Abstract
The termite gut is a highly structured microhabitat with physicochemically distinct regions. It is generally separated into the foregut, midgut and hindgut. The distribution of gut microbiota is greatly influenced by varying physicochemical conditions within the gut. Thus, each gut compartment has a unique microbial population structure. In this study, the bacterial communities of foregut, midgut and hindgut of wood-feeding higher termite, Bulbitermes sp. were analyzed in detail via metagenomic sequencing of the 16S rRNA V3-V4 region. While the microbiomes of the foregut and midgut shared a similar taxonomic pattern, the hindgut possessed more diverse bacterial phylotypes. The communities in the foregut and midgut were dominated by members of the group Bacilli and Clostridia (Firmicutes) as well as taxon Actinomycetales (Actinobacteria). The main bacterial lineage found in hindgut was Spirochaetaceae (Spirochaetes). The significant difference among the three guts was the relative abundance of the potential lignin-degrading bacteria, Actinomycetales, in both the foregut and midgut. This suggests that lignin modification was probably held in the anterior part of termite gut. Predictive functional profiles of the metagenomes using 16S rRNA marker gene showed that cell motility, energy metabolism and metabolism of cofactors and vitamins were found predominantly in hindgut microbiota, whereas xenobiotics degradation and metabolism mostly occurred in the foregut segment. This was compatible with our 16S rRNA metagenomic results showing that the lignocellulose degradation process was initiated by lignin disruption, increasing the accessibility of celluloses and hemicelluloses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Termites are efficient decomposers of lignocelluloses. They maintain a dense and diverse microbiota in their gut and are dependent on these gut microorganisms for their survival on a diet of wood or plant matter (Ohkuma 2003). The rapid mineralization of lignocelluloses by termites has encouraged the study of their gut microbial partners. Lignocellulose is the principle component of plant biomass and is recognized as one of the most abundant renewable resource for biofuels production. The understanding of lignocelluloses decomposition by termite may help to improve the efficiency of industrial conversion of lignocelluloses to biofuels and other valuable chemicals (Brune 2014).
The termite gut is a highly compartmentalized microenvironment (Ohkuma 2003). Each gut compartment has distinct physicochemical conditions (e.g. oxygen and hydrogen partial pressure, pH and redox potential) and is colonized by a particular microbial community (Kohler et al. 2012). Microorganisms live in termite gut are not evenly dispersed but occupy respective microniches within the gut (Ohkuma 2003). The termite gut is generally separated into three parts, i.e. foregut, midgut and hindgut. The diverse atmosphere within the gut has a significant impact on the distribution of the gut microbes and their metabolic capacities. Most previous studies on the symbiotic microbial community of the termite gut have focused on the whole gut or the hindgut compartment, with the perception that prokaryotes are primarily found in the hindgut. For a comprehensive understanding of the mechanisms underlying lignocelluloses digestion, we have examined the bacterial 16S amplicon of the foregut, midgut, and hindgut of a wood-feeding termite, Bulbitermes sp..
Most of the vast varieties of microorganisms residing in the termite gut are unculturable. These symbionts have tight associations with their host and are difficult to culture in vitro (Amann et al. 1995). Thus study of the symbiotic systems of termite guts has been restricted, including the predominant species within the gut community (Ohkuma 2003). However, the molecular approach employing metagenomics has made it possible to access the full spectrum of the microbial diversity in the gut of the termite. Most of the studies of microbial decomposition of lignocelluloses in the termite gut have focused on the isolated pure culture of microorganisms. Microorganisms discovered with pure culture are regularly characterized by unsatisfactory lignocellulolytic activities (Enroth-Cugell and Robson 1984). It appears that the lignocelluloses degradation process is carried out by a group of microorganisms that act synergistically on the lignocellulosic substrate. In this study, we target the functional profiles of the whole bacterial communities in foregut, midgut, and hindgut segments.
Amann et al. (1995) suggested that both host phylogeny and diet are the important factors in shaping the bacterial community structure in termite gut. Here, we have investigated the bacterial diversity of wood-feeding higher termite Bulbitermes sp.. Reports of the symbiotic system from this species are scarce. There is a lack of information about the symbiotic bacteria in the foregut and midgut of wood-feeding higher termites. In this study, we focused on the bacterial community structures in foregut, midgut and hindgut of Bulbitermes sp. using 16S rRNA metagenomic analysis and phylogenetically compared them to clarify in situ individual populations. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt), a bioinformatics software package was run to determine the functional potentials of the bacterial communities of termite foregut, midgut and hindgut using 16S rRNA marker gene. Functional profiles were described to provide a basis for exploration of efficient digestive system. The knowledge of the microbial composition in the termite gut is a prerequisite for the successive isolation of pure culture of a particular microorganism. Moreover, the understanding of the natural way of termite biomass degradation may contribute to optimization of the plant biomass bioconversion process.
