Abstract
An Ochrobactrum anthropi bacterial strain named as NC-1, capable of utilizing phenmedipham (PMP) herbicide as the sole of carbon source and energy for growth was isolated from pesticide-contaminated soil sample by enrichment culture technique. The isolated bacterial strain was identified as Ochrobactrum anthropi NC-1 (MH 796134) based on its morphological, cultural, biochemical characteristics and analysis of 16S rRNA gene sequence. The strain NC-1 could degrade more than 98.5% of PMP (2 mM) within 168 h. The optimal degradation pH and temperature were 7.0 and 30–35 °C, respectively. The strain NC-1 degraded PMP by a pathway involving its initial hydrolysis of their central amide carbamate linkage to yield m-aminophenol via methyl-N-(3-hydroxyphenyl) carbamate and m-toluidine were the major intermediates. However, m-aminophenol was not further metabolized, because they neither supported the growth of organism nor stimulated oxygen uptake. But m-toluidine released by dealkylation was followed by hydrolysis. Further, results also revealed that degradation of 4-methyl catechol proceeded via 2-hydroxy-5-methyl-6-oxohexa-2, 4-dienoate through meta cleavage ring processes. The formation of these compounds was confirmed by UV, TLC, HPLC, IR, NMR, and GC–MS spectral analysis. The cell-free extracts of O. anthropi NC-1 grown on PMP contained the activities of PMP hydrolase, toluidine dioxygenase, and 4-methyl catechol 1, 2-dioxygenase. These results demonstrate the biodegradation of PMP and promote the potential use of strain NC-1 to bioremediate PMP-contaminated environment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Phenmedipham [methyl 3-(3-methylcarbaniloyloxy) carbanilate] (PMP), is a commonly used broad-spectrum phenyl carbamate herbicide which is being produced increasingly at a large scale, due to its agricultural benefits. It is selective systemic post-emergence herbicide has been widely used for weed control in sugar beet crops (Beta Vulgaris, L.), garden (table) beet, sunflower (Helianthus Annuus) and in spinach (Lewis et al. 2006; Tomlin 2006). Initially, PMP absorbed through leaves in the plants with translocation primarily in the apoplast to inhibit photosynthesis. PMP disrupts photosynthetic electron transport at the photosystem II (PS II) receptor site and is sensitive to environmental stress factors and for some xenobiotics, it may lead to a reduction in its activity (Ikeda et al. 2003). The electron transfer is negatively influenced and simultaneously the production of intermediary energy components such as ATP and NADPH2 and disrupt CO2 fixation. The mode of action is electron transfer blocking between the primary and secondary quenons (QA and QB) by binding to the D1 protein of PS II in chloroplast (Jursík et al. 2011). PMP is poorly soluble in water (1.8–7 mg l−1), slightly mobile in soil (adsorption coefficient = 147) and has a high potential for bioaccumulation (log P = 2.7) (Kegley et al. 2009). The U.S. Environmental Protection Agency (EPA) has classified phenmedipham and its metabolites as a persistent bioaccumulative toxic compound distributed in the environment (Edwards 2005). Phenmedipham has an oral LD50 of 0.8 g kg−1 in rats, which shows that RBCs is the primary target that results in hemolytic anemia. Chronic dosing resulted in a reproducible and dose-dependent and reduction in body weight (Jursík et al. 2011). However, its use may adversely affect imperiled species of terrestrial, semi-aquatic plants and invertebrates including mollusks, fishes, and birds (Kamrin 1997). Further, it is highly toxic to Vibrio fischeri bacteria, Pseudokirchneriella subcapitata, Chlorella vulgaris, Chlamydomonas pseudostate microalgae, Lamna minor macrophyte and Cladocerans (Daphnia Magna and longispina) species (Vidal et al. 2012). PMP has been reported to be degraded by Arthrobacter P52 and Pseudomonas putida strain (Sonawane and Knowles 1971; Pohlenz et al. 1992). These organisms were shown to degrade phenmedipham by hydrolysis of their central carbamate linkage to yield methyl-N-(3-hydroxyphenyl) carbamate (MHPC), m-aminophenol and m-toluidine (Perret et al. 2001). But its complete degradative pathway has not been studied. Thus, the widespread use and release of such toxic metabolites lead to environmental pollution and loss of soil fertility. This is a growing cause of global concern. Hence, sustainable agriculture has become a priority issue in the twenty-first century (FAO 2008) and the remediation of contaminated soils and aquatic bodies is essential for the protection of the environment and conservation of natural resources. Therefore, the microorganisms play a significant role in detoxifying the toxic herbicides and their products in the environment (Ishag et al. 2017). Thus microbial degradation is an efficient, eco-friendly, and economical method to decontaminate the polluted ecosystem. In this paper, we have described the isolation, identification, characterization of PMP degrading organism and also the process of investigation into the biodegradative pathway of phenmedipham through the meta-cleavage via 4-methylcatechol by O. anthropi NC-1.
