Abstract
Banana bunchy top virus (BBTV) is a circular single-stranded DNA virus with multi-components. The knowledge about interaction between viral proteins and pathogenesis mechanism of BBTV remains unclear. In this study, the coat protein gene (CP, ORF 516 bp) and nuclear shuttle protein gene (NSP, ORF 465 bp) from BBTV B2 isolate of the Southeast-Asia group were cloned. The intracellular localization analysis showed the CP locates in the cell nucleus of tobacco cells, while the NSP distributes in the cell nucleus and cytoplasm. Co-localization analysis indicated the NSP itself does not change distribution, but CP re-distributes to the cell nucleus and cytoplasm, suggesting that NSP interacts with CP and re-locates the CP in the cell. The interaction between CP and NSP was further verified by co-immunoprecipitation (Co-IP) in tobacco protoplasts. The study will help us to understand the interaction between viral proteins and pathogenesis mechanism of BBTV in host plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana is one of the most important economic crops for developing countries in tropical and sub-tropical areas (Ghag and Ganapathi 2018). However, Banana bunchy top virus (BBTV), Cucumber mosaic virus (CMV) and Banana streak virus (BSV) are three major viral pathogens which have caused great economic losses in banana production (Kumar et al. 2015). The BBTV-induced banana bunchy top disease (BBTD) can cause whole plant dwarfing and inhibit the growth and development of host leading to no flowers and fruits (Yu and Liu 2011; Qazi 2016). BBTV belongs to the genus Babuvirus within the family Nanoviridae. Its genome contains at least six circular single-stranded DNA moleculars with 1.0–1.1 kb in size, namely DNA-R, -U3, -S, -M, -C and -N (Burns et al. 1995; Yu et al. 2012; Kumar et al. 2017). Some BBTV isolates may carry 1–3 distant satellite molecules which have similar function as DNA-R component (Stainton et al. 2016; Yu et al. 2011). Each component is encapsulated into an icosahedral particle of 17–20 nm that has no envelope (Gronenborn 2004). BBTV is transmitted by the banana aphid, Pentalonia nigronervosa (Watanabe and Bressan 2013; Bressan and Watanabe 2011).
DNA-S encodes a coat protein (CP) which encapsidates each DNA component inside (Wanitchakorn et al. 1997). Furthermore, CP is a viral silencing suppressor that inhibits the host resistance (Niu et al. 2009). DNA-N encodes a nuclear shuttle protein (NSP) which is predicted to be a virulence factor that suppresses transmembrane receptor kinase activity (Fontes 2004). Meanwhile, the NSP contains a “FNGSF” motif that inhibits the plant stress granules (SG) formation (Krapp et al. 2017). In vitro experiments showed that both CP and NSP proteins located in the cell cytoplasm and nucleus of a banana protoplast cell. Co-expression with movement protein (MP) revealed that MP is able to re-locate NSP or NSP–DNA complex around the cell periphery, but not for CP (Wanitchakorn et al. 2000).
Although some evidences revealed the partial functions of CP and NSP, the relationship between CP and NSP proteins are remaining unclear. In this study, intracellular localization and co-localization analyses of the CP and NSP proteins were investigated, and the interaction between the two proteins was also verified. This study extends the knowledge about interaction between BBTV proteins and the molecular mechanism of BBTV CP nuclear export.
Materials and methods
Materials and plasmids
Total DNA of BBTV-infected B2 sample (Haikou, China) was stored in Laboratory of Molecular Virology, Institute of Tropical Bioscience and Biotechnology (ITBB), Chinese Academy of Tropical Agricultural Sciences (CATAS). Wild-type Nicotiana benthamiana (N. benthamiana) (Ferox genus), GV1300-Flag and pRTL2-RFP vector were also kept at the above laboratory. The GV1300 plasmid (pCAMBIA1300: GFP) was provided by Professor Ming Peng at ITBB, CATAS (Li et al. 2018). Primers for amplifying the ORFs of CP and NSP were designed based on the nucleotide sequences of DNA-S (GenBank accession No. MG545612) and DNA-N (GenBank accession No. MG545615). All primers were purchased from Sangon Biotech (Shanghai, China) (Table 1).