2 Materials and methods
2.1 Termite sample preparation
Termites from one colony were collected from a rotten rubber tree in Hutan Rekreasi Universiti Teknologi Malaysia (UTM), Skudai, Johor. They were kept in a plastic container and fed with wood. The termite was sent for identification by an entomologist of Universiti Tun Hussein Onn Malaysia (UTHM), Malaysia. Termite identification was performed based on morphology characterization and the termite was confirmed as Bulbitermes sp. (Isoptera, Nasutitermitinae). Only worker caste termites were used in the experiments because only workers forage. The guts were dissected under aseptic conditions. Termite workers were surface sterilized with 70% ethanol and air-dried for 1 min prior to dissection. The whole gut was extracted from the abdomen of the termite using a needle with forceps holding the head. The intact gut was then separated into foregut, midgut and hindgut. Each individual gut was pooled from 450 worker termites and homogenized in a microcentrifuge tube.
2.2 Total genomic DNA extraction
Total genomic DNA extraction from the foregut (F12101121), midgut (M3132) and hindgut (H22b) of the termites were carried out using gDNA tissue DNA extraction kit, NucleoSpin® Tissue (Macherey- Nagel, Germany) following the manufacturer’s protocol. The kit uses column-based DNA extraction method to avoid co-extraction of interfering substances as environmental sample contains a lot of contaminants. Briefly, lysis of the gut tissue was achieved by incubation of the sample materials in the mixture of sodium dodecyl sulfate (SDS) and proteinase K solutions. For the adjustment of the appropriate binding condition, chaotropic salts and ethanol were added to the lysate. After that, lysate was applied to the silica membrane in the NucleoSpin® Tissue Column and centrifuged. The bound DNA on the silica membrane was washed twice to remove contaminants and salts. Genomic DNA was finally eluted with a low salt elution buffer (Qiagen, Germany). Purified DNA were evaluated by gel electrophoresis, NanoDrop® ND-1000 spectrophotometer (Thermo Scientific, USA) and Qubit® 2.0 Fluorometer (Invitrogen, Belgium) to determine the concentration and quality of the DNA.
2.3 16S rRNA metagenomic library preparation
For 16S rRNA metagenomic library preparation, PCR amplification was carried out using region of interest-specific primers with an attached Illumina overhang adapter. The primers used in the study which target the bacterial 16S V3 and V4 region were S-D-Bact-0341-b-S-17 (5′-CCTACGGGNGGCWGCAG-3′) and S-D-Bact-0785-a-A-21 (5'-GACTACHVGGGTATCTAATCC-3′) (Klindworth et al. 2013). Illumina overhang adapters were added to the primers and thus the full-length primer sequences were 16S Amplicon PCR Forward Primer = 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and 16S Amplicon PCR Reverse Primer = 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC. PCR product purification was performed using Agencourt AMPure XP (Beckman Coulter Inc., USA) to purify the 16S V3 and V4 amplicon from the amplification reaction mixture. Index PCR was performed to attach dual indices and Illumina sequencing adapters to PCR amplicons using Nextera XT Index Kit (Illumina, USA). The PCR products with indices were purified before quantification. 2100 Bioanalyzer (Agilent Technologies, USA) was used to verify the size of the PCR amplicon. The concentration of the metagenomic libraries were measured by quantitative PCR and Qubit 2.0 Fluorometer with the Qubit dsDNA HS Assay Kit (Invitrogen, Belgium). The libraries were normalized and pooled to a final concentration of 4 nM prior to sequencing. The samples were subjected to paired-end sequencing on a MiSeq sequencer (Illumina, USA) using MiSeq Reagent Kit (v3) according to manufacturer instructions.
2.4 Data analysis
The 16S rRNA metagenomic samples were sequenced with 2 × 300 base pairs (bp) paired-end reads. The resulting sequence reads underwent quality assessment using FastQC software followed by sequence filtration using FASTQ Quality Filter (q = 20, p = 80) of the FASTX Toolkit. The passed filter sequences were trimmed and merged using Pair-End Read Merger (PEAR). The merged fragments were then subjected to homology search against the National Center for Biotechnology Information (NCBI) 16S microbial database using BLASTN program (e-value = 1e−6). The BLAST results were further analysed with MEGAN, using the lowest common ancestor (LCA) assignment algorithm, to assign the sequence reads into taxa (parameter: Min Support = 5, Min Score = 50 and Top Percent = 10). Taxonomic tree and rarefaction analysis graph were generated by MEGAN to show the assignment of reads to the NCBI taxonomy and coverage obtained from sampling, respectively.
