Abstract
Diseases caused by begomoviruses are becoming the major limiting factors for the production of watermelon in India. Survey for the incidence of plants showing symptoms typical to begomovirus infection was conducted in watermelon fields. The study revealed that 40% of the watermelon plants were showing the yellowing and downward curling symptoms. Twenty infected samples were collected from the different farmer’s fields to know the association of begomoviruses. The PCR amplification using begomovirus-specific primers resulted in an expected 1.2 kb PCR product indicating the begomovirus association with the watermelon samples. The sequence comparison results of 1.2 kb representing partial genome revealed that all sequences obtained from watermelon samples have a nucleotide (nt) identity of more than 98% among them and are maximum homology with Tomato leaf curl New Delhi virus (ToLCNDV). One watermelon sample (WM1) was selected for complete genome amplification using RCA method (rolling-circle amplification). Amplification of DNA B and no amplification of betasatellites and alphasatellite indicated this virus as bipartite. Sequence Demarcation Tool (SDT) analysis of the DNA A component of the WM1 isolate showed the maximum nt identity of 94.6–97.9% and 85.2–95.8% with ToLCNDV infecting cucurbits. The recombinant analysis showed that the genome was likely to be derived from the recombination of already reported begomoviruses (ToLCNDV, ToLCPalV, and MYMIV) infecting diverse crops. The whitefly cryptic species predominant in the begomovirus-infected watermelon fields were identified as Asia-II-5 group. The LAMP assay developed based on coat protein gene sequence was able to detect the ToLCNDV in the infected samples. Visual detection of the LAMP-amplified products was observed with the hydroxy naphthol blue. LAMP assay was also validated with ToLCNDV infected sponge gourd, spine gourd, ivy gourd, ridge gourd, and cucumber. This is the first report of ToLCNDV association with leaf curl and yellowing disease of watermelon from India and World based on complete genome sequencing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Watermelon (Citrullus lanatus (Thunb.) Mastum and Nakai, synonyms: C. vulgaris) originated from Africa is an economically important fruit crop belongs to the family Cucurbitaceae. The family Cucurbitaceae consists of 100 genera and over 750 species, distributed widely in different parts of the world (Yamaguchi 1983). Watermelon being cultivated in more than 96 countries in the world. China is a leading producer, contributing to more than 70.30% share of world production. In India, the crop is being cultivated in different states viz., Assam, Haryana, Himachal Pradesh, Orissa, Karnataka, Rajasthan, Punjab, Uttar Pradesh, Tamil Nadu, and West Bengal throughout the year. The area under watermelon production in India is 101 thousand hectares with the production of 2480 metric tons (National Horticulture Board 2017). The watermelon fruit is a good source of vitamin A and C and contains 92% water, 0.2% protein, 3.0% minerals, and 7.0% carbohydrates.
Watermelon productivity in India is low due to the attack of pests and diseases. Among these, plant viral diseases are major constraints in its production. Across the world, more than ten different viruses causing diseases and resulting in poor yield in watermelon were reported (Provvidenti 1986). Sometimes, it is difficult to find even a single plant free from infection at the end of the crop season and resulting in severe yield reduction (Varma and Giri 1995). An outbreak of diseases caused by begomoviruses is becoming a major threat for the production of many crops including watermelon due to their wide host range including many weed hosts, population explosion, and invasive nature of their vector Bemisia tabci and rapid evolution of their genome through recombination, acquisition of new DNA components, and satellites (Melgarejo et al. 2013). Important begomoviruses known to infect watermelon are Watermelon chlorotic stunt virus (WmCSV) (Jones et al. 1988), Squash leaf curl virus (SLCV) (Abudy et al. 2010), Cucurbit leaf crumple virus (CuLCrV) (Guzman et al. 2000), Melon chlorotic mosaic virus (MeCMV) (Romay et al. 2010), Tobacco curly shoot virus (TbCSV) (Zhao et al. 2017), and Chilli leaf curl virus (Shahid et al. 2017).
The begomoviruses belong to the family, Geminiviridae, and have isometric twin particle morphology with single-stranded DNA as genome. Based on the genome organization, the begomoviruses are categorized into monopartite viruses having genome homologous to DNA A of bipartite viruses and bipartite viruses having two genome components DNA A and DNA B (Zaidi et al. 2017). The genome organization of begomoviruses differs based on their geographical origin. The evidence available so far clearly shows that the Old World (OW) harbors both mono and bipartite begomoviruses with majority being monopartite, while the New World (NW) harbors only bipartite begomoviruses (Nawaz-ul-Rehman et al. 2012). Both have genome coding for capsid protein (CP), replication-associated protein (Rep), transactivator protein (TrAP), replication enhancer protein (REn), and C4/AC4 proteins, which are essential for multiplication and infection (Harrison and Robinson 1999; Gutierrez 1999). However, the genome component of monopartite begomoviruses (homolog of DNA A of bipartite viruses) is bigger than NW bipartite begomoviruses. The CP is required for systemic infection of monopartite begomoviruses and not for many bipartite begomoviruses (Sudarshana et al. 1998). Furthermore, the CP of NW bipartite begomoviruses has a distinctive N-terminal PWRsMaGT motif, which is absent in OW begomoviruses. The AC4 is a pathogenicity determinant in monopartite begomoviruses, whereas in many NW bipartite begomoviruses, it does not play any role in pathogenicity (Rojas et al. 2005; Pooma and Petty 1996). In addition to these genomic components, the monopartite viruses are known to be associated with subgenomic components, viz., betasatellite, alphasatellite, and deltasatellites (Briddon et al. 2002; Briddon and Stanley 2006; Fiallo-Olive et al. 2016). However, some recent reports revealed the association of satellites with the bipartite begomoviruses also (Venkataravanappa et al. 2019).