Plasmids construction and intracellular localization of CP and NSP proteins
To obtain the recombinant plasmids GV1300-CP and GV1300-NSP, the ORF of CP gene was amplified using primers DNAS-SalI and DNAS-BamHI, while the ORF of NSP gene was amplified with the primers DNAN-SpeI and DNAN-BamHI. The amplified DNA fragments were gel-extracted using the DNA Gel Extraction Kit (Omega BioTek, Doraville, GA, USA) and subsequently cloned into plant expression vector GV1300 using T4 DNA ligase (Takara, Dalian, China). The ligated product was transformed into Escherichia coli (E. coli) Trans 5α competent cells (TransGen, Beijing, China) and three positive clones were randomly selected for bidirectional sequencing by primers 1300-F and 1300-R at Thermo Fisher (Guangzhou, China).
The plasmids GV1300, GV1300-CP and GV1300-NSP were separately transformed into Agrobacterium tumefaciens GV3101 competent cells by freeze–thaw method and positive clones were subsequently identified by PCR using primers 1300-F and 1300-R. Then, a rapid, transient expression method of fluorescent fusion proteins in tobacco plants was conducted according to the references (Sparkes et al. 2006; Li et al. 2018). To observe the intracellular localization of the fusion proteins, the transfected tobacco leaves were cut into pieces of 1 cm × 1 cm and fluorescence images were visualized at room temperature on a microscope (FluoView FV1000D IX81; Olympus, Tokyo, Japan) under 488 nm and 546 nm.
Co-localization of CP with NSP in tobacco cells
To obtain the recombinant plasmid GV1300-CP-RFP, the RFP (Red fluorescent protein gene) was amplified from pRTL2-RFP vector using primers RFP-BamHI and RFP-SacI. The GFP (Green fluorescent protein gene) in GV1300-CP was replaced by the RFP using BamHI and SacI, and GV1300-CP-RFP was constructed. The recombinant plasmid GV1300-CP-RFP was identified by bidirectional sequencing with primers 1300-F and 1300-R. The plasmids GV1300, GV1300-NSP and GV1300-CP-RFP were separately transformed into Agrobacterium tumefaciens GV3101 competent cells and the positive clones were subsequently identified by PCR. The tobacco leaves co-transfected by GV1300-NSP and GV1300-CP-RFP were described above. The images were also visualized on a microscope (FluoView FV1000D IX81; Olympus, Tokyo, Japan). The tobacco leaves co-transfected by GV1300 and GV1300-CP-RFP were used as negative control.
Identification of the interaction between CP and NSP by co-immunoprecipitation
To obtain the recombinant plasmid GV1300-GST-CP, GST and CP fragments were separately amplified using pair-primers GST-F/GSTDNAS-R and GSTDNAS-F/DNAS-BamHI. The two fragments were further used as templates for overlapping PCR by primers GST-F and DNAS-BamHI. The overlapped DNA fragment was gel-extracted and subsequently cloned into plant expression vector GV1300. In addition, to obtain the recombinant plasmid GV1300-Flag-NSP, the Flag sequence was added to the upstream of NSP gene using the primers DNAN-flagSpeI and DNAN-BamHI. The tobacco protoplasts were prepared according to Wu and Hanzawa (Wu and Hanzawa 2018). The eukaryotic expression plasmids GV1300-Flag-NSP and GV1300-GST-CP were co-transfected into tobacco protoplasts. The GV1300-Flag and GV1300-GST-CP vectors were used as the control.
After transfection for 72 h, the protoplasts were harvested for SDS–PAGE and western blot analysis (Yang et al. 2014). Collected cells were lysed in 2 ml lysis buffer, then centrifuged at 12 000 rpm at 4 °C for 10 min. The supernatant was saved as the input control. The remainder was incubated with GST (mouse monoclonal) antibody-conjugated magnetic beads (Takara) at 4 °C for 4 h. The beads were washed four times with 2 ml lysis buffer. The input and elution were analyzed by immunoblotting with anti-GST (1: 1000) and anti-Flag (1: 1000). Peroxidase-conjugated goat anti-mouse IgG (H + L) antibody was used as the secondary antibody. The signals were detected with chemiluminescent horseradish peroxidase (HRP) substrate (Millipore).