Materials and methods
Chemicals
Phenmedipham, chlorpropham, desmedipham, 4-methyl catechol, m-aminophenol, m-toluidine, 4-chlorocatechol, catechol, protocatechuate, 2-aminobiphenyl, 3-chloroaniline, and 4-aminobiphenyl were purchased from Sigma Aldrich Chemical Co. Bangalore, India. Methanol, acetonitrile, ethyl acetate, acetone, chloroform, butanol, and n-hexane (HPLC grade) were purchased from Himedia, India. All other chemicals were of pure analytical grade or highest grade available commercially. Stock solution (2 mM) of PMP was prepared with chloroform and stored in brown bottles at − 20 °C before use.
Organism and growth conditions
The soil samples were collected from the pesticide-contaminated field, pH of the soil sample was 7–8, air dried and stored in a polythene bag at − 20 °C in the laboratory. The organism was isolated by selective enrichment culture technique on PMP as the sole source of carbon and energy. It was grown on mineral salts medium (Seubert’s 1960) contained phenmedipham (2 mM) in 500-ml Erlenmeyer flasks on a rotary shaker (150 rpm) at room temperature. The growth of the isolated microorganism was measured turbidimetrically at 600 nm using photoelectric colorimeter (Systronic, India). The identification of PMP degrading organism was done on the basis of its morphological, cultural, and biochemical characteristic (Holt et al. 1994). The cultures were maintained on mineral salts agar slants, containing 0.02% PMP. DNA isolation and determination of its G + C content was done according to Marmur (1961) and Mandel and Marmur (1968).
Media
The Seubert’s (1960) mineral salts medium (MSM) was used for the cultivation of O. anthropi strain. The medium consisted of the following components (g l−1): K2HPO4 6.30, KH2PO4 1.83, (NH4)2NO3 1.00, MgSO4.H2O 0.10, Na2MoO4.2H2O 0.005, MnSO4.H2O 0.1 and FeSO4.7H2O 0.10. The pH of the medium was adjusted to 7.0 before autoclaving at 120 °C for 20 min. Nutrient agar broth medium contained beef extract 0.3 g, peptone 1.0 g, agar 1.0–1.7 g, NaCl 0.5 g in 100 ml distilled water with pH 7.0.
Utilization of phenmedipham herbicide by O. anthropi NC-1
O. anthropi NC-1 utilized various herbicides such as phenmedipham, desmedipham, chlorpropham and other aromatic compounds such as m-toluidine and 4-methyl catechol as a sole source of carbon and energy was determined by the measurement of the growth in MSM contained 2 mM of the compounds. Utilization of the PMP during growth of the strain NC-1 was determined at regular intervals. Uninoculated controls were used to determine any transformation of PMP affected by physical factors. The effect of PMP concentration on the growth of O. anthropi NC-1 and the rate of degradation of PMP at different pH values (5.0–9.0) and temperatures (20–40 °C) were measured at different time intervals.
