Abstract
The genetic diversity among native nodular rhizobia of Lima bean in soil from Piauí State, Brazil, was characterized and evaluated. The genotype UFPI-491 was used as a trap plant for rhizobia, with soil samples collected in Piaui State being used as the inoculum source. For isolation, nodules were collected at 45 days after seedling emergence—a period that presented the highest values for number and biomass of nodules evaluated in a previous experiment. In total, 50 isolates were obtained and placed into groups based on divergence from the morphological and physiological characterization. In general, the restriction patterns obtained with endonucleases MboI, HaeIII and NheI showed sufficient variability to discriminate between the isolates in this study. The main characteristics exhibited by isolates identified species from the genera Bradyrhizobium, Mesorhizobium and Rhizobium.
Similar content being viewed by others
Introduction
Phaseolus lunatus, the lima bean, is the second most economically important species of Phaseolus and 1 of the 12 primary grain legumes (Fofana et al. 1999). This crop shows high rusticity and the capacity to resist long dry periods. These characteristics are important for success in semi-arid regions and increase the economic and social importance of the crop (Azevedo et al. 2003).
Among the legumes cultivated throughout the world, P. lunatus is one that has the ability to perform biological nitrogen fixation (BNF) due to symbiosis between this legume plant and nitrogen-fixing bacteria (Schultze and Kondorosi 1998). These bacteria, commonly known as rhizobia, invade root tissues and induce the formation of specialized structures known as nodules, where they differentiate and fix atmospheric nitrogen that is then supplied to the plant (Santos et al. 2007). The agronomic implications of this symbiosis have promoted research on the diversity of these bacteria, mainly in tropical soils brackets (Ormeno-Orrillo et al. 2006).
In recent years, several papers have reported the diversity of rhizobia isolated in a variety of host legumes (Giongo et al. 2008; Grossman et al. 2005; Loureiro et al. 2007; Mahdhi et al. 2008; Musiyiwa et al. 2005). The results showed a great diversity of rhizobia in soil around the world. In association with the genus Phaseolus, the literature has shown that studies of rhizobial diversity refer mostly to symbiosis of the common bean (P. vulgaris) (Martínez-Romero et al. 1991; Martínez-Romero 2003; Souza et al. 1994).
By contrast, rhizobia associated with P. lunatus have been far less studied. Ormeno-Orrillo et al. (2006) studied genetic diversity of native rhizobia isolated from the lima bean in Peru. They observed that lima beans are nodulated mainly by the genus Bradyrhizobium. However, a recent study conducted in the same location (Ormeño-Orrillo et al. 2007) observed that naturally growing species of nodulating P. lunatus belonged more commonly to the genus Rhizobium than to Sinorhizobium.
Zilli (2004) analyzed the diversity of rhizobium populations in recently clear-cut areas of the Cerrado, during a rice-cowpea and soybean rotation, using genotypic [amplified ribosomal DNA restriction analysis (ARDRA) Heyndrickx et al. 1996] and morphologic/phenotypic characterization methods, and concluded that understanding native rhizobial population diversity will ultimately contribute to the selection of inoculants for soybean and cowpea in the Brazilian Cerrado. Analysis of gene sequences coding for 16S rRNA (16S rDNA) have been used to infer phylogenetic relationships among species of rhizobia—homology between the sequences of 16S rRNA being the criterion most frequently used to estimate phylogenetic relationships among bacteria. One advantage of this approach is that DNA sequences and gene products can be compared in an evolutionary context (molecular systematics) (Van Berkun et al. 2000). Typing by DNA-based PCR can also be combined with restriction enzymes in methods such as ARDRA. The gene for 16S or 23S rDNA with or without the intergenic region is amplified by PCR with universal primers and then digested with different combinations of restriction enzymes. In contrast to other methods of analysis of fragments, such methods generate species-specific patterns (Gürtler et al. 1991) considering the conserved nature of rRNA genes.
However, Fofana et al. (1999) obtained isolates from areas where this legume is native. Peru is one of the known centers of origin and diversity of P. lunatus. There have been no studies on rhizobial diversity associated with lima beans in areas where this legume is not native, such as Brazil. Thus, we studied the genetic diversity among nodulating rhizobia of lima beans in soils from Piauí State, Northwest Brazil.