Among the begomoviruses reported across the world, ToLCNDV is emerging as a single major threat in the production of many crops in India (Brown et al. 2015) and drawn attention in Europe (Zaidi et al. 2017). The host range of ToLCNDV is expanding very quickly aided by its evolution to adopt new hosts (Juarez et al. 2019). The ToLCNDV is having genome typical to the bipartite begomovirus originated from the New World (Moriones et al. 2017). Even though ToLCNDV was reported to infect many crops (Brown et al. 2015; Fortes et al. 2016; Juarez et al. 2019), there are no reports available about complete genomes of ToLCNDV infecting watermelon from India.
Evidence based on the partial genome sequence showed an association of ToLCNDV with watermelon leaf curl from Pakistan (Mansoor et al. 2000) and India (Jyothsna et al. 2013). Apart from these, CP gene sequences of ToLCNDV (KM269350 and KM098111) associated with watermelon from Tamil Nadu and Punjab are available at NCBI database. For assigning the begomovirus species to a particular disease, full-length sequence of its genome is essential. In the present study, the association of ToLCNDV with leaf curl and yellowing disease of watermelon and its detection using LAMP assay are discussed.
Materials and methods
Survey for the incidence of leaf curl and yellowing disease in watermelon, collection of virus-infected watermelon samples, and whiteflies
During summer 2017–18, yellowing and downward curling symptoms on watermelon were observed in the farmer’s fields at different locations of Bengaluru rural district, Karnataka State, India. Ten fields [Hessaraghatta (13.1500°N, 77.4900°E), Doddaballapura (3.2957°N, 77.5364°E, 13.1870°N, 77.5502°E), Devanahalli, (13.2437°N, 77.7172°E), Hosakote (13.0693°N, 77.7982°E), Nelamanagala (13.0982°N, 77.3948°E), and Vijapura (13.2955°N, 77.8010°E)] were surveyed for the prevalence of the disease. The percent incidence was estimated in each field by counting the infected plants over the non-symptomatic plants. To know the status of begomovirus, totally 20 infected watermelon plant samples (two infected samples from each field) along the non-symptomatic samples (one from each field) were collected. Whitefly samples were also collected from each field, which is a natural vector of begomoviruses. Both plant and whitefly samples were subjected to the PCR analysis to know the virus associated with them. Each infected sample was designated as one virus isolate (WM-1 to 20).
DNA isolation, detection of virus, and identification vector
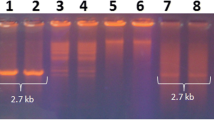
The total nucleic acid was isolated from the 20 symptomatic and non-symptomatic watermelon leaf samples by CTAB method (Doyle and Doyle 1990). The pellet was suspended in Tris–EDTA buffer. The quality of the nucleic acid was estimated on 1% agarose gel and stored at – 20 ℃ until further use. Initially, the begomovirus infection in watermelon samples was detected by Polymerase Chain Reaction (PCR) using begomovirus-specific primers pair OY2395F 5′-GCTCCCTGAATGTTCGGATGGA- 3′/OY680R 5′-GTTCTCRTCCATCCATATCTTAC-3′ designed to amplify genome segment of DNA A (Venkataravanappa et al. 2012). The partial amplified genome (1.2 kb) was cloned and sequenced by primer walking strategy (Medauxin PVT LTD, Bengaluru, India). One watermelon sample (WM-1) was processed for amplification of complete genome of the virus using the RCA method. The 50 ng of total nucleic acid was added to 5 µl of sample buffer and denatured at 95 °C for 3 min. Subsequently cooled down for 1 min on ice followed by the addition of 5 µl reaction buffer and 10U of enzyme mix. The amplification was carried by incubating samples at 30 °C for 20 h and stopped by incubating for 10 min at 65 °C. The amplified RCA product was digested with restriction endonucleases BamH1 and XbaI to release monomeric units of DNA A and DNA B, respectively. The monomeric units of the genomes were isolated. The 2.7-kb fragments represent complete genomic components of the virus were purified and ligated into a linearized pUC18 vector (BamH1 or XbaI) with the ligating enzyme as per the manufacturer’s instructions (Fermentas, Germany). The recombinant positive clones were identified by PCR amplification and restriction digestion. The five positive clones were selected and the viral genome insert was sequenced by primer walking strategy (Medauxin PVT LTD, Bengaluru, India). Furthermore, detection of betasatellite (DNAβ) and alphasatellite in the infected watermelon samples by PCR using beta (Briddon et al. 2002) and alphasatellite (Kumar et al. 2010) specific primers was carried out to identify their possible association with the disease.
Similarly, total genomic DNA isolated from the whiteflies collected from surveyed fields to know the status of cryptic species of B. tabaci present and detection of the begomovirus in the vector (Ashwathappa et al. 2020). PCR amplification using mtCOI gene-specific primers was carried out for cryptic species identification (Simon et al. 1994). For begomovirus identification of virus detection, PCR amplification was carried out as described by Venkataravanappa et al. (2017b). The amplified PCR products of mtCOI gene were cloned and sequenced. Genome sequences were analyzed as described below.