Identification of bacterial isolates by 16S rRNA gene analysis
The total genomic DNA was isolated from O. anthropi strain NC-1and amplification of 16S rRNA gene using specific primers 785F 5′-GGA TTA GAT ACC CTG GTA-3′ and 907R 5′-CCG TCA ATT CCT TTR AGT TT-3′ which yielded a product of approximately 1475 base pairs. The polymerase chain reaction (PCR) was carried out with forward and reverse primers using BDT V 3.1 cycle sequencing kit. The PCR conditions were 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 45 s and extension at 72 °C for 2.5 min. The PCR product was purified by PEG–NaCl precipitation (Sambrook et al. 1989). Briefly, the PCR product was mixed with 0.6 volumes of PEG–NaCl solution (20% PEG 6000, 2.5 M NaCl) and incubated for 10 min at 37 °C. The precipitate was collected by centrifugation at 12,000 r min−1 for 10 min. The pellet was washed twice with 70% ethanol and dried under vacuum, which was then resuspended in distilled water at a concentration of more than 0.1 pmol ml−1. The purified product was directly sequenced using a Big Dye Terminator kit (Applied Biosystems, Inc., Foster City, CA) (Pidiyar et al. 2004). The sequencing reactions were made to run on the ABI-PRISM automated sequencer (ABI-373xl genetic analyzer). The nucleotide sequence analysis was performed at the BLASTn program at the NCBI server (http://www.ncbi.nlm.nih.gov/BLAST). The alignment of the sequences was performed using CLUSTALW program VІ.82 at the European Bioinformatics site (http://www.ebi.ac.uk/clustalw). The analysis of the 16S rRNA gene sequence was performed at Ribosomal Database Project (RDP) IІ (http://rdp.cme.msu.edu) (Cole et al. 2004). The sequence was refined manually after cross-checking with raw data to remove ambiguities. The phylogenetic tree was constructed using the aligned sequences by the neighbor-joining (NJ) method by Kimura 2-parameter distances in the MEGA 7 software (Molecular Evolutionary Genetic Analysis version) and Molecular Biology and Evolution 30:2725–2729 (Tamura et al. 2013). The phylogenetic analysis of 16S rRNA gene sequence was performed in Euro fins Genomics Pvt. Ltd. Bangalore, India. The sequence of strain NC-1 was deposited in the GenBank sequence database.
Isolation and identification of metabolites
The metabolites were extracted from the culture filtrates of O. anthropi NC-1 after 96 and 144 h grown on 2 mM of PMP with ethyl acetate, acidification to pH 2.0 with 2M HCl. The residues were dissolved in methanol and analyzed for metabolites by thin layer chromatography (TLC) on silica gel G plates using the following solvent systems. (A) chloroform–acetonitrile (4:1 v/v), (B) benzene–methanol–acetic acid (45:8:4 v/v) and (C) chloroform–acetic acid (95:5 v/v).
The metabolites were visualized under UV light at 254 nm or by exposure to iodine vapors and also by spraying with 1% FeCl3–K3Fe(CN)6 solution in water. Aromatic compounds gave their characteristic color on spraying with diazotized p-nitroaniline or with Gibbs reagent (2% solution of 2, 6-dichloroquinone-4-chlorimide in methanol). o-Dihydroxy compounds were detected by spraying with Arnow’s reagent (1937). Primary amines such as m-aminophenol and m-toluidine were detected by spraying orcinol reagent on the TLC plate to form yellow colored derivatives (Janghel et al. 2005). Phenolic compounds gave blue color on spraying with Folin–Ciocalteu’s phenol reagent and bluish-green color with 100 mM FeCl3, which turned red on exposure to ammonia.
PMP concentrations were determined by High-Performance Liquid Chromatography (HPLC) as reported by Loncar et al. (2004). At regular intervals, 5 ml samples were withdrawn and centrifuged at 10,000 rpm for 10 min. The supernatants were extracted with ethyl acetate and the residue obtained after evaporation (Buchi Rotavapor R 210) was dissolved in methanol (HPLC grade) and used for HPLC analysis. 10 µl of each residual samples of PMP were analyzed by HPLC (Shimadzu, Japan) equipped with UV detector using 5 µm Spherisorb-ODS (C18) column (25 cm × 4.6 mm) acetonitrile–phosphate buffer (0.05M, pH 7.0) in the ratio 70:30 (vol/vol) as the mobile phase at a flow rate of 1 ml per min. Peaks were detected at 235 nm or scanned 220–400 nm in a spectrophotometer. The retention time of PMP under the condition tested was 7.68 min. The absorbance of the substrate (PMP) was measured at 235 nm. The mass spectra were recorded with gas chromatography–mass spectra (GC–MS) Shimadzu QP 2010S spectrometer. The temperature programme for the experimentation was held at 50 °C for one minute with 20 °C increase per minute to a final temperature at 28 °C for 14.5 min and the injector temperature was kept at 250 °C. The injection volume was 1 µl and helium as carrier gas. The mass spectrometer was operated at electron ionization energy of 70 eV. Nuclear magnetic resonance spectra (NMR) of metabolites were recorded using 400 MHz spectrometers (JNM-ECZ 400S, 2017 Model) with tetramethylsilane (TMS) as the internal standard at room temperature. NMR spectra were done in SAIF, NMR Research Centre, Indian Institute of Science, Bangalore, India. UV–visible absorbance spectra were recorded with the Hitachi 3100 spectrophotometer. Fourier transmission-infrared spectrometer (IR) (Nicolet Impact-410, USA) was recorded in the range 4000–400 cm−1. Ammonium ions released during PMP degradation were assayed according to the method of Park et al. (2009). Nessler’s test for ammonium ions and lime water test for CO2 were carried out according to standard protocols (Vogel and Svehla 1996).
