised and un-optimised basal medium, respectively. The ratio of enzyme production to cell growth rate was 29-fold higher in our statistically optimised medium than in the basal medium, indicating that the enzyme production could be classified as mixed type of growth. Cell-bound COX accounted for 90.68 ± 2 % of the total enzymatic activity of the growth medium. Interactions between the COX-inducing substrate cholesterol and medium growth substrates yeast extract and (NH4)2HPO4 significantly enhanced the production of cell-bound COX. Our results validate the statistical approach as a potential technique for achieving the large-scale production of cell-bound COX from Rhodococcus sp. NCIM 2891.
Similar content being viewed by others
Introduction
Cholesterol oxidase (COX, EC 1.1.3.6) is widely used in clinical settings to detect cholesterol (MacLachlan et al. 2000) and steroids (Ahmad et al. 1992) in biological samples. It also has potential applications for the biotransformation of sterols and non-steroidal compounds, optical resolution of non-steroidal compounds, production of steroid hormone precursors, biosynthesis of antifungal antibiotics, among others (Doukyu 2009). COX is a flavoenzyme that catalyses the oxidation and isomerisation of cholesterol to cholest-4-en-3-one (Kreit and Sampson 2009). The enzyme is mainly generated in bacteria, although there have been a few reports of COX production in fungi (Fukuyama and Miyake 1979). This family of enzymes occurs in both the membrane-bound and secreted forms (Inouye et al. 1982; Kreit et al. 1994; Sojo et al. 1997). COX enzymes from different microbial sources have been purified and characterised (Doukyu 2009); however, reports on COX production methods are limited (Aihara et al. 1986; Kreit et al. 1994; Lee et al. 1998, 1999; Yazdi et al. 2001; Chang et al. 2005; Chauhan et al. 2009) even though the applications of COX have been rapidly expanding.
COX produced from Rhodococcus strains has gained importance due to its high stability and activity over a wide range of pH values (Wang et al. 2008) and its high yields (Aihara et al. 1986; Watanabe et al. 1989). Rhodococcus species can produce both secreted and cell-bound forms of COX (Watanabe et al. 1989; Kreit et al. 1992; Kreit et al. 1994; Sojo et al. 1997). In the study reported here, we first studied the effect of different nutritional factors on the production of total COX by a Rhodococcus sp. NCIM 2891 strain using the classical ‘one-variable-at-a-time’ method. The medium components were then analysed for their significant effects on enzyme production using a statistical Plackett–Burman factorial design (Plackett and Burman 1946). We then used a full-factorial central composite design (CCD) for response surface methodology (RSM) to derive important information on the optimal level of each variable, along with its interactions with other variables and its effects on product yield. A detailed account on our findings is presented here.
Materials and methods
Microorganism and culture conditions
Rhodococcus sp. NCIM 2891 was obtained from the National Collection of Industrial Micro-organisms (NCIM) (National Chemical Laboratory, Pune, India). This strain was cultured in modified culture medium (basal medium) based on Yazdi et al. (2001), which contains the following additives (in g/l): (NH4)2HPO4, 2.0; yeast extract, 5.0; K2HPO4, 0.25; FeSO4 · 7H2O, 0.1; NaCl, 1.0; cholesterol, 2.0; Tween 80, 2.0 (v/v). The pH was adjusted to 7.0. For the fermentation experiments, 1-ml aliquots of the active seed culture were added to 50 ml of sterilised basal medium in 250-ml Erlenmeyer flasks and incubated at 30°C on a rotary shaker (200 rpm). For bacterial cell count measurements, 1-ml samples were withdrawn from the flask at 12-h intervals during the culture period and diluted tenfold with phosphate buffered saline [(in g/l): NaCl, 8.0; KCl, 0.2; Na2HPO4, 1.44; KH2PO4, 0.24; pH 7.4]. Nutrient agar plates were uniformly streaked with 100 μl of the diluted broth sample using a cell spreader and incubated for a maximum of 6 days at 30 °C. The colonies formed on each plate were then counted to estimate the colony forming unit (CFU) per millilitre of the original sample (CFU/ml).