Sequence analysis
Similarity searches for the nucleotide (nt) sequence of DNA A and DNA B genome components were performed with all the sequences available in the GenBank database using BlastN (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences showing maximum identity score with watermelon isolate [DNA A (Table S1a) and DNA B (Table S1b)] were retrieved and aligned using the Muscle method implemented in Sequence Demarcation Tool (SDT) version 1.2 (Muhire et al. 2014). The retrieved sequences used in the analysis included, 21 Tomato leaf curl New Delhi virus (ToLCNDV) isolates, 8 Tomato leaf curl Palampur virus (ToLCPalV) isolates, 4 Squash leaf curl China virus (SLCCNV) isolates, and 2 Mungbean yellow mosaic Indian virus (MYMIV) isolates. The percent pairwise identities of the watermelon isolate with the representative sequences were calculated using SDT (Muhire et al. 2014). A phylogenetic tree was constructed by MEGA 7 (Kumar et al. 2016) using the Maximum-Likelihood Method with 1000 bootstrapped replications. Recombination analysis was carried out using the Recombination Detection Program (RDP), GENECOV, Bootscan, Max Chi, Chimaera, Si Scan, and 3Seq which are integrated in RDPv 4.46 (Martin et al. 2015). Default RDP4.46 settings were used throughout (P value cut-off = 0.05 and the standard Bonferroni correction). The sequences were considered as circular. Consensus daughters were found and breakpoints were polished. More than one method was considered to infer and evaluate the performance of these recombination detection events.
LAMP assay
Primer designing
The specific LAMP primers were designed based on the coat protein (AV1) gene sequence of ToLCNDV, which is highly conserved and essential for insect transmission using the Primer Explorer V.5 software (https://primerexplorer.jp/e) program. The specificity of the primers was confirmed with BLAST search (Standard nt BLAST available at https://blast.ncbi.nlm.nih/gov/blast.cgi) (Fig. 1). The primers were synthesized at Sigma-Aldrich (Table 1). A forward inner primer (FIP) consisted of the complementary sequence of F1c and F2, and a backward inner primer (BIP) consisted of B1c and B2. The outer primers F3 and B3 used for the initiation of the LAMP reaction.
PCR assay
The DNA amplification was performed with 35 cycles of denaturation for 1 min at 94 °C, primer annealing for 45 s at 55 °C, and primer extension for 1 min 30 s at 72 °C, with an initial denaturation at 94 °C for 3 min and a final extension for 10 min at 72 °C. The PCR reactions were carried out in a Gene Amp PCR system 9700 (PE Applied Biosystems, Foster City, CA) thermocycler. All amplifications were performed in volumes of 25 μl PCR mix containing 2 μl DNA template, 1.5 U Taq DNA polymerase (Fermentas Life Sciences, USA), 25 mM Mgcl2, 2 mM dNTPs (Fermentas Life Sciences, USA), and 10 pmol of each F3 and B3 primers. PCR products were electrophoresed (1 h at 80 V) on 1.2% agarose gels in Tris–borate-EDTA buffer, pH 8. Gels were stained with ethidium bromide (10 mg/ml) and viewed in Gel documentation system (Alpha Innotech, USA).
LAMP assay
The DNA from leaf samples (three symptomatic and one non-symptomatic) of watermelon weighing 5 mg each were extracted with 1 ml of extraction buffer (CTAB) using a pre-cooled mortar and pestle. LAMP assay was performed in a total volume 25 μl LAMP reaction mixture contains 0.5 μl (10 μM) of ToLCNDV-F3, ToLCNDV-B3, 1.0 μl of (10 μM) ToLCNDV-FIP, ToLCNDV-BIP, ToLCNDV-LF and ToLCNDV-LB primers, 1.5 μl of 10 mM dNTPs, 1.0 μl of 5 M betaine, 6.5 μl of sterile double-distilled water, 2.5 μl of 1X ThermoPol Reaction buffer (20 mM Tris–HCl, 10 mM (NH4)SO4, 10 mM KCl, 2 mM MgSO4, 144 0.1% Triton X-100, pH 8.8 @ 25 °C), 0.5 μl of 100 mM MgSO4, and 1.0 μl of 8U Bst DNA Polymerase (New England Biologicals, USA). The reaction mixture was incubated at 63 °C for 45 min followed by 80 °C for 10 min to terminate the reaction in water bath. The DNA from the healthy sample and distilled water were included as negative controls. The LAMP reaction products were analyzed on 1.5% agarose gel with 65 V for 3 h and then visualized using ethidium bromide stain viewed under Gel documentation system (Alpha Innotech, USA).
Visual detection
For the visual detection of the LAMP products, 1 μl (3 mM) of hydroxy naphthol blue (HNB) coloring agent (Lemongreen, Shanghai, China) was separately added to the LAMP master mix prior to amplification. All positive reactions can be easily identified visually by change in the color from violet to sky blue. Furthermore, the LAMP amplification was confirmed by running the product on 1.5% agarose gel stained with ethidium bromide.