Atomic force microscopy (AFM)
The isolated O. anthropi NC-1 was grown on nutrient agar media at 30–35 °C. A layer of the strain was clearly seen on the surface of the nutrient agar plates. Prior to imaging, cultures were gently scratched on the agar surface with a metallic wire loop and dispersed in deionized water. The sample was centrifuged at 10,000 rpm for 12 min at 4 °C to harvest bacterial pellets. The pellets were repeatedly washed 3–4 times with deionized water to remove culturally bound salts that may interfere with bacterial cell binding. The suspension was filtered with the help of syringe fitted with glass wool. The final purified pellet was resuspended in deionized water. A 10 µl of 10−2 mol polylysine solution was applied to a freshly cleaved mica surface and left to dry for 5 min. 5 µl of strain NC-1 cell suspension was immobilized on a polylysine-coated mica surface and allowed to dry under laminar air flow cabinet. The air-dried slides were subjected to contact and tapping modes of operation using Nanosurf Easy scan Atomic Force Microscope (Nanosurf Easy scan 2 AFM, Switzerland) system in applied force with air mode (Greif et al. 2010).
Enzyme assay
Cell-free extracts were prepared from the washed cells suspended in three volumes 50 mM phosphate buffer, pH 8.0 by sonication (Ultrasonic processor model XL 2010) for 5 min and centrifugation at 10,000 rpm for 35 min at 4 °C. The clear supernatant was used as a crude extract for enzyme assays. PMP hydrolase activity was assayed spectrophotometrically by measuring the increase in absorbance at 237 nm due to the disappearance of the substrate PMP. The whole cells of cell-free extract of strain NC-1 grown on PMP contained the activities of PMP hydrolase, toluidine dioxygenase, and 4-methylcatechol 1, 2 dioxygenase (Williams and Murray 1974). Glucose-grown cells did not contain any of these enzyme activities. Protein concentration was determined using the Lowry method (1951) using bovine serum albumin (BSA) as standard. Protein samples were standardized to 0.65 mg per ml. One unit of enzyme activity was defined as the amount required to catalyze the formation or consumption of 1 µmol of product or substrate per min. Specific activity was expressed as units per mg of proteins.
Statistical analysis
All experiments were carried out in three independent replicates. Analysis of data was done by one-way ANOVA variance SPSS version 7.5 software and data were denoted as means ± standard errors. The difference between each means was located using Duncan’s multiple range tests (Duncan 1955).
Results
Characterization of organism able to metabolize PMP
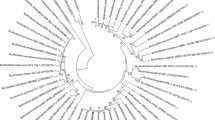
Phenmedipham degrading strain NC-1 is aerobic, Gram-negative, rod-shaped, motile by means of peritrichous flagella that was catalase, urease, and oxidase positive. The organism reduced nitrate, hydrolyzed gelatine, starch test were positive and there is no utilization of citrate. There was no hydrolysis of casein and no DNase activities. According to Bergey’s manual of determinative bacteriology, NC-1 strain belonged to genus Ochrobactrum anthropi. However, this genus was further identified to species level by 16S rRNA gene sequence analysis. The nucleotide sequence of 16S rRNA of NC-1 (1475 bp) was submitted to GenBank (Accession Number MH 796134). The isolated strain has been clustered with O. anthropi strain NC-1 as shown in Fig. 1. The G + C content of DNA from the bacterial strain was found to be 56.22 mol%. Atomic force microscopy (AFM) is only the technique that can image the surface of a living cell at high resolution. The average width and length of the O. anthropi strain NC-1 were shown to be 235 nm and 117.20 nm.