2.5 Functional profile prediction
The functional composition of the gut microbiota was analysed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) bioinformatics software package (Langille et al. 2013). The analysis was performed by default PICRUST analysis according to Langille et al. 2013 and the sequenced reference used is Greengenes (greengenes_13_8 with 202,421 bacterial and archaeal sequences) (McDonald et al. 2012). PICRUSt analysis was done by first picking OTU against the Greengene database. The precalculated files including 16S copy number normalization and KEGG ortholog (KO) predictions were downloaded from PICRUSt official website (http://picrust.github.io/picrust/install.html#install). The resulting OTU counts were normalized by dividing with their predicted 16S copy number. The normalized OTU tables were then underwent metagenome functional predictions with KO. The output file was further analyzed using statistical analysis of metagenomic profiles (STAMP) software package (Parks et al. 2014).
Data accessibility
The original sequencing output files have been deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database under accession number SRP074305.
3 Results
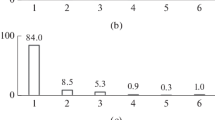
The bacterial taxonomic compositions of foregut (F12101121), midgut (M3132) and hindgut (H22b) of wood-feeding termite Bulbitermes sp. were analysed in detail by Illumina sequencing of V3-V4 region of the 16S rRNA genes. A total of 2,612,387, 3,871,743, and 6,374,587 sequence reads were generated for samples F12101121, M3132 and H22b, respectively, using Illumina Miseq sequencer. After the quality assessment and sequence reads merging process, the number of sequence reads (merged fragments) obtained for samples F12101121, M3132 and H22b were 2,185,744, 2,933,905 and 5,552,945, respectively. Diversity coverage of each of the samples was analysed via rarefaction analysis. In this study, rarefaction curves were computed at family-level (Electronic Supplementary Material, Fig. S1). The rarefaction curves of all three samples started with a steep slope, and at some point began to flatten as the samples reached near-saturation for the taxonomic assignment. The highest diversity was found in the hindgut, while foregut was least diverse. Each gut section contained sequences from 15 to 20 different taxa (phylum-level).
3.1 Community diversity and identification of the core microbiota
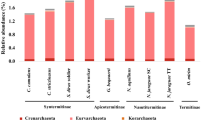
The most abundant group colonizing both the foregut and midgut of Bulbitermes sp. was Firmicutes, comprising mainly Clostridia and Bacilli (Figs. 1 and 2). Other lineages found in this bacterial group included Erysipelotrichia and Veillonellaceae (Selenomonadales, Negativicutes) (Electronic Supplementary Data, Fig. S2 and S3). Of the foregut bacterial population, a majority of the Clostridia were members of the taxon Clostridiales with the prevalent lineages including Clostridiaceae and Gracilibacteraceae. Clusters of Bacilli, on the other hand, were dominated by Enterococcaceae and Streptococcaceae (Lactobacillales) (Fig. 1). The predominant members of the Firmicutes group found in midgut were fairly similar with those in the foregut, yet midgut’s Bacilli cluster comprised a portion of Bacillaceae (Bacillales) but a low density of Streptococcaceae (Fig. 2). According to Figs. 1 and 2, another prevalent group observed in both the foregut and midgut microbiota was Actinobacteria, comprised mainly taxon Actinomycetales with Actinomycineae and Micrococcineae as the major bacterial lineages.
As shown in Fig. 3 and the Electronic Supplementary Data (Fig. S4), termite hindgut microbiota was dominated by Spirochaetes, comprised mainly Spirochaetaceae. Members of the taxon Synergistaceae (Synergistetes) were also found abundantly in the hindgut community compare to the other two gut segments. Firmicutes, represented mainly by Clostridia, contributed a portion to the hindgut bacterial population. 16S rRNA metagenomic data showed that the major representatives for this Clostridia cluster included Clostridiaceae, Ruminococcaceae, Gracilibacteraceae and Peptococcaceae, all belonged to the taxon Clostridiales. Another lineage of Firmicutes, Bacilli, consisting mainly of Streptococcaceae and Enterococcaceae (Lactobacillales), was also present in a small fraction in the hindgut community. Besides, Fibrobacteres, Proteobacteria and Bacteroidetes were also found as minor groups in the hindgut microbiota. In the group of Fibrobacteres, the main bacterial lineages discovered were Fibrobacteraceae and Holophagaceae. Bacteroidetes in the hindgut consisted mainly of Porphyromonadaceae (Bacteroidales, Bacteroidia), while the Proteobacteria group was dominated by Desulfovibrionaceae (Desulfovibrionales, Deltaproteobacteria).