Results
Intracellular localization of CP and NSP proteins
The specific DNA bands with the expected sizes of CP and NSP were amplified (data not shown) and the two DNA fragments were used to construct the plasmids GV1300-CP and GV1300-NSP using Sal I and BamH I. To analyze the intracellular localization of CP and NSP proteins, the GV1300-CP or GV1300-NSP was transformed into tobacco leaves by Agrobacterium tumefaciens GV3101. The fluorescence signals were visualized by a microscopy at 72 h per injection (hpi). The results showed the green fluorescence from CP-GFP fusion protein presented in the cell nucleus, while the green fluorescence of NSP-GFP was observed both in the cell cytoplasm and nucleus. These results indicated that BBTV CP protein localizes in the cell nucleus but the NSP protein localizes in the cell cytoplasm and nucleus in tobacco cells (Fig. 1). As a control, the vector expressed only GFP protein was localized in the cell cytoplasm and nucleus.
Subcellular localization of GFP, CP-GFP and NSP-GFP in tobacco cells. Schematic diagram of GV1300-CP and GV1300-NSP plasmids construction (a) and Subcellular localization of GFP, CP-GFP and NSP-GFP in tobacco cells under the light of 488 nm and 546 nm (b). 35S, a constitutive promoter from the cauliflower mosaic virus; GFP, green fluorescent protein; NOS, nopaline synthase terminator; Bar represents 20 µm
Co-localization of CP with NSP
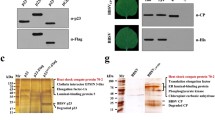
In order to assure the co-localization of CP and NSP in tobacco cells, the GV1300-CP-RFP and GV1300-NSP were co-transfected into tobacco leaves by Agrobacterium tumefaciens GV3101. At 72 hpi, green fluorescence of NSP-GFP fusion protein was observed in the cell cytoplasm and nucleus. Meanwhile, red fluorescence of CP-RFP was also observed in the cell cytoplasm and nucleus, but the most was present in the nucleus (Fig. 2a). The co-localization results indicated that the NSP itself does not change distribution, but affects the CP distribution in tobacco cells. To further assure the GFP does not affect CP distribution, GV1300-CP-RFP and GV1300 were co-transfected into tobacco leaves as a control group. The results showed that CP-RFP is localized in the cell nucleus, which is not affected by GFP protein (Fig. 2a). Therefore, these results indicated that NSP interacts with CP and re-locates the CP in tobacco cells.
Co-localization and CoIP analysis of CP and NSP in tobacco cells. Co-localization analysis of CP and NSP in tobacco cells (a). Tobacco cells transfected with plasmids GV1300-CP-RFP and GV1300-NSP were observed at 72 hpt. NSP-GFP fusion protein distributed in the cell cytoplasm and nucleus, while CP-RFP protein distributed in the cell cytoplasm and nucleus. The yellow color indicated the co-localization. GV1300-CP-RFP and GV1300 were used as a negative control. Bar represents 20 µm. Co-IP analysis of CP and NSP in tobacco protoplasts (b). Tobacco protoplasts were co-transfected with the plasmids GV1300-GST-CP and GV1300-Flag-NSP and collected at 72 hpt. GV1300-GST-CP and GV1300-Flag were used as a negative control. Cell lysates were subjected to immunoprecipitation using anti-GST (mouse monoclonal) magnetic beads. The whole cell lysates (Input) and the immuoprecipitation (IP) fractions were immuoblotted with indicated antibodies to detect their interactions
Confirmation of the interaction between CP and NSP by Co-IP
To further verify the interaction between the CP and NSP, the constructed plasmids GV1300-GST-CP and GV1300-Flag-NSP were co-transfected into tobacco protoplasts, while the plasmids GV1300-GST-CP and GV1300-FLAG co-transfection were used as a negative control. The results showed that GST-CP co-precipitates with Flag-NSP, but does not co-precipitate with Flag in tobacco protoplasts (Fig. 2b). Therefore, the interaction between CP and NSP was confirmed by Co-IP, suggesting NSP has ability to re-locate the CP distribution in tobacco cell.