Material and methods
The soil samples used in this study were collected at 0.0 to 0.2 m depth in ten areas with a history of lima bean cultivation. The selected areas are located in two regions of Piaui state (07°61′970″S and 93°43′538″W). The main soil type is an Orthic Acrisol (Typic Hapludult, US taxonomy). These soils samples were analyzed in the Soil Fertility Laboratory of the Federal University of Piauí. The values of pH ranged from 6.4 to 7.2. Soil organic matter content ranged from 24 to 30 g kg−1 soil. The phosphorus and potassium contents ranged from 60.2 to 75.3 and 50.1 to 62.5 mg kg−1 soil, respectively. The sand, silt and clay content ranged from 31.4 to 43.7, 20.1 to 30.0 and 10.4 to 25.6%, respectively.
The lima bean genotype was UFPI-491 (“Fava miúda”), obtained from the Active Germplasm Bank of Fava Bean, Universidade Federal do Piauí (BAG of fava bean of UFPI). This genotype was selected for being relatively well cultured in the States of Piauí and Maranhão. According to the data from BAG, the UFPI-491 genotype, originating from Varzea Grande City, PI, has seeds with white skin, and average length and width of 17.53 and 18.18 mm, respectively.
The experiment was conducted in a greenhouse at the Plant Production Department of the Agricultural Sciences Center, UFPI, Campus of Socopo, from 24 March to 6 June 2007. The genotype was sown in plastic bags containing 5 kg soil originating from the selected area, following a completely randomized design with four replications. The release of free-N mineral fertilizer was applied at planting, according to the technical recommendations for the crop (Vieira 1992). At sowing, seeds were placed four per bag and thinning was performed 15 days after emergence, leaving one plant per bag. The bags were irrigated daily to maintain soil moisture close to field capacity (gravimetric method).
The nodules used for isolation and characterization of rhizobia strains were collected 45 days after seedling emergence—a period that yielded the highest values for number and biomass of nodules in a preliminary experiment. Immediately after collection, nodules were allowed to desiccate in test tubes with gel silica and a thin cotton layer screw cap-sealed.
Rhizobia isolation, as well as morphological, physiological and biochemical characterization of isolates, was performed at the Soil Biology Laboratory of Agronomical Institute of Pernambuco (IPA) according to the methodology used by Hungria (1994). The isolation was performed in Petri dishes containing YMA culture medium, pH 6.8, with Congo red indicator, and incubated at 28°C for the period necessary to highlight the colonies’ growth characteristics (Vincent 1970). Morphological characteristics recorded included colony shape (CS: R round, E ellipsoid), elevation (EL: C convex, F flattened) and color (CC: WM white/milky, T transparent).
The physiological characteristics were evaluated for isolates grown in bromothymol blue medium measuring growth time (GT: F fast, 1–3 days; I intermediate, 4–5 days), formation of acids and alkalis (FAA: AC acid, AL alkaline), formation of mucus (FML: A absent, P present), volume of mucus (VM: MU much, M Medium, L little, D dry) (Melloni et al. 2006); mucus elasticity (ME: P presence of wire, A absence of wire) and production of bubbles (PB: P present, A absent). Gram tests were performed in all isolates. All isolates presented pink staining (Gram negative).
The native isolates were characterized by ARDRA having as references the type strains: Rhizobium sp NGR234 (ER1), Mesorhizobium mediterraneanus BR523 (ER2), Rhizobium etli CFN42 (ER3), Ensifer fredii USDA205 (ER4), Bradyrhizobium japonicum BR111 (ER5) and Rhizobium tropici CIAT899 (ER6). Type strains were provided by EMBRAPA-Agrobiologia, RJ, Brazil (Dra. Rosa Pittar) and were preserved at −80°C in the Laboratório de Genoma (IPA). The DNA from isolates was extracted using the Invitek Invisorb kit (http://www.invitek.de) as recommended by the manufacturer. 16S rDNA genes were amplified using primers fD1 (5′-AGA GTT TGA TCC TGG CTC AG-3′) and rD1 (5′-AAG GAG GTG ATC CAG CC-3′) and subsequently digested with endonucleases MboI, NheI and HaeIII.