Results
Symptomatology and incidence of leaf curl and yellowing disease in watermelon
Survey in the watermelon fields revealed watermelon plants exhibiting the symptoms, yellowing and downward curling, dwarfing of entire veins, and deformation of fruits, which are typical to the begomovirus infection (Fig. 2). The incidence of plants showing these symptoms was around 40% in the surveyed fields. Furthermore, a large number of whitefly (B. tabaci) infestation were noticed on both healthy and infected plants. Based on the occurrence of whiteflies and symptoms, it was suspected that the infected watermelon plants might be associated with the begomovirus and further analysis was carried out.
Begomovirus genome organization and sequence analysis
The PCR amplification of the virus genome using begomovirus-specific primers from the DNA samples extracted from the 20 infected watermelon leaf samples resulted in the expected amplicon of 1.2 Kb confirming the association of begomovirus. The sequence analysis of the amplicons comprising the partial genome showed that sequences derived from watermelon isolates (WM1-20) are having more than 98% nt identity between themselves and are related to Tomato leaf curl New Delhi virus (ToLCNDV).
Complete genome amplification of one virus isolate from watermelon (WM1) was done using RCA method and genome sequence data were obtained. The DNA A and DNA B components of watermelon (WM1) isolate were determined to be 2737 nt and 2692 nt in length, respectively. The sequences were submitted to the NCBI database (Accession no. MK087116 and MK087117). The attempts to amplify the subgenomic components, betasatellite (DNAβ) and alphasatellite, resulted in no amplification, indicating that the virus is not associated with the satellites. The organization of genome (DNA A and DNA B) of ToLCNDV associated with the watermelon leaf curl and yellowing disease was same as Old World bipartite begomoviruses containing four conserved ORFs: precoat protein (AV2, nt 121–459/coding 112 aa), coat protein (AV1, nt 281–1051/coding 256 aa) in sense orientation; and replication enhancer protein (AC3, nt 1048–1458/coding 136 aa), transcriptional activator protein (AC2, nt1178-1597/coding 139 aa), replication-associated protein (AC1, nt 1500–2585/coding 361 aa) and AC4 (nt 2135–2428/coding 97 aa) in the complementary sense strand. The sense and antisense of the viral strands are separated by a noncoding intergenic region (IR) between 2586 and 2737 and 1 and 120 nt. Which contained highly conserved nonanucleotide TAATATTAC sequence nicked by the rolling-circle initiator protein (Rep protein) for viral strand replication. The DNA B component is having two open-reading frames, one on sense strand BV1 (nuclear shuttle protein, nt 479–1285/ coding 268 aa) and the other on the antisense strand BC1 (movement protein, nt 1340–2185/coding 281 aa).
The complete genome of watermelon (WM1) isolate of begomovirus was compared with the sequences of other begomoviruses retrieved from the database using SDT (Table S1). The SDT analysis of the complete genome of ToLCNDV infecting watermelon shared the maximum nt identity of 94.6–97.9% with ToLCNDV infecting cucurbits, 88.3–89.5% with ToLCNDV infecting tomato and chilli, 88.3–88.8% with ToLCNDV infecting potato available in the NCBI database (Table S1, Table 2). While DNA B component showed the maximum level of nt identity of 85.2–95.8% with ToLCNDV infecting cucurbits, 86.2–93.9% with ToLCNDV infecting tomato and chilli, and 88.3–88.6% with ToLCNDV infecting potato (Table 3). The threshold value for begomoviruses classification was set at 91% nt sequence identity (Brown et al. 2015) and sequence data indicate that the watermelon isolate in the current study is an isolate of ToLCNDV. The phylogenetic analysis also revealed watermelon isolate closely clustered with ToLCNDV infecting cucurbits (Fig. 3a, b).
Dendrograms were constructed using the maximum-likelihood method. a DNA A and (b) DNA B sequences of begomovirus isolated from watermelon. The begomoviruses acronyms given are: tomato leaf curl New Delhi virus (ToLCNDV), tomato leaf curl Palampur virus (ToLCPalV), squash leaf curl China virus (SLCCNV), and mungbean yellow mosaic Indian virus (MYMIV)
Furthermore, the ORF wise sequence identities of ToLCNDV associated with watermelon leaf curl and yellowing disease at protein level showed the maximum aa sequence identities with ToLCNDV infecting cucurbits in all regions, viz., precoat protein (AV2), coat protein (CP), REn (C3), TrAP (C2), Rep (C1), and C4 regions of DNA A component (Table 2) and BC1 (movement protein) and BV1 (nuclear shuttle protein) of DNA B component. Among these, the maximum aa identity of 83.5–97.7% and 85.2–95.8% with the begomoviruses infecting tomato and cucurbits, respectively, in the BV1 and BC1 regions (Table 3). The length of intergenic region (IR) is 272 nt and has more homology with IRs of other bipartite begomoviruses reported so far. The IR has a highly conserved hairpin-like region containing nonanucleotide sequence (TAATATTAC), which initiate Rep virion-strand DNA replication. Furthermore, between the IR of DNA A and DNA B of watermelon isolate, 110 nt are common and the sequence identities between these two components are more than 89%.