3.2 Comparative analysis of foregut, midgut and hindgut bacterial communities
Sequences data sets retrieved from termite foregut, midgut, and hindgut were compared. Figure 4 summarizes the relative abundance of the 50 major families represented in different gut segments. The details of the bacterial taxonomic structure (up to family-level) of foregut, midgut and hindgut are available at Electronic Supplementary Data (Fig. S2, S3 and S4). Of the Firmicutes in the foregut community, members of the taxon Eubacteriaceae, Lachnospiraceae and Oscillospiraceae (Clostridiales) found in both midgut and hindgut were absent whereas Thermoactinomycetaceae (Bacillales) was only detected in the foregut sample. In contrast, midgut bacterial population showed a higher density of Bacillaceae while taxon Paenibacillaceae, Planococcaceae and Bacillales Family XI. incertae sedis (Bacillales) were only found in the midgut. The hindgut sample showed a diverse lineage of Proteobacteria, which is outnumbered by numerous sulfate- and sulfur-reducing bacteria. The members of the Proteobacteria group only found in hindgut community include Kordiimonadales, Nitrosomonadales, Rhodocyclales, Bacteriovoracaceae, Desulfarculaceae, Desulfovibrionaceae, Desulfobulbaceae, Desulfurellaceae, Desulfuromonadales, Cystobacterineae, Syntrophobacterales and Acidithiobacillaceae. The foregut showed a relatively high abundance of Enterobacteriaceae (Enterobacteriales, Gammaproteobacteria) compared to the other two segments. There was also a diverse bacterial lineage of Preoteobacteria found in foregut, with the presence of Caulobacteraceae, Magnetococcaceae, Rhodospirillaceae, Rickettsiales genera incertae sedis, Sphingomonadaceae, Alcaligenaceae, Halothiobacillaceae, Coxiellaceae, Sinobacteraceae and Xanthomonadaceae specific to the foregut compartment.
The members of the group Actinobacteria present in the foregut and midgut communities largely differed with those in the hindgut population. The foregut and midgut samples showed a higher diversity and density of Actinobacteria, with members of the Actinomycetales as the predominant lineage. There was a relatively higher abundance of Actinomycineae in the midgut community and Corynebacteriaceae and Micrococcaceae were only present in the midgut. Hindgut community lacked several bacterial lineages that were found in both foregut and midgut, i.e. Acidimicrobineae, Nocardioidaceae, Microbacteriaceae, Pseudonocardiaceae and Thermomonosporaceae, however, Rubrobacterineae were only detected in the hindgut.
3.3 Predictive functional profiles
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis was carried out to predict metagenomes based on the Greengenes 16S rRNA database and KO.
The number of sequences of sample F12101121, M3132 and H22b that matched the greengenes database are 364,312.0, 832,611.0 and 1,803,291.0, respectively at threshold 0.97. Principal component analysis (PCA) was performed to demonstrate the correlation of the functional profiles of the bacterial communities in foregut (F12101121), midgut (M3132) and hindgut (H22b) of termite. PCA plot (Fig. 5) showed that metabolic profile of the hindgut was significantly differed from the metabolic profiles of the foregut and midgut. The deviation of sample H22b was revealed on the principal component (PC) 1 which carries 92.3% variation. Heat map plots (Figs. 6 and 7) were constructed to compare the abundance of the metabolic features among different gut segments. Due to differences in the abundance scale, the subsystems ABC transporters, phosphotransferase system (PTS) and transporters (functional class: membrane transport), which show high redundancy compare to other metabolic features, were plotted separately. From the results of this study, energy metabolism and cell motility were found to be overrepresented in the hindgut community. The subsystems included in the energy metabolism were carbon fixation pathways in prokaryotes, methane metabolism, nitrogen metabolism, oxidative phosphorylation and sulfur metabolism. Cell motility which includes bacterial chemotaxis, bacterial motility proteins and flagellar assembly showed higher abundance in the sample H22b than M3132 and least abundant in F12101121. ABC transporters and transporters were also represented the hindgut community. In contrast, xenobiotics biodegradation and metabolism, which includes benzoate degradation, bisphenol degradation, drug metabolism by cytochrome P450, ethylbenzene degradation, naphthalene degradation and polycyclic aromatic degradation, were observed to have higher proportions in the foregut and midgut community. Additionally, metabolism of carbohydrate such as amino acid and nucleotide sugar, butanoate, fructose and mannose as well as starch and sucrose were detected to be more represented in the foregut and midgut microbiome. The citrate cycle (TCA cycle), on the other hand, showed a higher proportion in the hindgut community.