Phylogenetic relationships established by the neighbor-joining method (Felsenstein 1985; Saitou and Nei 1987) based on 16S rRNA gene sequence (1475 bp) of Ochrobactrum anthropi NC-1 strain (GenBank accession No. MH 796734) and its number of nucleotide changes/sequence positions (Kimura 1980). The number of nodes shows the bootstrap values obtained with 1,000 resampling analysis
Utilization of PMP and other compounds by O. anthropi NC-1
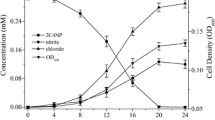
The strain NC-1 utilized herbicide PMP and other compounds as a sole source of carbon and energy for its growth. The growth of NC-1 strain in the mineral salts medium contained 2 mM of PMP as shown in Fig. 2. The cells of the strain NC-1 were able to utilize desmedipham, m-toluidine, catechol and 4-methyl catechol as growth substrates (Table 1). However, 4-aminobiphenyl, 3-chloroaniline, m-aminophenol and 3-aminobenzoic acid did not serve as a sole source of carbon for their growth of the organism.
Utilization of PMP (red square) during growth (orange triangle) of O. anthropi NC-1 and release of ammonium ions (green diamond) in the culture supernatant. Uninoculated controls (blue circle) in the mineral salts medium contained 2 mM PMP. Data values represent the average of three replicate determinations
Effect of pH and temperature on PMP degradation
The effect of pH and temperature on the degradation of PMP in MSM has been shown in Fig. 3a, b. In the pH range of 5.0–9.0, the degradation rate of PMP increases with the increase of pH of the medium and the optimum degradation rate of about 98.5% was achieved at pH 7.0. This indicates that strain NC-1 degraded more efficiently under neutral conditions. However, at lower (5.0) and higher (9.0) values of pH, it impeded the degradation rate (25.5 and 32%) of PMP. In addition, the degradation rate gradually increases in the temperature range of 20–35 °C and the highest degradation rate has been achieved (98.2%) at 30–35 °C. At 40 °C, the degradation rate drastically decreases to less than 18%. All the values that represent the mean ± standard error (SE) for each experiment are significantly different (P ≤ 0.05) from others according to Duncan’s test. These results indicate that the optimum pH and temperature for the growth and degradation of PMP by the strain NC-1 is 7.0 and 30–35 °C.
Identification of metabolites
The analysis of culture extracts of Ochrobactrum anthropi strain NC-1 grown on PMP by TLC revealed the presence of three metabolites I, II, and III. The Rf and λmax values of isolated metabolites corresponded well with that of authentic m-aminophenol, m-toluidine, and 4-methylcatechol (Table 2). These metabolites were purified by preparative TLC and analyzed by HPLC. Culture filtrates were collected at regular intervals of time (24, 48, 72, 96, 120, 144 and 168 h) and analyzed by HPLC to detect their retention time. In 72 h sample, a single peak was detected with a retention time of 7.68 min corresponded to PMP and decreased over a time and completely disappeared within 168 h. In 96 h sample, two metabolite peaks were observed with a retention time of 0.77 and 2.56 min corresponded to m-toluidine and m-aminophenol. After 144 h incubation of sample, another metabolite peak was observed with retention time of 1.43 min corresponded to that of authentic 4-methyl catechol as shown in Fig. 4a, b.
The mass spectrum of isolated metabolite I (Fig. 5) showed molecular peak M+ at m/z 109 is in good agreement with empirical formulae C6H7NO. The main mass fragments observed at m/z 41, 53, 63, 80, 91 and 109 were in identical with that of m-aminophenol. The IR spectrum of metabolite I showed characteristic absorption bands of –NH stretching at 3360–3296 cm−1, –CH stretching at 2928 cm−1, C=C stretching at 1493–1596 cm−1, –OH stretching at 3360 cm−1and C–N stretching at 1250 cm−1. The NMR of metabolite I showed deshielded region at δ 8.80 ppm, one singlet corresponds to –OH, followed by another singlet at δ 6.80 ppm assigned for –C2–H. Whereas one triplet was observed for –C2–H at δ 6.775 ppm (J = 6.2 Hz) and two protons at 6.438 (Jo&p = 7.6 and 3.2 Hz) and 6.398 (Jo&p = 7.2 and 4.0 Hz) ppm as doublets of doublets. –NH2 protons appeared at δ 4.85 ppm (J = 4.4 Hz). Thus the isolated metabolite of PMP was identified as m-aminophenol.