After digestion, cleavage was confirmed by visualizing the banding pattern obtained upon agarose gel electrophoresis (2.5%) in 0.5X TBE buffer for 4 h at 80 V with dye SybrGold (Invitrogen; http://www.invitrogen.com) and photographed using a Loccus LPIX-HE (http://www.loccus.com.b). The restriction patterns (ribotypes) were characterized by the number and size of the fragments obtained. Dendrograms were constructed with the program NTSYS-pc v.2.1 (http://www.exetersoftware.com), the similarity matrices used the correlation coefficient Simple Matching (SM), and the unweighted pair group method with arithmetic mean (UPGMA) algorithm was used according to the electrophoretic profiles obtained.
Results and discussion
Upon cultivation of the P. lunatus UFPI-491 genotype used as a trap plant, 50 pure isolates with phenotypic characteristics different from bacterial colonies were obtained (Table 1). The morphological characteristics, in general, showed low variation in colony form among the isolates. All isolates presented with a rounded shape, except ISOL15 and ISOL8, which presented with ellipsoid shapes. The isolates obtained yielded convex and flattened colonies. Most isolates showed white colonies with milky appearance, while a few isolates showed transparent colonies. According to Jordan (1984), colors found in colonies of rhizobia include white, yellow or pink, although it is unusual to find rhizobia forming yellow or pink colonies (Hungria 1994; Santos et al. 2007; Soares et al. 2006).
Results indicate morphological diversity between native nodular rhizobia of fava in Piauí soils. Morphological diversity among other groups of rhizobia was also found by Silva et al. (2007) and Martins et al. (1997) for isolates obtained in soils from Pernambuco and Mato Grosso do Sul, respectively.
The isolates showed rapid and intermediate growth. The growth patterns observed indicate that the isolates may be more adapted to the edaphoclimatic conditions of the region, since rapid and intermediate growth favor greater survival in the soil as compared to slow-growing isolates (Medeiros et al. 2007).
Concerning the production of acid and alkali compounds, a prevalence of acid reactions was observed, although some isolates presented alkaline reactions. The presence of intermediate-growth isolates with production of alkali, characteristics that exemplify the genus Bradyrhizobium (Vargas et al. 2007) suggests that this genus performs symbiosis with P. lunatus. Similar results were observed by Ormeno-Orrillo et al. (2006) in isolates obtained from lima bean nodules grown in the soil of Peru, where the genus Bradyrhizobium is the predominant symbiont. The bacteria from the genus Bradyrhizobium are able to form symbiosis with several species, such as Phaseolus vulgaris (Parker 2002), Glycine max (Hungria 1993), Vigna unguiculata (Zilli et al. 2006) and Arachis hypogaea (Gerin et al. 1996).
Following the classification proposed by Melloni et al. (2006), 50% of isolates presented colonies with high production of mucus. However, isolated colonies varying among moderate, low and no production of mucus (ISOL1) were observed. The production of mucus (exopolysaccharides) in most isolates is an important feature related to the initial infection of roots (Freitas et al. 2007). Thus, isolates that show this characteristic possess a greater competitive advantage in infection, colonization and formation of nodules.
The phenotypic characteristics observed for isolates suggest that they were subject to different selective environmental pressure. Additionally, according to Hartwig (1998), in addition to edaphoclimatic conditions, genotypic characteristics of the macrosymbiont influence symbiotic specificity. Some studies in Brazil have shown variability in nodulation by rhizobial strains in soybeans (Bohrer and Hungria 1998), common-bean (Franco et al. 2002) and cowpea (Xavier et al. 2006).