4 Discussion
While the foregut and midgut of Bulbitermes sp. are relatively small, the voluminous hindgut paunch accommodates a large portion of gut microbiota. A previous study of the wood-feeding higher termite, Nasutitermes corniger, found that hindgut segment contained the highest density of bacteria, with Spirochaetes as the dominant bacterial lineage (Kohler et al. 2012). In representatives of higher termites, Nasutitermitinae and Termitinae, which have cellulose-rich diets, the hindguts were colonized by Spirochaetes and Fibrobacteres as well as the related candidate phylum TG3 (Hongoh et al. 2005, 2006; Warnecke et al. 2007). Brune reported that the high motility feature of spirochaetes enables them to strive in the dynamic environment in hindgut segment (Brune 2014). Our results showed that cell motility was highly represented in the hindgut microbiome. This is consistent with the previous report of metaproteomic analysis which found that the most represented proteins observed in the hindgut paunch of Nasutitermes sp. were flagellin-related hook-associated proteins and methyl-accepting chemotaxis proteins (Burnum et al. 2011). According to the metagenomic and metatranscriptomic analysis conducted by He et al., the most highly expressed functional categories of the hindgut microbiota in wood- and dung-feeding higher termites, N. corniger and Amitermes wheeleri, were cell motility and carbohydrate transport and metabolism. Higher abundances of the cell motility and chemotaxis genes were found in N. corniger, in which Spirochaetes and Fibrobacteres dominated. The study also hypothesized that the difference in motility-related gene abundance reflects the community structural differences, particularly the dominance of spirochaetes (He et al. 2013). There were also substantial chemotaxis genes (Fraser et al. 1997) and methylaccepting chemotaxis protein (MCP) genes (Bellgard et al. 2009) detected in the spirochaetes isolate genomes. Cell motility and associated chemotaxis are required by gut microorganisms to actively access their substrates and respond to the steep physicochemical gradients found in the termite hindgut (He et al. 2013).
According to Brune (2014), the diverse microorganisms in the hindgut compartment not only contribute to the hydrolysis and subsequent fermentation of plant biomass but also compensate for the nutritional deficits of the lignocellulosic diet. The composition of short-chain fatty acids and other fermentation products in the hindgut fluid of termites suggested a variety of microbial fermentations occur simultaneously in this gut segment (Schultz and Brezbak 1979; Odelson and Breznal 1983; Anklin-Mühlemann et al. 1995; Tholen and Brune 2000). The dilated hindgut paunch is the only anoxic gut region in which large amounts of hydrogen accumulate. Kohler et al. reported that molecular hydrogen was produced by a dense community of Spirochaetes and Fibrobacteres (Kohler et al. 2012). Hydrogen acts as the central intermediate that drives the reduction of CO2 and methanogenesis (Brune 2014). This was consistent with the high rates of reductive acetogenesis observed in the hindgut homogenates of several Nasutitermes sp. (Brauman et al. 1992). CO2-reductive acetogenesis yields acetate which serves as the major carbon and energy source for the termite host (Warnecke et al. 2007). Our results demonstrated that the TCA cycle which consumes acetate (in the form of acetyl-CoA) as substrate is present in a higher proportion in hindgut microbiota. Metagenomic and functional analysis of hindgut microbiota of a Nasutitermes sp. suggested that CO2-reductive acetogenesis was dominated by spirochaetes, particularly Treponema sp.. In the study, the genes encoding for formyltetrahydrofolate synthase (FTHFS) and carbon monoxide dehydrogenase (CODH) acetyl-CoA synthase, marker genes of CO2-reductive acetogenesis, were detected. Both of the functional genes were predicted to be encoded by treponemes (Warnecke et al. 2007).
Energy metabolism, including carbon fixation pathways, methane metabolism, nitrogen metabolism, oxidative phosphorylation and sulfur metabolism as well as membrane transport system, was also found to be more represented in the hindgut microbiota. Since lignocellulosic materials are poor in nitrogen, termites rely on the nitrogen-fixing symbionts in their guts to acquire the essential amino acids and vitamins (Kohler et al. 2012). Warnecke et al. identified 12 nifH homologues as well as other nitrogenase components within the hindgut microbiome of Nasutitermes sp. (Warnecke et al. 2007). Brune suggested that the various nifH genes observed in termite guts were affiliated with Spirochaetes, Clostridia, Bacteroidetes, and possibly Fibrobacteres. The fermentation process of plant fibre yields varying amounts of methane. Methanogens present in the hindgut of termites are engaged in removing H2 and CO2 during lignocelluloses degradation, promoting the digestion process (Kudo 2009). In this study, sulfur metabolism overrepresented in the hindgut microbiome was consistent with the 16S metagenomic analysis that the hindgut community was outnumbered by numerous sulfate- and sulfur-reducing bacteria. In addition, metaproteomic analysis of the bacterial community resident in the hindgut paunch of Nasutitermes sp. have showed redundancy of several metabolic pathways including carbohydrate transport and metabolism, nitrogen fixation and assimilation, energy production and amino-acid synthesis. According to the study, the transportation and fermentation pathways were important with regard to possible energy substrates for the microbiota (Burnum et al. 2011).