The mass spectrum of isolated metabolite II (Fig. 6) showed molecular peak M+ at m/z 107, is in good agreement with empirical formulae C7H9N. The main mass fragments observed at m/z 41, 43, 57, 73, 97 and 107 were in identical with that of m-toluidine. The IR spectrum of metabolite II showed characteristic absorption bands of –NH stretching at 3430–3360 cm−1, –CH stretching at 2921 cm−1, C=C stretching at 1493 cm−1. The NMR spectrum of metabolite II, –C5–H as a triplet at δ 7.05 ppm (J = 7.2 Hz), one proton at δ 6.78 ppm as a doublet for –C4–H, one proton at δ 6.62 ppm for –C2–H. One more doublet appeared at 6.3 ppm (J = 8 Hz) was assigned to –C6–H. –NH2 resonated at δ 3.85 ppm with J = 4.8 Hz. Finally, in the shielded region at δ 2.13 ppm for –CH3 were assigned. Thus the isolated metabolite of PMP was identified as m-toluidine.
The mass spectrum of isolated metabolite III (Fig. 7) showed molecular peak M+ at m/z 124, is in good agreement with empirical formulae C7H8O2. The main mass fragments observed at m/z 41, 51, 78, 106 and 124 were in identical with that of 4-methyl catechol. The IR spectrum of metabolite III showed characteristic absorption bands of –OH stretching at 3413–3340 cm−1,–CH stretching at 2925–2858 cm−1, C=C stretching at 1508 cm−1 and C=C stretching at 1508. The NMR spectrum of metabolite III, one proton at δ 6.82 ppm (J = 8 Hz) for –C6–H, –C3–H at δ 6.63 ppm and one doublet at δ 6.59 ppm (J = 8 Hz) assigned to –C5–H. One broad singlet peak appeared at δ 5.83 ppm which was assigned to two –OH protons. Finally, one peak at δ 2.19 ppm to –CH3 was observed. Thus the isolated metabolite of PMP was identified as 4-methyl catechol. During the degradation of 4-methylcatechol by meta ring cleavage, yellow transient intermediate accumulated in the culture supernatant. It showed a λmax at 382 nm and was identified as 2-hydroxy-5-methyl-6-oxohexa-2, 4-dienoate (McClure and Venables 1986). AFM technique provides a three-dimensional image of O. anthropi NC-1 under physiological conditions was found to be rod-shaped with an average length of 235 nm and a height of 114 nm (Fig. 8).
The release of ammonium ions
There was the release of ammonium ions in the culture medium during the degradation of PMP by strain NC-1. About 0.12 mM of ammonium ions were released after 120 h of incubation during the breakdown of m-toluidine by toluidine dioxygenase to 4-methylcatechol through oxidative deamination process as shown in Fig. 2. However, Nessler’s reagent and lime water tests were positive for NH4+ ions and CO2.
Enzymatic activities in cell-free extracts
In the crude extracts of PMP-induced cells of strain NC-1, we have detected the enzyme activities of PMP hydrolase, m-toluidine dioxygenase and 4-methyl catechol 1, 2-dioxygenase activity, but not 4-methylcatechol 2, 3-dioxygenase activity. These results have stipulated that the degradative enzymes were induced by growth of the organism on PMP, but not by glucose-grown cells. Specific activities (Table 3) of the enzymes are the mean ± SD (standard deviation) of assays from triplicate cell-free extracts.