Isolation and assay of enzyme
After 60 h of cultivation, bacterial cultures were centrifuged at 10,000 rpm for 15 min at 4 °C. The supernatant fluid was used as the source of the extracellular enzyme. The cell pellets were re-suspended in 5 ml of 50 mM potassium phosphate buffer, pH 7.0, containing 1% Triton X-100 (extraction buffer) and mixed vigorously for 5 min with a vortex (MS 3 digital; IKA, Staufen, Germany). The cell homogenate was then centrifuged (12,000 rpm for 15 min) to collect the cell-free supernatant, which was used for measuring cell-bound COX. COX activity was assayed using an horseradish peroxidase (HRP)-coupled assay that monitored H2O2 production by measuring 2, 2′-azino-bis [3-ethylbenzothiazoline-6-sulphonic acid] diammonium salt (ABTS) radical formation at 405 nm and 35°C (ε405 = 18400/M/cm) (Werner et al. 1970). The enzyme was assayed by kinetic assay method in a final volume of 1 ml containing 50 mM potassium phosphate buffer (pH 7.0), 5 μl of 1,080 U/ml HRP 50 μl of 14 mg/ml ABTS and 50 μl of 3 mM cholesterol. A stock solution of cholesterol (20 mM) was prepared by dissolving cholesterol in 10 % Triton X-100 in distilled water at approximately 165 °C for 5 min. The solution was then diluted with water containing 1 % Triton X-100 to a final cholesterol concentration of 3 mM. The amount of enzyme activity (U/ml) was calculated using Eq. 1.
where 36.8 is the millimolar extinction coefficient for two molecules of oxidised ABTS because the reduction of one molecule of H2O2 requires two molecules of ABTS. One enzyme unit was defined as the amount of enzyme that forms 1 μmol hydrogen peroxide/min at 35 °C.
Optimisation of medium components by experimental design
The culture medium was optimised using the Plackett–Burman design and CCD. Each factor was examined at two levels: −1 and +1 for low and high levels, respectively. A focus of the study was to evaluate the linear and curvature effects of the variables. The Plackett–Burman experimental design is based on a first-order polynomial model, as shown in Eq. 2.
where Y is the response (productivity), β o is the model intercept, β i is the linear coefficient and x i is the level of the independent variable. This model was used to screen and evaluate the medium components influencing COX production. Seven assigned variables were screened in 12 experimental trials, with an additional two runs made at their centre points, as shown in Table 1. Based on regression analysis of the variables, factors with greater than 95 % significance (P < 0.05) were considered to have a significant effect on COX production and were further optimised by RSM.
A CCD with five settings for each factor was used to evaluate the quadratic effects and two-way interactions among these variables, and a full factorial CCD was used to optimise the key variables for COX production in shaking cultures. The variables used were cholesterol, (NH4)2HPO4, and yeast extract, each at five coded levels (−α, −1, 0, +1, +α), as shown in Table 2. The relation between the coded forms and the actual value of the variables is described in Eq. 3.
where x i is a coded value and X i is the actual value of the variable, X 0 is the actual value of the same variable at the centre point and ΔX i is the step change of the variable.
These three factors, each with five coded levels, consisted of 20 experimental runs that included six replicates at the centre point with α = 1.682, were used to measure the entire range of variable combinations, as shown in Table 3. The optimal point was predicted using a second-order polynomial model (Eq. 4) to correlate the relationship between the variables and the response.
where Y is the predicted response, k is the number of factor variables, β o is the model constant, β i is the linear coefficient, β ii is the quadratic coefficient and β ij is the interaction coefficient.
Statistical analysis was performed using Minitab ver. 15 (Minitab, State College, PA) which was used to evaluate the analysis of variance (ANOVA) to determine the significance of each term in the fitted equations and to estimate the goodness of fit in each case. The fit of the model was checked by the coefficient of determination R 2. The closer the values of R 2 to 1, the better the model explains the variability between the experimental and predicted values. To validate the conditions optimised with CCD, we used the newly optimised medium to study COX production.