Members of the group Synergistetes were previously reported in the guts of wood-feeding termites (Hongoh et al. 2005; Kohler et al. 2012; Makonde et al. 2013). As they are mainly inhabits anaerobic environment (Vartoukian et al. 2007), members of Synergistetes are most probably exist in the anaerobic termite hindgut. Dahle and Birkeland (2006) have shown that some of the Synergistetes species were implicated in amino acid degradation, which is an important process in termite gut (Makonde et al. 2013). Clone-based metagenomic analysis revealed that Clostridia and Bacilli were dominant in the first proctodeal (P1) segment in the hindgut of higher termites examined (Thongaram et al. 2005). The anterior hindgut of higher termite showed high alkalinity and thus alkali-tolerant Firmicutes-related bacteria represent the majority of the P1 community. Clostridium sp. belongs to the taxon Clostridiaceae has been recognised as an anaerobic cellulose-degrading bacteria that plays a role in degrading plant biomass (Makonde et al. 2013), suggesting cellulose decomposition capability among Clostridia group members. Apart from that, many clostridia may involve in the fermentation and nutrition processes in termite gut by their ability to degrade polysaccharides to produce acetone, alcohol, lactate, acetate, carbon dioxide and hydrogen (Tokuda et al. 2000).
The findings of high abundance of Firmicutes in the termite midgut are in agreement with the findings of another wood-feeding termite, albeit a lower termite, Reticulitermes santonesis. The most abundant phylogenetic group in the midgut microbiota belonged to Firmicutes, particularly members of Clostridiales, followed by Lactobacillales, with the majority affiliated with the lineage Streptococcus (Yang et al. 2005). According to Kohler et al. (2012), the bacterial community in the midgut of N. corniger was dominated by Firmicutes, particularly members of Lachnospiraceae which was comprised of many species with high cellulolytic, xylanolytic and proteolytic activities. Our study of termite midgut microbiota, however, showed that the prevalent lineage under Firmicutes included Clostridiaceae and Enterococcaceae. Firmicutes (Clostridales) has proved to participate in the fibre- digestion, which contributes to the cellulose hydrolysis process (Mikaelyan et al. 2014). In a study of the influence of nutritional components on bacterial community structure in the gut of N. takagoensis, Miyata et al. (2007) demonstrated that Firmicutes was predominant in the clone library from xylan- and xylose-fed termites, suggesting that members of the group Firmicutes may have the ability to metabolize hemicelluloses. On the other hand, the high abundance of Firmicutes (Clostridiales and Lactobacillales) observed in the foregut bacterial community in this study was consistent with a previous study showing that Firmicutes, mainly Streptococcaceae, as the major bacterial phyla in the crop section of the gut of N. corniger (Kohler et al. 2012). The functional analysis in our study indicated that metabolism of carbohydrate such as amino acid and nucleotide sugar, butanoate, fructose, and mannose, as well as starch and sucrose, has a relatively high proportion in the foregut and midgut microbiome, suggesting that the hydrolysis process of wood polysaccharides occurs in these gut segments.
Another prevalent group observed in both the foregut and midgut microbiota was Actinobacteria, comprised mainly of Actinomycetales, with Actinomycineae, Microbacteriaceae and Promicromonosporaceae as the major bacterial lineages. (Pasti et al. 1990) successfully isolated actinomycete strains with specific peroxidase activity from the gut of higher termites. A diverse collection of actinomycete bacteria have been shown to possess extracellular peroxidase activity (Mercer et al. 1996), suggesting the metabolic capabilities of actinomycetes in lignin degradation. (Godden et al. 1992) showed the degradation of lignin model compounds by actinomycetes with the detection of extracellular peroxidase and catalase activity. Tuomela et al. (2001) reported that actinomycetes form multicellular filaments and are thus able to solubilize and modify the lignin structure extensively. According to Bugg et al. (2011), the bacterial strains isolated from the guts of various termites and wood-boring beetles that show lignin breakdown activity fall into three classes, i.e. Actinomycetes, α-Proteobacteria, and γ-Proteobacteria. Additionally, a wide range of species of the taxon Actinomycetales, including the lineages Mycobacterium, Nocardia, Rhodococcus, Pseudonocardia, and Streptomyces, have been shown to metabolize aromatic compounds, benzoate (B) and p-hydroxybenzoate (pHB) (Hammann and Kutzner 1998). In this study, the phylogenetic results were consistent with the PICRUSt analysis that xenobiotics biodegradation and metabolism showed higher proportions in the foregut and midgut microbiome.