Recombination analysis
The recombination breakpoint analysis indicates inter-specific recombination in genomes of ToLCNDV-WM1 infecting watermelon. A recombination breakpoint was detected at 1639 nt in DNA A of ToLCNDV-WM1 with major and minor parents resembling ToLCPalV (FJ931537) and ToLCNDV (AM747291) infecting pumpkin. The recombination breakpoints were detected at 126 and 1765 nt [regions of precoat protein (V2), coat protein (V1), and replication enhancer protein (C3)] with an average probability value of 3.53 × 10–30. Similarly, another breakpoint was detected at 633 nt in DNA A component of ToLCNDV-WM1 with major and minor parents resembling ToLCNDV (GQ865546, EF068246) infecting tomato. The recombination was predicted between 302 and 935 nt [(regions of precoat protein (V2), coat protein (V1)] with the probability value of 9.31 × 10–01. The overall results showed that more recombination in precoat protein (V2) and coat protein (V1) and replication enhancer protein (C3) of the DNA A component of the virus was most significant as the average P value is less than 0.05.
Furthermore, recombination fragment of 273 nt was detected in DNA B component of ToLCNDV-WM1 with the major and minor parents resembling ToLCPalV (FJ660425) and MYMIV (AY271894) infecting cucumber and green gram, respectively. The recombination was predicted between 509 and 782 nt [(regions of BV1 (nuclear shuttle protein)] with a probability value of 1.316 × 10–02. Another breakpoint at 1074 nt was detected in the DNA B component of ToLCNDV-WM1 with the major and minor parents resembling ToLCNDV (HM989846, AM778833) infecting cucurbits and the breakpoints were predicted between 1176 and 2250 nt [((regions of BC1 (movement protein))] with a probability value of 6.387 × 10–38. The overall results showed that more significant recombinations were observed in both the ORFs BV1 and BC1 of DNA B component of the virus as the average P value is less than 0.05.
Identification of cryptic species of B. tabaci from watermelon fields
PCR amplification using mtCOI gene-specific primers from the DNA isolated from B. tabaci adults (five samples) collected during survey resulted in the expected 600 nt in size amplicon. No amplification was observed in the negative control (Fig. 4). The amplified PCR products were cloned, sequenced, and used for B. tabaci identification (Frohlich et al. 1999; Dinsdale et al. 2010). The analysis showed that sequences obtained from all samples have more than 98% nt similarity among themselves. Therefore, one sample sequence was used for analysis (Acc. No. MK894588). The phylogenetic analysis showed that the mtCOI gene amplified from the whitefly collected on watermelon was closely clustered with whitefly belongs to the Asia-II-5 group (Fig. 5).
Phylogenetic tree showing the relationship of cryptic species of B. tabaci (Asia-II-5) sequence collected in this study to the Dinsdale et al. (2010) consensus sequences
Detection of begomovirus in whiteflies collected in watermelon field
DNA isolated from the whitefly samples (five) collected from watermelon resulted in 1.2 kb in size amplicon in the PCR carried out using begomovirus DNA A component-specific primers (Fig. 6). No amplification was noticed in the negative control. The sequences obtained from the amplicons were subjected to similarity check at the NCBI using the BLASTn. The analysis showed that the virus sequences amplified from the whitefly are having more than 97% nt similarity with ToLCNDV infecting cucurbits.
PCR assay for detection of begomovirus in watermelon using LAMP primers (ToLCNDV-F3 and ToLCNDV-B3)
For detection of ToLCNDV infecting watermelon, 100 ng of total DNA extracted from begomovirus-infected watermelon leaf sample was subjected to PCR using ToLCND-F3 and ToLCNDV-B3 primers as described above. PCR amplicon of 180 nt was obtained in infected samples, which was observed on agarose gel (Fig. 7c).
LAMP assay for detection of ToLCNDV in watermelon. Visualization: (a) On 1.5% agarose gel electrophoresis, (b) Hydroxy naphthol blue, (c) Detection of ToLCNDV by F3 and B3 LAMP primers. Lane M1: 1 kb DNA marker; Lane W: water control, H: healthy watermelon, Lanes 1-–3; ToLCNDV infected watermelon samples, (d) Detection of ToLCNDV infecting watermelon through PCR
LAMP assay
Attempt to detect ToLCNDV in the watermelon samples by LAMP assay using different LAMP primers specific to CP gene showed that ToLCNDV was successfully detected in the begomovirus-infected watermelon samples (Fig. 7a). No amplification was observed from the DNA of negative sample and water control. The LAMP amplicons (a ladder-like pattern) were observed on 1.5% agarose gel from positive samples, but not from the negative sample and water control. The positive reactions can be visualized directly by the naked eye with samples showing sky blue color pattern, whereas a violet color change is observed in negative and water control (Fig. 7b).
Validation of LAMP assay
DNA isolated from the ToLCNDV infected watermelon, sponge gourd, spine gourd, ivy gourd, ridge gourd, and cucumber was subjected to LAMP assay using LAMP primers. The leaf samples collected from the different cucurbit species infected with begomovirus and positive samples gave amplification in the LAMP assay with a ladder-like pattern, and no amplification in negative controls (Fig. 8a). Samples positive for amplification showed sky blue color pattern, whereas a violet color was observed in negative or healthy samples and water control (Fig. 8b). To confirm this, these samples (watermelon, sponge gourd, spine gourd, ivy gourd, ridge gourd, and cucumber) were subjected to the conventional PCR using DNA A component-specific primers of the begomovirus. The resulted PCR amplicon of 1.2 kb in size in all samples and no amplification was noticed in the negative control (Fig. 8c), supporting the accuracy of LAMP assay for the detection of plant viruses.