Discussion
The bacterial strain Ochrobactrum anthropi NC-1 utilized the herbicide phenmedipham (2 mM) as the sole source of carbon and energy. AFM technique gave topography of the living surface cell of NC-1 strain. PMP is the most toxic ingredient, reached the aquatic compartment and bioaccumulation in biota with repercussions for the ecosystem. It also acts by inhibition of photosynthesis at photosystem II, which is an early target site for this herbicide (Casida 2009). The photosynthetic inhibitor PMP binds to D-1, quinone-binding proteins blocking the photosynthetic electron transport system in plants (Duke 1997) and photosynthetic organisms. The strain NC-1 degraded the herbicide PMP by initial hydrolysis of central carbamate ester linkages followed by dealkylation to yield m-aminophenol and m-toluidine as major intermediates. The other product of PMP hydrolysis was carbon dioxide. Due to the hydrolysis of PMP by PMP hydrolase, it destroys the herbicidal activity leading to detoxification. However, m-aminophenol formed by hydrolysis via methyl-N-(3-hydroxyphenyl) carbamate (MHPC) was not further metabolized, because of it neither supported the growth of the organism nor stimulated oxygen uptake. There are reports of formation of m-aminophenol and m-toluidine by the hydrolysis of PMP (Kossmann 1970; Knowles and Benezet 1981). Sonawane and Knowles (1971) reported that MHPC subsequently decomposes into m-aminophenol and the presence of PMP and its metabolites in the soil were determined using LCMS (Perret et al. 2001). But the further degradation of its hydrolyzed product by microorganism has not been studied. However, m-toluidine was further metabolized by the isolated NC-1 strain with the formation of an intermediate 4-methylcatechol. Toluidine dioxygenase catalysis the utilization of molecular oxygen and simultaneously removes the amino group present in m-toluidine by oxidative deamination process. During this process, 0.12 mM of ammonia was released. Similar results were reported in Pseudomonas testosterone and were shown to degrade m-toluidine via 4-methyl catechol (Raabe et al. 1984) followed by meta cleavage pathway. High activity of 4-methylcatechol 1,2-dioxygenase in the cell-free extract of strain NC-1 grown on PMP suggested that 4-methylcatechol was further oxidized through meta cleavage product of methylated derivative of 2-hydroxy semialdehyde. Pseudomonas picketti pk01 was also degraded 4-methylcatechol via meta-cleavage pathway reported by Kukor and Olsen (1991). However, further metabolism of 4-methyl catechol by NC-1 strain, yellow transient intermediate accumulated in the culture medium. The yellow intermediate showed an absorption maximum at 382 nm and was identified as 2-hydroxy-5-methyl-6-oxohexa-2, 4-dienoate (McClure and Venables 1986) and entered into the TCA cycle. The enzyme was induced by the growth of the organism on PMP, but a cell-free extract of glucose-grown cells did not contain any of these activities. Thus there was a complete degradation of PMP by O. anthropi NC-1 as shown in Fig. 9. Such bacterial strains could be potentially useful in the bioremediation of soil and water contaminated (Ishag et al. 2017) with toxic phenyl carbamate herbicides. It may have an adverse effect on soil microflora especially on microorganisms that are involved in nitrogen cycling of agricultural soils.
References
Arnow LE (1937) Colorimetric determination of the components of 3, 4-dihydroxyphenylalanine-tyrosine mixtures. J Biol Chem 118:531–537
Casida JE (2009) Pest toxicology: the primary mechanisms of pesticide action. Chem Res Toxicol 22:609–619. https://doi.org/10.1021/tx8004949
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM (2004) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33: D294–D296. https://doi.org/10.1093/nar/gki038
Duke SO (1997) Overview of herbicide mechanisms of action. Environ Health Perspect 87:263–271
Duncan DB (1955) Multiple Range and Multiple F Tests. Biometrics 11:1. https://doi.org/10.2307/3001478
Edwards D (2005) Reregistration eligibility decision (RED) for phenmedipham list A case no. 0277. 89
FAO (2008) Biofuels: prospects, risks and opportunities. FAO, Rome
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Greif D, Wesner D, Regtmeier J, Anselmetti D (2010) High resolution imaging of surface patterns of single bacterial cells. Ultramicroscopy 110:1290–1296. https://doi.org/10.1016/j.ultramic.2010.06.004
Holt JG, Sneath PH, Krieg NR, Staley JT (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams and Wilkins, Baltimore
Ikeda Y, Ohki S, Koizumi K (2003) Binding site of novel 2-benzylamino-4-methyl-6-trifluoromethyl-1,3,5-triazine herbicides in the D1 protein of photosystem II. Photosynth Res 77:35–43. https://doi.org/10.1023/A:1024982804013