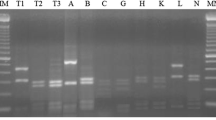
According to the classification scheme proposed by Wang et al. (2008), to date six genera have been proposed: Allorhizobium, Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium. Based on the sequences of 16S rRNA genes, all defined rhizobia are members of the subdivision of the Proteobacteria. This indicates that, like other tropical legumes (Santos et al. (2007), P. lunatus also exhibits symbiosis with a wide range of rhizobia. Generally, the restriction patterns obtained with endonucleases MboI (Fig. 1), NheI (Fig. 2) and HaeIII (Fig. 3) showed sufficient variability to discriminate between the isolates in this study. The enzyme MboI showed the most discrimination among isolates, giving a banding pattern very different between each. The simple matching coefficient (SM) was set to 65% genetic similarity in all the dendrograms formed, and we observed four clusters for this enzyme. Cluster 1 was composed of 28 strains grouping with reference strains ER6 (Rhizobium tropici CIAT899) and ER1 (Rhizobium sp NGR234). In cluster 2, ten isolates clustered with strains ER2 (Mesorhizobium mediterraneus BR523) and ER3 (Rhizobium etli CFN42). In cluster 3, seven isolates clustered with ER5 (Bradyrhizobium japonicum BR11). Finally, in cluster 4, only four isolates grouped and the ISOL45 and ER4 (Ensifer fredii USDA205) formed monophyletic branches.
With endonucleases NheI and HaeIII, the variability was lower, showing that these enzymes are not sufficient to detect the genetic variability in these isolates. When we compiled the data matrix with two enzymes, MboI + NheI (Fig. 4), and NheI + HaeIII showed that genetic variability was greater among isolates in all dendrograms compiled, and that the ER4 reference behaved as an outgroup. The phylogenetic tree constructed from the compilation of the matrices of the three endonucleases MboI, NheI and HaeIII showed three major clusters and strain ER4 with the same behavior mentioned above (Fig. 5). In cluster 1 and 2, 18 isolates were grouped together with B. japonicum BR11). In cluster 3 and 4, 21 isolates were clustered and almost all seem to be different isolates, except isol14 and isol15, which were identical and appear raw, monophyletic to the ER6 reference strain of Rhizobium tropici CIAT899. In cluster 5, the three reference strains M. mediterraneus BR523, R. etli CFN42, and Rhizobium sp NGR234 were grouped with ten isolates. As in this study, the fragment used for amplification and subsequent ARDRA analysis of 16S rDNA was isolated and showed a great diversity and complexity of trees.
Conclusion
We can conclude that most of the isolates may be bacteria forming symbiosis with P. lunatus and have not been characterized genetically since, according Rosado et al. (1999), phylogenetically more distant species are the most isolated and using the 16S rDNA fragment may be useful for classification. The complexity of the phylogenetic trees indicates that the 16S rDNA gene studied in isolates proved to have genetic characteristics quite distinct from the bean culture. This may indicate that different soils and different geographic regions may have interfered with this diversity.
References
Azevedo JN, Franco JD, Araújo ROC (2003) Composição química de sete variedades de feijão-fava. Embrapa Meio-Norte, Teresina, p 4
Bohrer TRJ, Hungria M (1998) Avaliação de cultivares de soja quanto à fixação biológica do nitrogênio. Pesq Agropec Bras 33:937–952
Fofana B, Baudoin JP, Vekemans X, Debouck DG, Du Jardin P (1999a) Molecular evidence for an Andean origin and a secondary gene pool for the Lima bean (Phaseolus lunatus L.) using chloroplast DNA. Theor Appl Gen 98:202–212