In addition, a previous study on lignin modification during passage through the three gut segments of lower subterranean wood-feeding termites, Coptotermes formosanus and Reticulitermes flavipes, using pyrolysis gas chromatography/mass spectrometry (Py-GC/MS), showed an increase of 1,2- diformyloxyethane, dimethyl 3,8-dioxodecanedioate and p-cresol in the foregut and midgut. Phenol and guaiacol, however, were found to have increased in the midgut. These findings suggest the occurrence of lignin ring demethylation, decarboxylationand side-chain oxidation, reactions that pretreat wood for cellulose utilization, are initiated in the foregut, and occur mainly in the midgut of termites (Ke et al. 2011). This is consistent with the relatively high levels of oxygen observed in the midgut that could facilitate lignin oxidation (Ke et al. 2010). Moreover, in-vivo degradation of aromatic compounds with different substructures by C. formosanuswas also investigated. The results revealed that the wood feeding termite is able to metabolize the conjugated structures of aromatic compounds by side-chain addition, ring hydroxylation, ring/side-chain oxidation, ring mineralization, as well as β-O-4 and 5–5 linkage cleavage. The degradation efficiency of these conjugated aromatic structures was found to be higher in the foregut and midgut sections than in the hindgut (Ke et al. 2011). Another study reported laccase transcript and phenoloxidase activity in the salivary gland and foregut tissue, suggesting the sites of aromatic compound metabolism (Coy et al. 2010).
In the 16S metagenomic and functional analysis of bacterial communities in the foregut, midgut and hindgut of higher wood-feeding Bulbitermes sp., the core microbiota of each gut segments were determined. The relative abundance of Actinobacteria group and the xenobiotics degradation and metabolism in both foregut and midgut microbiome suggest a high possibility of lignin degradation in these gut segments. It has been shown that the symbiotic metabolic process in the hindgut is likely dominated by spirochaetes. The anaerobic conditions in the hindgut may promote the fermentation of the products derived from cellulose depolymerization. Our work demonstrates the complex interaction among diverse gut microbiota and their symbiotic interaction between termites, which are critical for the effective degradation of lignocelluloses.
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59(1):143–169
Anklin-Mühlemann R, Bignell DE, Veivers PC, Leutold RH, Slaytor M (1995) Morphological, microbiological and biochemical studies of the gut flora in the fungus-growing termite Macrotermes subhyalinus. J Insect Physiol 41(11):929–940. https://doi.org/10.1016/0022-1910(95)00062-Y
Bellgard MI, Wanchanthuek P, La T, Ryan K, Moolhuijzen P, Albertyn Z, Shaban B, Motro Y, Dunn DS, Schibeci D, Hunter A, Barrero R, Phillips ND, Hampson DJ (2009) Genome sequence of the pathogenic intestinal spirochete brachyspira hyodysenteriae reveals adaptations to its lifestyle in the porcine large intestine. PLoS One 4(3):e4641. https://doi.org/10.1371/journal.pone.0004641
Brauman A, Kane MD, Labat M, Breznak JA (1992) Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science (New York, NY) 257(5075):1384–1387. https://doi.org/10.1126/science.257.5075.1384
Brune A (2014) Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12(3):168–180. https://doi.org/10.1038/nrmicro3182
Bugg TD, Ahmad M, Hardiman EM, Singh R (2011) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22(3):394–400. https://doi.org/10.1016/j.copbio.2010.10.009
Burnum KE, Callister SJ, Nicora CD, Purvine SO, Hugenholtz P, Warnecke F, Scheffrahn RH, Smith RD, Lipton MS (2011) Proteome insights into the symbiotic relationship between a captive colony of Nasutitermes corniger and its hindgut microbiome. ISME J 5(1):161–164. https://doi.org/10.1038/ismej.2010.97
Coy MR, Salem TZ, Denton JS, Kovaleva ES, Liu Z, Barber DS, Campbell JH, Davis DC, Buchman GW, Boucias DG, Scharf ME (2010) Phenol-oxidizing laccases from the termite gut. Insect Biochem Mol Biol 40(10):723–732. https://doi.org/10.1016/j.ibmb.2010.07.004