LAMP assay for detection of ToLCNDV in different cucurbits. Visualization: (a) On agarose gel, (b) Hydroxy naphthol blue. Lane M1 1 kb DNA marker, Lane W water control, H healthy watermelon, Lanes 1–6 ToLCNDV infected watermelon, sponge gourd, spine gourd, ivy gourd, ridge gourd and cucumber samples, (c) Detection of ToLCNDV infecting different cucurbits through PCR
Discussion
Around the world, more than ten viruses (includes RNA and DNA viruses) are reported on watermelon and are main constraints for the production of watermelon worldwide (Provvidenti 1986). The study revealed the prevalence of leaf curl and yellowing disease in all the watermelon fields surveyed. The leaf samples from watermelon plants showing the symptom of yellowing and downward curling collected from the farmer’s field were confirmed with the association of begomovirus by PCR analysis. Furthermore, the whole-genome sequence and phylogenetic analysis revealed that the virus isolate in this study is a variant of ToLCNDV as per the ICTV guidelines for species demarcation (Brown et al. 2015). ToLCNDV is the important emerging virus infecting many cultivated and non-cultivated plant species belongs to Solanaceae, Malvaceae, Cucurbitaceae, Euphorbiaceae, and Fabaceae families throughout the world and poses a serious threat for their cultivation (Zaidi et al. 2017; Fortes et al. 2016; Moriones et al. 2017; Juarez et al. 2019). The literature showed that ToLCNDV infects many cucurbits in India (Sohrab et al. 2003; Shorab et al. 2006; Tiwari et al. 2010; Patil et al. 2017; Venkataravanappa et al. 2017a, 2018, 2019a, b). One report from each from India (Jyothsna et al. 2013) and Pakistan (Mansoor et al. 2000) is available about ToLCNDV infecting watermelon, which was based on the partial genome information. This is the first report of ToLCNDV associated with the watermelon leaf curl and yellowing, adding to the list of host range of ToLCNDV based on complete genome sequencing.
Recombination among strains and/or species due to the high frequency of mixed infections under natural conditions is playing a critical role in the evolution of novel geminiviruses leading to new pathogenic phenotype, which are often more virulent than the parent species. The recombination analysis of ToLCNDV infecting watermelon showed that the DNA A component has obtained at least some of its sequences from ToLCNDV and ToLCPalV. The DNA B component might have been evolved from the recombination of two cucurbits infecting viruses (ToLCPalV & ToLCNDV) and one leguminous infecting virus (MYMIV), which are reported from different parts of India. Overall, the results of the recombination analysis, phylogenetic analysis, and the deletions/insertions within the genome suggest that isolate ToLCNDV evolved from ToLCNDV and ToLCPalV (the major parent) with some contribution from MYMIV genome components and adapted to the watermelon in India.
The whitefly is the only known vector transmitting the begomovirus in a circulative persistent manner. The transmission efficiency of the virus is depending on the type of cryptic species population in the field (Chowda-Reddy et al. 2012; Venkataravanappa et al. 2017b). In the present study, the cryptic species of whitefly associated with watermelon crop was identified as Asia II-5 group population based on mtCOI gene analysis. Furthermore, the begomovirus was detected in the whiteflies using the begomovirus-specific primers by PCR analysis. Even though MEAM species having more proven efficiency in transmission of begomoviruses was reported from several crops from this region and different parts of the world (Rekha et al. 2005; Shankarappa et al. 2007; Chowda-Reddy et al. 2012; Masood et al. 2017; Venkataravanappa et al. 2017b; Kanakala and Ghanim, 2019), there was no evidence in this regard in the present study. More sampling based on crop ecosystem may provide vital information on crop-specific association of whitefly cryptic species.
Detection of the virus in the infected plant sample is essential for the management and understanding the epidemiology of the disease. LAMP assay developed is able to detect ToLCNDV infection in watermelon and is more useful than the PCR in terms of simplicity, safety, time, cost, and being user friendly (Naganur et al. 2019; Panno et al. 2020). LAMP-positive amplicons were observed with the naked eye after the addition of fluorescent or metal dyes to the reaction tubes (Goto et al. 2009) overcoming the limitation of conventional PCR.
In conclusion, this is the first report of ToLCNDV associated with leaf curl and yellowing of watermelon from India based on the complete genome sequencing of the virus. The LAMP assay developed can be used for the detection of the ToLCNDV infecting the different crops.