Franco MC, Cassini STA, Oliveira VR, Vieira C, Tsai SM (2002) Nodulação em cultivares de feijão dos conjuntos gênicos andino e meso-americano. Pesq Agropec Bras 37:1145–1150
Freitas ADS, Vieira CL, Santos CEES, Stamford NP, Lyra MCCP (2007) Caracterização de rizóbios isolados de Jacatupé cultivado em solo salino do Estado de Pernambuco, Brasil. Bragantia 66:497–504
Gerin MAN, Feitosa CT, Rodrigues Filho FSO, Pereira JCVNA, Nogueira SSS, Igue T (1996) Adubação do amendoim (Arachis hypogaea L.) em área de reforma de canavial. Sci Agric 53:84–87
Giongo A, Ambrosini A, Vargas LK, Freire JRJ, Bodanese-Zanettini MH, Passaglia LMP (2008) Evaluation of genetic diversity of bradyrhizobia strains nodulating soybean [Glycine max (L.) Merrill] isolated from South Brazilian fields. Appl Soil Ecol 38:261–269
Grossman JM, Sheaffer C, Wyse D, Graham PH (2005) Characterization of slow-growing root nodule bacteria from Inga oerstediana in organic coffee agroecosystems in Chiapas, Mexico. Appl Soil Ecol 29:236–251
Gürtler V, WIlson AV, Mayall BC (1991) Classification of medically important clostridia using restristion endonuclease site differences of PCR-amplified 16S rDNA. J Gen Microbiol 137:2673–2679
Hartwig UA (1998) The regulation of symbiotic N2 fixation: a conceptual model of N feedback from the ecosystem to the gene expression level. Perspect Plant Ecol Evol Syst 1:92–120
Heyndrickx M, Vauterin L, Vandamme P, Kersters K, Vos P (1996) Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Methods 26:247–259
Hungria M (1993) Efeito das temperaturas elevadas na exsudação de compostos fenólicos que atuam como sinais moleculares na nodulação da soja e do feijoeiro. Rev Bras Fisio Veg 5:67–74
Hungria M (1994) Coleta de nódulos e isolamento de rizóbios. In: Hungria M, Araujo RS (eds).Manual de métodos empregados em estudos de microbiologia agrícola. EMBRAPA-SPI, Brasília, DF, pp 45–61
Jordan DC (1984) Family III Rhizobiaceae. CONN. 1938. In: Krieg NR (ed) Bergey’s manual of systematic bacteriology. Williams and Wilkins, Baltimore, pp 234–256
Loureiro MF, Kaschuk G, Alberton O, Hungria M (2007) Soybean [Glycine max (L.) Merrill] rhizobial diversity in Brazilian oxisols under various soil, cropping, and inoculation managements. Biol Fertil Soils 43:665–674
Mahdhi M, Nzoue A, Lajudie P, Mars M (2008) Characterization of root nodulating bacteria on Retama raetam in arid Tunisian soils. Progr Nat Sci 18:43–49
Martínez-Romero E (2003) Diversity of Rhizobium-Phaseolus vulgaris symbiosis: overview and perspectives. Plant Soil 252:11–23
Martínez-Romero E, Segovia L, Mercante FM, Franco AA, Graham P, Pardo MA (1991) Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int J System Evol Microbiol 41:417–426
Martins CM, Loureiro MF, Souto SM, Franco AA (1997) Caracterização morfológica e avaliação da tolerância em níveis crescentes de NaCl e altas temperaturas de isolados de rizóbio de nódulos de raiz e caule de Discolobium spp. nativas do Pantanal Mato-Grossense. Rev Agríc Tropic 3:7–14
Medeiros EV, Silva KJP, Martins CM, Borges WL (2007) Tolerância de bactérias fixadoras de nitrogênio provenientes de municípios do Rio Grande do Norte à temperatura e salinidade. Rev Biol Cien Terra 7:160–168
Melloni R, Moreira FMS, Nóbrega RSA, Siqueira JO (2006) Eficiência e diversidade fenotípica de bactériasdiazotróficas que nodulam caupi [Vigna unguiculata(L.) Walp] e feijoeiro (Phaseolus vulgaris L.) em solos de mineração de bauxita em reabilitação. Rev Bras Cien Solo 30:235–246
Musiyiwa K, Mpeperekia S, Giller KE (2005) Symbiotic effectiveness and host ranges of indigenous rhizobia nodulating promiscuous soybean varieties in Zimbabwean soils. Soil Biol Biochem 37:1169–1176
Ormeno-Orrillo E, Vinuesa P, Zúñiga-Davila D, Martinez-Romero E (2006) Molecular diversity of native bradyrhizobia isolated from (Phaseolus lunatus L.) in Peru. System Appl Microbiol 29:253–262