Results and discussion
Optimisation of COX production using classical methods
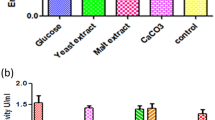
The effects of pH, temperature, cultivation time and different types of carbon and nitrogen sources on COX production by Rhodococcus sp. NCIM 2891 were first studied in the basal medium using the ‘one-variable-at-a-time’ method. The optimal conditions for COX production in the basal medium were 30°C, pH 7.0 and a 60-h cultivation time. Enzyme production under these optimal conditions was 0.335 U/ml. Cell-bound COX contributed half of the total enzymatic activity in the basal medium. To determine the effect of different carbon sources on COX production, we tested different steroidal compounds, a readily assimilable carbon substrate (glucose) and a non-steroidal hydrophobic substrate (n-hexanoate). COX was produced with all of the carbon sources tested; however, only the steroidal substrates (β-sitosterol, cholesterol and stigmasterol) produced a significant increase in COX levels. Of the tested sterols, cholesterol produced the most COX (Fig. 1a). We also tested the effects of different combinations of inorganic and organic nitrogen sources on cell growth and COX production and found that the combination of (NH4)2HPO4 and yeast extract supported the highest enzyme production (0.353 U/ml) (Fig. 1b). When grown in the control medium containing 0.2 % cholesterol, COX activity reached 0.346 U/ml. Increasing the cholesterol content from 0.2 % to 0.35 % also increased COX activity by 24.15 %, to 0.433 U/ml. COX production reached saturation at 0.4 % cholesterol (Fig. 2). Adding (NH4)2HPO4 to the medium stimulated the production of COX (Fig. 2), with maximum COX activity (0.397 U/ml) observed for media containing 0.25 % (NH4)2HPO4. However, higher concentrations of (NH4)2HPO4 caused enzyme production to decline. The effects of adding yeast extract to the medium on COX activity were also concentration dependent, with a noticeable effect first observed at 0.1 % concentration and maximum activity (0.483 U/ml) reached at 0.9 %. Further increases in yeast extract concentrations did not improve COX production (Fig. 2). Optimal concentrations of (NH4)2HPO4, yeast extract and cholesterol in our classically optimised medium were 2.5, 9.0 and 3.5 g/l, respectively. Under these optimal conditions the enzyme production was 0.75 U/ml, with cell-bound COX accounting for 65.71 % of the total enzyme activity.
Effects of carbon sources (a; 0.2 % w/v) and inorganic (b; 0.2 % w/v) and organic (0.5 % w/v) nitrogen sources on cholesterol oxidase (COX) production by Rhodococcus sp. NCIM 2891. A.A CH3COONH4,, A.C NH4Cl, A.N NH4NO3, A.S (NH4)2SO4, S.N NaNO3, DAHP (NH4)2HPO4, B.E beef extract, M.E malt extract, Y.E yeast extract, Pep peptone, Try tryptone
Evaluation of significant components by the Plackett–Burman design
Cholesterol oxidase activity varied widely (0.157 ± 0.01 to 1.248 ± 0.02 U/ml) in the experiments, indicating the potential for optimising COX production. The analysis of regression coefficients, t values and P values for these seven variables is shown in Table 4. Both the t value and the P value (<0.05) were used to confirm the significance of the parameters studied. Cholesterol was significant to a 99.4 % confidence level, while (NH4)2HPO4 and yeast extract had significance levels of 99.9 and 98.9 %, respectively. Based on P values, cholesterol, (NH4)2HPO4, and yeast extract were chosen for further optimisation. Because NaCl, K2HPO4, Tween 80 and FeSO4 7H2O did not have significant effects on COX production, they were added to the cultivation medium at their median values for subsequent experiments.
Optimisation of medium components by CCD
The significant variables from the Plackett–Burman factorial design were further examined by CCD. The fitting of the second order polynomial model was verified by the coefficient of determination, R 2. The model shows a high determination coefficient (R 2 = 99.82), which explains almost 100 % of the variability in the response. The regression coefficients were calculated, and the regression equation obtained is shown the Eq. 5:
where Y, X 1, X 2, and X 3 represent the response, and the amounts of cholesterol, (NH4)2HPO4, and yeast extract, respectively. The F value of the model is 612.62 (P < 0.05), which indicates that the model is a good fit and explains most of the observed response variation. All of the model terms X 1, X 2, X 3, \( X_1^2 \), \( X_2^2 \), \( X_3^2 \), X 1 X 2, X 1 X 3 and X 2 X 3, exhibited confidence levels of >95 % (P < 0.05), which indicates that our chosen variables (cholesterol, (NH4)2HPO4, yeast extract and their interaction effects) were significant in our model equation for both linear and quadratic fits (Table 5). The regression equation is graphically represented by the contour plots to visualise the effect of the component concentration on the expected response (Fig. 3). It is evident that the interactions of cholesterol with yeast extract (Fig. 3a) cholesterol and (NH4)2HPO4 (Fig. 3b) are highly significant, while the interaction between yeast extract and (NH4)2HPO4 (Fig. 3c) is only moderately significant.