References
Abudy A, Sufrin-Ringwald T, Dayan-Glick C, Guenoune-Gelbert D, Livneh O, Zaccai M, Lapidot M (2010) Watermelon chlorotic stunt and Squash leaf curl begomoviruses- New threats to cucurbit crops in the Middle East. Isr J Plant Sci 58:33–42
Ashwathappa KV, Venkataravanappa V, Reddy CNL, Krishna Reddy M (2020) Association of Tomato leaf curl New Delhi virus with mosaic and leaf curl disease of chrysanthemum and its whitefly cryptic species. Indian Phytopathol. https://doi.org/10.1007/s42360-020-00214-1
Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG (2002) Universal primers for the PCR- mediated amplification of DNA beta-a molecule associated with some monopartite begomoviruses. Mol Biotechnol 20:315–318
Briddon RW, Stanley J (2006) Subviral agents associated with plant single-stranded DNA viruses. Virology 344:198–210
Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Jose CF, Silva Fiallo-Olive E, Briddon RW, Hernandez-Zepeda C, Idris A, Malathi VG, Martin DP, Rivera-Bustamante R, Ueda S, Varsani A (2015) Revision of begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol 160(6):1593–1619
Chowda-Reddy RV, Kiran kumar M, Seal SE, Muniyappa V, Girish BV, Govindappa MR, Colvin J (2012) Bemisia tabaci phylogenetic groups in India and the relative transmission efficacy of Tomato leaf curl Bangalore virus by an indigenous and an exotic population. J Integr Agric 11(2): 235-248
Dinsdale A, Cook L, Riginos C, Buckley Y, De Barro PJ (2010) Refined global analysis of Bemisia tabaci (Gennadius) (Hemiptera: Sternorrhyncha: Aleyrodoidea) mitochondrial CO1 to identify species level genetic boundaries. Ann Entomol Soc Am 103:196–208
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fiallo-Olive E, Tovar R, Navas-Castillo J (2016) Deciphering the biology of deltasatellites from the New World:maintenance by New World begomoviruses and whitefly transmission. New Phytol 212:680–692
Fortes IM, Sanchez-Campos S, Fiallo-Olive E, Diaz-Pendón JA, Navas-Castillo J, Moriones E (2016) A novel strain of tomato leaf curl New Delhi virus has spread to the Mediterranean basin. Viruses 8(11):307
Frohlich DR, Torres-Jerez I, Bedford ID, Markham PG, Brown JK (1999) A phylogeographical analysis of Bemisia tabaci species complex based on mitochondrial DNA markers. Mol Ecol 8:1683–1691
Goto M, Honda E, Ogura A, Nomoto A, Hanaki K (2009) Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46:167–172
Gutierrez C (1999) Geminivirus DNA replication. Cell Mol Life Sci 56:313–329
Guzman P, Sudarshana MR, Seo YS, Rojas MR, Natwick E, Turini T, Mayberry K, Gilbertson RL (2000) A new bipartite geminivirus (Begomovirus) causing leaf curl and crumpling in cucurbits in the Imperial valley of California. Plant Dis 84:488
Harrison BD, Robinson DJ (1999) Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (begomoviruses). Annu Rev Phytopathol 37:369–398
Jones P, Sattar MHA, Al Kaff N (1988) The incidence of virus disease in watermelon and sweetmelon crops in the peoples democratic republic of Yemen and its impact on cropping policy. Ann Appl Biol 17:203–207
Juarez M, Rabadan MP, Martínez LD, Tayahi M, Grande-Perez A, Gomez P (2019) Natural hosts and genetic diversity of the emerging tomato leaf curl New Delhi virus in Spain. Front Microbiol 10:140
Jyothsna P, Haq QMI, Priyanka S, Sumiya KV, Praveen S, Ramaveer R, Briddon RW, Malathi VG (2013) Infection of tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. Appl Microbiol Biotechnol 97(12):5457–5471
Kanakala S, Ghanim M (2019) Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS ONE 14(3):e0213946
Kumar J, Kumar A, Roy JK, Tuli R, Khan JA (2010) Identification and molecular characterization of begomovirus and associated satellite DNA molecules infecting Cyamopsis tetragonoloba. Virus Genes 41:118–125
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Mansoor S, Khan SH, Hussain M, Mushtaq N, Zafar Y, Malik KA (2000) Evidence that watermelon leaf curl disease in Pakistan is associated with Tomato leaf curl virus-India, a Bipartite Begomovirus. Plant Dis 84:102
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. https://doi.org/10.1093/ve/vev003
Masood M, Amin I, Hassan I, Mansoor S, Brown JK, Briddon RW (2017) Diversity and distribution of cryptic species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex in Pakistan. J Econ Entomol 110(6):2295–3230
Melgarejo TA, Kon T, Rojas MR, Lenin PC, Zerbini FM, Gilbertsona RL (2013) Characterization of a New World Monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in Geminivirus evolution. J Virol 87:5397–5413
Moriones E, Praveen S, Chakraborty S (2017) Tomato leaf curl New Delhi virus: an emerging virus complex threatening vegetable and fiber crops. Viruses 9(10):264
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9(9):e108277. https://doi.org/10.1371/journal.pone.0108277
Naganur P, Premchand U. Shankarappa KS, Mesta RK, Manjunatha C, Patil CV (2019). Development of a loop-mediated isothermal amplification assay for detection of Tomato leaf curl New Delhi virus in ridge gourd [Luffa acutangula (L.) Roxb.]. Int J Curr Microbiol Appl Sci 8:2282–2295
National Horticulture Board (2017) Indian horticulture database 2017. NHB Department of Agriculture and Cooperation, Government of India, New Delhi
Nawaz-ul-Rehman MS, Briddon RW, Fauquet CM (2012) A Melting pot of Old World Begomoviruses and their satellites infecting a collection of Gossypium species in Pakistan. PLoS ONE 7(8):e40050
Panno S, Matic S, Tiberini A, Caruso AG, Bella P, Torta L, Stassi R, Davino S (2020) Loop mediated isothermal amplification: principles and applications in plant virology. Plants 9:461
Patil CV, Ramdas SV, Premchand U, Shankarappa KS (2017) Survey, symptomatology, transmission, host range and characterization of begomovirus associated with yellow mosaic disease of ridge gourd from southern India. Virus Dis 28(2):146–155
Pooma W, Petty ITD (1996) Tomato golden mosaic virus open reading frame AL4 is genetically distinct from its C4 analogue in monopartite geminiviruses. J Gen Virol 77:1947–1951
Provvidenti R (1986). Viral disease of cucurbits and source of resistance. Food and Fertilizer Technology Center Technical Bulletin No. 93.