Ormeño-Orrillo E, Torres R, Mayo J, Rivas R, Peix A, Velázquez E, Zúniga D (2007) Phaseolus lunatus is nodulated by a phosphate solubilizing strain of Sinorhizobium meliloti in a Peruvian soil. In: Velázquez El, Rodríguez-Barrueco C (eds) Developments in Plant and Soil Sciences. Springer, Dordrecht, pp 243–247
Parker MA (2002) Bradyrhizobia from wild Phaseolus, Desmodium and Macroptilium species in Northern Mexico. Appl Environ Microbiol 68:2044–2048
Rosado AS, Duarte GF, Mendonça-Hagler LC (1999) A moderna microbiologia do solo: Aplicação de técnicas de biologia molecular. In: Siqueira JO, Moreira FMS, Lopes AS, Guilherme LRG, Faquin V, Frutini-Neto AE, Carvalho JG (eds) Inter-relação fertilidade, biologia do solo e nutrição de plantas. SBCS, Lavras: UFLA/DCS, Viçosa, pp 429–448
Santos CERES, Stamford NP, Neves MCP, Runjanek NG, Borges WL, Bezerra RV, Freitas ADS (2007) Diversidade de rizóbios capazes de nodular leguminosas tropicais. Rev Bras Cien Agr 2:249–256
Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32:33–57
Silva VN, Silva LESF, Figueiredo MVB, Carvalho FG, Silva MLRB, Silva AJN (2007) Caracterização e seleção de populações nativas de rizóbios de solo da região semi-árida de Pernambuco. Pesq Agropec Trop 37:6–21
Soares ALL, Pereira JPAR, Ferreira PAA, Vale HMM, Lima AS, Andrade MJB, Moreira FMS (2006) Agronomic efficiency of selected rhizobia strains and diversity of native nodulating populations in Perdões (MG, Brazil): I—cowpea. Rev Bras Cien Solo 30:795–802
Souza V, Eguiarte L, Ávila G, Capello R, Gallardo C, Montoya J, Pinero D (1994) Genetic structure of Rhizobium etli biovar phaseoli associated with wild and cultivated bean plants (Phaseolus vulgaris and Phaseolus coccineus) in Morelos, Mexico. Appl Environ Microbiol 60:1260–1268
Van Berkun P, Fuhrmann JJ, Eardly BD (2000) Phylogeny of Rhizobia. In: PEdrosa FO, Hungria M, Yates G, Newton WE (eds) Nitrogen fixation: from molecules to crop productivity. Kluwer, Dordrecht, pp 3–8
Vargas LK, Lisboa BB, Scholles D, Silveira JRP, Jung GC, Granada CE, Neves AG, Braga MM, Negreiros TA (2007) Diversidade genética e eficiência simbiótica de rizóbios noduladores de acácia-negra de solos do Rio Grande do Sul. Rev Bras Cien Solo 31:647–654
Vieira RF (1992) A cultura do feijão-fava. Inf Agropec 16:30–37
Vincent JM (1970) A manual for practical study of the root-nodule bacteria. Blackwell, Oxford
Wang ET, Martínez-Romero J, López I (2008) Rhizobium y su destacada simbiosis con plantas. Microbios en linea. Disponível em: <www.biblioweb.dgsca.unam.mx/libros/microbios/Cap8/>. Accessed 5 January 2008
Xavier GR, Martins LMV, Rumjanek NG, Ribeiro JRA (2006) Especificidade simbiótica entre rizóbios e acessos de caupi de diferentes nacionalidades. Caatinga 19:25–33
Zilli JE (2004) Assessment of cowpea rhizobium diversity in Cerrado areas of northeastern Brazil. Braz J Microbiol 35:281–287
Zilli JE, Valicheski RR, Rumjanek NG, Araújo JLS, Freire Filho FR, Neves MCP (2006) Eficiência simbiótica de estirpes de Bradyrhizobium isoladas do solo de cerrado em caupi. Pesq Agropec Bras 41:811–818
Acknowledgments
We acknowledge the National Council for Scientific and Technological Development (CNPq) for granting a Master scholarship to Jardel O. Santos. and funding the research project with P. lunatus. Ademir S. F. Araújo, Angela C. A. Lopes and Márcia V.B. Figueiredo are supported by personal grants from CNPq-Brazil.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Santos, J.O., Antunes, J.E.L., Araújo, A.S.F. et al. Genetic diversity among native isolates of rhizobia from Phaseolus lunatus . Ann Microbiol 61, 437–444 (2011). https://doi.org/10.1007/s13213-010-0156-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-010-0156-7