Experimental validation of the medium composition optimised by CCD
The CCD model predicted that maximum COX activity of 3.36 U/ml would occur when the medium contained cholesterol, (NH4)2HPO4 and yeast extract at concentrations of 8.95, 10.57 and 26.75 g/l while keeping the other components at the median tested concentration determined from our optimisation studies. The model was experimentally validated by RSM for the statistically optimised medium. COX production in the statistically optimised medium is shown in Fig. 4a. COX production reached a maximum of 3.25 U/ml at nearly 60 h of cultivation, which agrees with the predicted value. Cell-bound COX accounted for 90.68 % of the total enzyme activity in the statistically optimised broth. A comparison of cell-bound and extracellular COX production revealed that, compared to the COX level in the basal medium, the optimised medium increased cell-bound COX levels by 40.52 %, while only marginally increasing the extracellular COX by 15.15 %.
The growth rate and enzyme production rate within the exponential phases (36–60 h) of growth were calculated from the slope of the lines shown in Fig. 4b. The growth rates (CFU/ml/h) in the statistically optimised, classically optimised and basal media were 3 × 107, 3 × 107 and 2 × 10 7, respectively, while the corresponding enzyme production rates (U/ml/h) were 0.087, 0.011 and 0.002. Thus, the ratio (×10-10) of enzyme production rate to growth rate in the statistically optimised, classically optimised and basal media are 29, 3.6 and 1, respectively. These results imply that the ratio of enzyme production rate to growth rate in the statistically optimised medium is 8.05- and 29-fold greater than that in the classically optimised medium and basal medium, respectively. These results demonstrate that although the production of cell-bound COX is linked to the growth of cell mass, the level of its production (i.e. activity per unit cell mass) can be increased by adjusting the critical nutrient components identified in our study to the statistically determined ratio. The cell mass obtained here using the growth stimulating nutrients, namely yeast extract and (NH4)2HPO4, supports the accumulation of COX induced by the cholesterol substrate in the cell wall.
Little data has been reported in the literature on the optimisation of COX production by microorganisms using statistical methods. Various compounds, such as cholesterol, yeast extract (Lee et al. 1998; Yazdi et al. 2001), glycerol, malt extract, soyabean meal (Chauhan et al. 2009), potato starch, yeast extract, malt extract and peptone (Varma and Nene 2003) have been reported to be substrates for an enhanced production of COX. The level of COX production obtained in our study (3.25 U/ml) following the statistical method is comparable or higher than many of the reported values (Lee et al. 1998; Yazdi et al. 2001; Chauhan et al. 2009; Varma and Nene 2003).
Conclusions
Our results demonstrate that statistical methods were more effective than the classical method in determining the optimised concentration of medium components for COX production. Statistically optimised medium can drastically increase COX production in Rhodococcus sp. NCIM 2891 compared to production in the classically optimised medium. Enhanced COX production was largely due to an increase in cell-bound COX. The enzyme production rate far exceeded the organism’s intrinsic growth, thus implying that although linked with cell growth, the level of production of the cell-bound COX in terms of activity per unit cell mass could be increased by using critical nutrient components at statistically determined ratios. The high production of cell-bound COX by Rhodococcus sp. NCIM 2891 in a simple medium under suitable environmental conditions is therefore a potential source of COX for industrial applications.