Discussion
Previous study by Wanitchakorn et al. (Wanitchakorn et al. 2000) showed that both BBTV CP and NSP are located at the cell nucleus and cytoplasm in banana embryogenic cells. In this study, CP protein was located in the cell nucleus of tobacco cells, while NSP was distributed in the cell nucleus and cytoplasm. Although the localization of NSP in tobacco cells was similar to that of banana embryogenic cells, CP localization was different. The difference is probably caused by a single banana protoplast in vitro and a whole plant in vivo. Therefore, subcellular localization in intact plant leaf cells might be better to understand the function of viral proteins and their distributions. Furthermore, co-localization analysis indicated that the NSP itself does not change distribution in tobacco cells, but affects the CP distribution. By interaction with NSP, the CP re-distributes into the cytoplasm and nucleus, suggesting that NSP interacts and re-locates the CP in tobacco cells. The interaction was further verified by the method of co-immunoprecipitation (Co-IP) in tobacco protoplasts.
The product of the BBTV CP gene is an important structural protein that constitutes the virion. Through the nuclear pore (80–120 nm), CP protein goes into the cell nucleus to wrap the virus genome inside and complete the virus assembly. NSP-GFP fusion protein was distributed both in the cell nucleus and cytoplasm of tobacco cells. NSP is a viral nuclear shuttle protein, which would help viral proteins or viral nucleic acid transport outside of the cell nucleus by interaction with movement protein (MP) (Wang 2015). Meanwhile, MP can redirect the NSP protein or NSP–DNA complex into the cell periphery (Fig. 3) (Wanitchakorn et al. 2000). Geminivirus, also the single-stranded circular DNA virus, encode the BV1 protein (NSP) that mediates protein (or protein and DNA complex) from nuclear to cytoplasmic transportation (McGarry et al. 2003). Furthermore, geminiviruses NSPs promote the transportation of viral DNA between cells and cells by cooperation with movement protein. It is speculated that BBTV NSP protein has similar functions (Frischmuth et al. 2007). Therefore, BBTV CP and NSP would participate in viral protein (or viral protein and DNA complex) transportation and cell to cell infection, but further experiments should be verified. The study extends the knowledge about interaction between BBTV proteins and the molecular mechanism of BBTV CP nuclear export.
A schematic illustration for BBTV CP and NSP roles in virus assembly and infection. CP protein was located in the cell nucleus. The product of the BBTV CP gene is an important structural protein that constitutes the virion. Through the nuclear pore (80–120 nm), CP protein goes into the cell nucleus to wrap the virus genome inside and complete the virus assembly. NSP protein was distributed both in the cell nucleus and cytoplasm of the cell. NSP is a viral nuclear shuttle protein, which would help viral proteins or viral nucleic acid transport outside of the cell nucleus by interaction with MP (Wang 2015). Then the virion is formed and transports outside of cell nucleus. MP can redirect the NSP protein or NSP–DNA complex into the cell periphery (Wanitchakorn et al. 2000)
References
Bressan A, Watanabe S (2011) Immunofluorescence localisation of Banana bunchy top virus (family nanoviridae) within the aphid vector, pentalonia nigronervosa, suggests a virus tropism distinct from aphid-transmitted luteoviruses. Virus Res 155(2):520–525
Burns TM, Harding RM, Dale JL (1995) The genome organization of Banana bunchy top virus: analysis of six ssDNA components. J Gen Virol 76(6):1471–1482
Fontes EPB (2004) The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev 18(20):2545–2556