Rekha AR, Maruthi MN, Muniyappa V, Colvin J (2005) Occurrence three genotypic clusters of B. tabaci and the rapid spread of B-biotype in south India. Entomol Exp Appl 117:221–233
Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43:361–394
Romay G, Chirinos D, Geraud-Pouey F, Desbiez C (2010) Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch Virol 155:1843–1847
Shahid MS, AL-Sadi AM, Briddon RW (2017). First report of Chilli leaf curl virus and tomato leaf curl betasatellite infecting watermelon (Citrullus lanatus) in Oman. Plant Disease, 101:1063
Shankarappa KS, Rangaswamy KT, Aswatha Narayana DS, Rekha AR, Raghavendra N, Lakshminarayana Reddy CN, Chancellor TCB, Maruthi MN (2007) Development of silverleaf assay, protein and nucleic acid-based diagnostic techniques for the quick and reliable detection and monitoring of biotype B of the whitefly, Bemisia tabaci (Gennadius). Bull Entomol Res 97:503–513
Shorab SS, Mandal B, Ali A, Varma A (2006) Molecular diagnosis of emerging begomovirus diseases in cucurbits occurring in northern India. Indian J Virol 17:88–95
Simon C, Frati F, Beckembach A, Crespi B, Liu B, Flook P (1994) Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701
Sohrab SS, Mandal B, Pant RP, Varma A (2003) First reports of association of Tomato leaf curl New Delhi virus with yellow mosaic disease of Luffa cylindrica in India. Plant Dis 87:1148
Sudarshana MR, Wang HL, Lucas WJ, Gilbertson RL (1998) Dynamics of bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol Plant Microbe Interact 11:277–291
Tiwari AK, Sharma PK, Khan MS, Snehi SK, Raj SK, Rao GP (2010) Molecular detection and identification of Tomato leaf curl New Delhi virus isolate causing yellow mosaic disease in bitter gourd (Momordica charantia), a medicinally important plant in India. Med Plants 2:117–123
Varma A, Giri BK (1995). Virus diseases of cucurbits in India. In: Nayar C, N.M, More TA (eds) Oxford and IBH Publishing House Private Limited, New Delhi.
Venkataravanappa V, Kodandaram MH, Reddy CNL, Shankarappa KS, Krishna Reddy M (2017b) Comparative transmission of Bhendi yellow vein mosaic virus by two cryptic species of the whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). 3 Biotech 7:331
Venkataravanappa V, Reddy CNL, Chauhan N, Bhardwaj DR, Krishna Reddy M (2017) Association of Tomato leaf curl New Delhi virus a bipartite begomovirus with mosaic disease of snake gourd in India. Int J Pure App Biosci 5(5):558–570
Venkataravanappa V, Reddy CNL, Jalali S, Krishna Reddy M (2012) Molecular characterization of distinct bipartite begomovirus infecting bhendi (Abelmoschus esculentus L.) in India. Virus Genes 44:522–535
Venkataravanappa V, Reddy CNL, Saha S, Shankarappa KS, Krishna Reddy M (2018) Detection and characterization of tomato leaf curl New Delhi virus association with mosaic disease of ivy gourd (Coccinia grandis (L.) Voigt) in North India. Arch Biol Sci 70(2):339–347
Venkataravanappa V, Reddy CNL, Shankarappa KS, Krishna Reddy M (2019) Association of Tomato leaf curl New Delhi virus, betasatellite, and alphasatellite with mosaic disease of spine gourd (Momordica dioica Roxb. Willd) in India. Iranian J Biotech 17(1):e2134
Venkataravanappa V, Reddy CNL, Shankarappa KS, Jayappa J, Pandey S, Krishna Reddy M (2019) Characterization of Tomato leaf curl New Delhi virus and DNA- satellites association with mosaic disease of cucumber. Int J Biotech Bioeng 5(6):93–109
Yamaguchi M (1983) World vegetables. AVI, Westport
Zaidi SSEA, Martin DP, Amin I, Farooq M, Mansoor S (2017) Tomato leaf curl New Delhi virus: a widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol Plant Pathol 18(7):901–911
Zhao LL, Ding M, Zhang XY, Yin YY, Li TT, Zhang ZK (2017) First report of tobacco curly shoot virus (TBCSV) and its associated satellites from watermelon in China. J Plant Pathol 99(3):761–764
Acknowledgements
The research was supported by the project “Consortium platform on Vaccines and diagnostics” (Centre Grant No. 16-10/PP/ICAR/17-18/21) funded by Indian Council of Agricultural Research, Government of India, New Delhi, India.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
V. Venkataravanappa and M. Krishna Reddy have received research grants from Indian Council of Agricultural Research, Government of India, New Delhi, India. C. N. Lakshminarayana Reddy, K. S. Shankarappa, and K. V. Ashwathappa are associated with compilation of manuscript. All the authors declare that they have no competing interests.
Research involving human and/or animal participants
This article does not contain any studies with human or animal subjects performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Venkataravanappa, V., Ashwathappa, K.V., Reddy, C.N.L. et al. Characterization of Tomato leaf curl New Delhi virus associated with leaf curl and yellowing disease of Watermelon and development of LAMP assay for its detection. 3 Biotech 10, 282 (2020). https://doi.org/10.1007/s13205-020-02245-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02245-x