Ishag AESA, Abdelbagi AO, Hammad AMA, Elsheikh EAE, Elsaid OE, Hur JH (2017) Biodegradation of endosulfan and pendimethalin by three strains of bacteria isolated from pesticides-polluted soils in the Sudan. Appl Biol Chem 60(3):287–297. https://doi.org/10.1007/s13765-017-0281-0
Janghel EK, Rai JK, Rai MK, Gupta VK (2005) New analytical technique for the simultaneous determination of aromatic amines in environmental samples. J Sci Ind Res 64:594–597
Jursík M, Soukup J, Venclová V, Holec J (2011) Post herbicide combinations for velvetleaf (Abutilon theophrasti) control in sugarbeet. Weed Technol 25:14–18. https://doi.org/10.1614/WT-D-10-00059.1
Kamrin MA (1997) Pesticide profiles: toxicity, environmental impact, and fate, 3rd edn. CRC/Lewis, Boca Raton, 676p
Kegley SE, Orme S, Choi AH (2009) An overview of the PAN pesticide chemical database. Pesticide Action Network, San Francisco, p 94102
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Knowles CO, Benezet HJ (1981) Microbial degradation of the carbamate pesticides desmedipham, phenmedipham, promecarb, and propamocarb. Bull Environ Contam Toxicol 27:529–533
Kossmann K (1970) Rate of decomposition and distribution of phenmedipham in the soil. Weed Res 10:342–359
Kukor JJ, Olsen RH (1991) Genetic organization and regulation of a meta cleavage pathway for catechols produced from catabolism of toluene, benzene, phenol, and cresols by Pseudomonas pickettii PKO1. J Bacteriol 173:4587–4594. https://doi.org/10.1128/jb.173.15.4587-4594
Lewis K, Tzilivakis J, Green A, Warner D (2006) Pesticide properties database (PPDB), dataset/database. University of Hertfordshire, Hertfordshire
Loncar E, Sekulic P, Zeremski-Skoric T, Malbasa R, Kolavar L (2004) Determination of phenmedipham and desmedipham in a commercial herbicide by high performance liquid chromatography. Acta Period Technol 193–198. https://doi.org/10.2298/APT0435193L
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Mandel M, Marmur J (1968) Use of ultraviolet absorbance-temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12:195–206
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3:208–218
McClure NC, Venables WA (1986) Adaptation of Pseudomonas putida mt-2 to growth on aromatic amines. J Gen Microbiol 132:2209–2218
Park G, Oh H, Ahn S (2009) Improvement of the ammonia analysis by the phenate method in water and wastewater. Bull Korean Chem Soc 30:2032–2038. https://doi.org/10.5012/bkcs.2009.30.9.2032
Perret D, Gentili A, Marchese S, Bruno F (2001) Liquid chromatographic/mass spectrometric determination of desmedipham and phenmedipham and their metabolites in soil. J AOAC Int 84(5):1407–1412
Pidiyar VJ, Jangid K, Patole MS, Shouche YS (2004) Studies on the cultured and uncultured microbiota of wild Culex quinquefasciatus mosquito midgut based on 16 s ribosomal RNA gene analysis. Am J Trop Med Hyg 70:597–603
Pohlenz HD, Boidol W, Schüttke I, Streber WR (1992) Purification and properties of an Arthrobacter oxydans P52 carbamate hydrolase specific for the herbicide phenmedipham and nucleotide sequence of the corresponding gene. J Bacteriol 174:6600–6607. https://doi.org/10.1128/jb.174.20.6600-6607.1992
Raabe T, Appel M, Lingens F (1984) Degradation of p-toluidine by Pseudomonas testosteroni. FEMS Microbiol Lett 25:61–64
Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sambrook J, Maniatis T, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Seubert W (1960) Degradation of isoprenoid compounds by microorganisms i. Isolation and characterization of an isoprenoid-degrading, Pseudomonas citronellolis, new species. J Bacteriol 79:426–434
Sonawane BR, Knowles CO (1971) Comparative metabolism of two carbanilate herbicides (EP-475 and phenmedipham) in rats. Pestic Biochem Physiol 1:472–482. https://doi.org/10.1016/0048-3575(71)90181-7
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tomlin CDS (2006) The pesticide manual: a world compendium, 14th edn. British Crop Protection Council, Alton, pp 186–187
Vidal T, Abrantes N, Gonçalves AMM, Gonçalves F (2012) Acute and chronic toxicity of Betanal® Expert and its active ingredients on nontarget aquatic organisms from different trophic levels. Environ Toxicol 27:537–548. https://doi.org/10.1002/tox.20671
Vogel AI, Svehla G (1996) Vogel’s qualitative inorganic analysis, 7th edn. Longman, Harlow
Williams PA, Murray K (1974) Metabolism of benzoate and the methyl benzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol 120(1):416–423
Acknowledgements
Authors are thankful to the DST, New Delhi, for providing financial assistance (Grant no. PURSE-Phase-2/3 (G), SR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no conflicts of interest exist.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Pujar, N.K., Laad, S., Premakshi, H.G. et al. Biodegradation of phenmedipham by novel Ochrobactrum anthropi NC-1. 3 Biotech 9, 52 (2019). https://doi.org/10.1007/s13205-019-1589-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1589-8