References
Ahmad S, Garg SK, Johri BN (1992) Biotransformation of sterols: selective cleavage of the side chain. Biotechnol Adv 10:1–67
Aihara H, Watanabe K, Nakamura R (1986) Characterization of production of cholesterol oxidases in three Rhodococcus strains. J Appl Bacteriol 61:269–274
Chang YC, Yu RC, Yang HY, Chou CC (2005) Physical factors and reuse affect the production of cholesterol oxidase in reactors containing calcium alginate-immobilized Rhodococcus equi No. 23. World J Microbiol Biotechnol 21:1129–1134
Chauhan AK, Survase SA, Kishenkumar J, Annapure US (2009) Medium optimization by orthogonal array and response surface methodology for cholesterol oxidase production by Streptomyces lavendulae NCIM 2499. J Gen Appl Microbiol 55:171–180
Doukyu N (2009) Characteristics and biotechnological applications of microbial cholesterol oxidases. Appl Microbiol Biotechnol 83:825–837
Fukuyama M, Miyake Y (1979) Purification and some properties of cholesterol oxidase from Schizophyllum commune with covalently bound flavin. J Biochem 85:1183–1193
Inouye Y, Taguchi K, Fujii A, Ishimaru K, Nakamura S, Nomi R (1982) Purification and characterization of extracellular 3β- hydroxysteroid oxidase produced by Streptoverticillium cholesterolicum. Chem Pharm Bull 30:951–958
Kreit J, Sampson S (2009) Cholesterol oxidase: physiological functions. FEBS J 276:6844–6856
Kreit J, Germain P, Lefebvre G (1992) Extracellular cholesterol oxidase from Rhodococcus sp. cells. J Biotechnol 24:177–188
Kreit J, Lefebvre G, Germain P (1994) Membrane-bound cholesterol oxidase from Rhodococcus sp. cells. Production and extraction. J Biotechnol 33:271–282
Lee MT, Chen WC, Chou CC (1998) Maximization of cholesterol oxidase production by Rhodococcus equi no. 23 by using response surface methodology. Biotechnol Appl Biochem 28:229–233
Lee MT, Chen WC, Chou CC (1999) Optimization and kinetic analysis of cholesterol oxidase production by Rhodococcus equi no.23 in submerged culture. Enzyme Microb Technol 25:598–604
MacLachlan J, Wotherspoon ATL, Ansell RO, Brooks CJW (2000) Cholesterol oxidase: sources, physical properties and analytical applications. J Steroid Biochem Mol Biol 72:169–195
Plackett RL, Burman JP (1946) The design of optimum multifactorial experiments. Biometrika 33:305–325
Sojo M, Bru R, Lopez-Molina D, Garcia-Carmona F, Argüelles J (1997) Cell-linked and extracellular cholesterol oxidase activities from Rhodococcus erythropolis. Isolation and physiological characterization. Appl Microbiol Biotechnol 47:583–589
Varma R, Nene S (2003) Biosynthesis of cholesterol oxidase by Streptomyces lavendulae NCIM 2421. Enzyme Microb Technol 33:286–291
Wang C, Cao Y, Sun B, Ji B, Nout MJR, Wang J, Zhao Y (2008) Preparation and some properties of cholesterol oxidase from Rhodococcus sp. R 14-2. World J Microbiol Biotechnol 24:2149–2157
Watanabe K, Aihara H, Nakagawa Y, Nakamura R, Sasaki T (1989) Properties of the purified extracellular cholesterol oxidase from Rhodococcus equi No. 23. J Agric Food Chem 37:1178–1182
Werner W, Rey HG, Weilenger H (1970) Uber die eigenschaften eine neuen chromogens fur die blutzuckerbestimmung nach der GOD/POD methode. Z Anal Chem 254:224–228
Yazdi MT, Malekzadeh F, Zarrini G, Faramarzi MA, Kamranpour N (2001) Production of cholesterol oxidase by a newly isolated Rhodococcus sp. World J Microbiol Biotechnol 17:731–737
Acknowledgements
The authors thank the Council of Scientific and Industrial Research (CSIR), India (Grant No. 37/1319/07/EMR-II) for providing financial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ahmad, S., Goswami, P. Enhanced production of cell-bound cholesterol oxidase from Rhodococcus sp. NCIM 2891 by the statistical method. Ann Microbiol 63, 199–205 (2013). https://doi.org/10.1007/s13213-012-0461-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-012-0461-4