Frischmuth S, Wege C, Hülser D et al (2007) The movement protein bc1 promotes redirection of the nuclear shuttle protein bv1 of abutilon mosaic geminivirus to the plasma membrane in fission yeast. Protoplasma 230(1–2):117–123
Ghag SB, Ganapathi TR (2018) Banana and Plantains: Improvement, Nutrition, and Health. In: Mérillon JM, Ramawat K (eds) Bioactive Molecules in Food. Reference Series in Phytochemistry. Springer, Cham, pp 1–20
Gronenborn B (2004) Nanoviruses: genome organisation and protein function. Vet Microbiol 98(2):103–109
Krapp S, Greiner E, Amin B et al (2017) The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus Pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res 227:6–14
Kumar PL, Selvarajan R, Iskra-Caruana ML et al (2015) Chapter seven—biology, etiology, and control of virus diseases of banana and plantain. Adv Virus Res 91(1):229
Kumar P, Arun V, Lokeswari TS (2017) Cloning of BBTV (Banana bunchy top virus) components and screening of BBTV using functionalized gold nanoparticles. 3 Biotech 7(3):225
Li B, Yang Y, Luo Z et al (2018) Quantitative Screening of secretory protein genes in Candidatus Liberibacter Asiaticus. Am J Plant Sci 9(12):2408–2419
McGarry RC, Barron YD, Carvalho MF et al (2003) A novel arabidopsis acetyltransferase interacts with the geminivirus movement protein NSP. The Plant cell 15(7):1605
Niu S, Wang B, Guo X et al (2009) Identification of two RNA silencing suppressors from Banana bunchy top virus. Arch Virol 154(11):1775
Qazi J (2016) Banana bunchy top virus and the bunchy top disease. J Gen Plant Pathol 82(1):2–11
Sparkes IA, Runions J, Kearns A et al (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protocols 1(4) 2019
Stainton D, Martin DP, Collings DA et al (2016) Comparative analysis of common regions found in babuviruses and alphasatellite molecules. Arch Virol 162(3):1–7
Wang X (2015) Protein-protein interactions between NSP and proteins of Banana bunchy top virus. Dissertation, Hainan University
Wanitchakorn R, Harding RM, Dale JL (1997) Banana bunchy top virus DNA-3 encodes the viral coat protein. Arch Virol 142(8):1673–1680
Wanitchakorn R, Hafner GJ, Harding RM et al (2000) Functional analysis of proteins encoded by Banana bunchy top virus DNA-4 to -6. J Gen Virol 81(Pt 1):299–306
Watanabe S, Bressan A (2013) Tropism, compartmentalization and retention of Banana bunchy top virus (Nanoviridae) in the aphid vector Pentalonia Nigronervosa. J Gen Virol 94(Pt1):691–697
Wu F, Hanzawa Y (2018) A simple method for isolation of soybean protoplasts and application to transient gene expression analyses. J Visual Exp, 131, https://doi.org/10.3791/57258
Yang JW, Fu JX, Li J et al (2014) A novel co-immunoprecipitation protocol based on protoplast transient gene expression for studying protein–protein interactions in rice. Plant Mol Biol Rep 32(1):153–161
Yu NT, Liu ZX (2011) New research advance of Banana bunchy top virus. Microbiol China 38(3):396–404
Yu NT, Feng TC, Zhang YL et al (2011) Bioinformatic analysis of BBTV satellite DNA in Hainan. Virol Sinica 26(4):279–284
Yu NT, Zhang YL, Feng TC et al (2012) Cloning and sequence analysis of two Banana bunchy top virus genomes in Hainan. Virus Genes 44(3):488–494
Funding
This work was supported by the National Natural Science Foundation of China (31401709), the Hainan Provincial Natural Science Foundation (20153130) and the Young Elite Scientists Sponsorship Program by CSTC (CSTC-QN201704).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Rights and permissions
About this article
Cite this article
Ji, Xl., Yu, Nt., Qu, L. et al. Banana bunchy top virus (BBTV) nuclear shuttle protein interacts and re-distributes BBTV coat protein in Nicotiana benthamiana. 3 Biotech 9, 121 (2019). https://doi.org/10.1007/s13205-019-1656-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1656-1