Dahle H, Birkeland NK (2006) Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int J Syst Evol Microbiol 56(7):1539–1545. https://doi.org/10.1099/ijs.0.63894-0
Enroth-Cugell C, Robson JG (1984) Functional characteristics and diversity of cat retinal ganglion cells. Basic characteristics and quantitative description. Invest Ophthalmol Vis Sci 25(3):250–267
Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390(6660):580–586. https://doi.org/10.1038/37551
Godden B, Ball AS, Helvenstein P, Mccarthy AJ, Penninckx MJ (1992) Towards elucidation of the lignin degradation pathway in actinomycetes. J Gen Microbiol 138(11):2441–2448. https://doi.org/10.1099/00221287-138-11-2441
Hammann R, Kutzner HJ (1998) Key enzymes for the degradation of benzoate, m- and p- hydroxybenzoate by some members of the order Actinomycetales. J Basic Microbiol 38(3):207–220. https://doi.org/10.1002/(SICI)1521-4028(199807)38:3<207::AID-JOBM207>3.0.CO;2-R
He S, Ivanova N, Kirton E, Allgaier M, Bergin C, Scheffrahn RH, Kyrpides NC, Warnecke F, Tringe SG, Hugenholtz P (2013) Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One 8(4):e61126. https://doi.org/10.1371/journal.pone.0061126
Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Vongkaluang C, Noparatnaraporn N, Kudo T (2005) Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol 71(11):6590–6599. https://doi.org/10.1128/AEM.71.11.6590-6599.2005
Hongoh Y, Deevong P, Hattori S, Inoue T, Noda S, Noparatnaraprn N, Kudo T, Ohkuma M (2006) Phylogenetic diversity, localization, and cell morphologies of members of the candidate phylum TG3 and a subphylum in the phylum Fibrobacteres, recently discovered bacterial groups dominant in termite guts. Appl Environ Microbiol 72(10):6780–6767. https://doi.org/10.1128/AEM.00891-06
Ke J, Sun J-Z, Nguyen HD, Singh D, Lee KC, Beyenal H, Chen S-L (2010) In-situ oxygen profiling and lignin modification in guts of wood-feeding termites. Insect Sci 17(3):277–290. https://doi.org/10.1111/j.1744-7917.2010.01336.x
Ke J, Singh D, Chen S (2011) Aromatic compound degradation by the wood-feeding termite Coptotermes formosanus (Shiraki). Int Biodeterior Biodegrad 65(6):744–756. https://doi.org/10.1016/j.ibiod.2010.12.016
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41(1):e1. https://doi.org/10.1093/nar/gks808
Kohler T, Dietrich C, Scheffrahn RH, Brune A (2012) High-resolution analysis of gut environment and bacterial microbiota reveals functional compartmentation of the gut in wood-feeding higher termites (Nasutitermes spp.) Appl Environ Microbiol 78(13):4691–4701. https://doi.org/10.1128/AEM.00683-12
Kudo T (2009) Termite-microbe symbiotic system and its efficient degradation of lignocellulose. Biosci Biotechnol Biochem 73(12):2561–2567. https://doi.org/10.1271/bbb.90304
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9):814–821. https://doi.org/10.1038/nbt.2676
Makonde HM, Boga HI, Osiemo Z, Mwirichia R, Mackenzie LM, Goker M, Klenk HP (2013) 16S- rRNA-based analysis of bacterial diversity in the gut of fungus-cultivating termites (Microtermes and Odontotermes species). Antonie Van Leeuwenhoek 104(5):869–883. https://doi.org/10.1007/s10482-013-0001-7
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6(3):610–618. https://doi.org/10.1038/ismej.2011.139
Mercer DK, Iqbal M, Miller P, McCarthy AJ (1996) Screening actinomycetes for extracellular peroxidase activity. Appl Environ Microbiol 62(6):2186–2190
Mikaelyan A, Strassert JFH, Tokuda G, Brune A (2014) The fibre-associated cellulolytic bacterial community in the hindgut of wood-feeding higher termites (Nasutitermes spp.) Environ Microbiol 16(9):2711–2722. https://doi.org/10.1111/1462-2920.12425
Miyata R, Noda N, Tamaki H, Kinjyo K, Aoyagi H, Uchiyama H, Tanaka H (2007) Influence of feed components on symbiotic bacterial community structure in the gut of the wood-feeding higher termite Nasutitermes takasagoensis. Biosci Biotechnol Biochem 71(5):1244–1251. https://doi.org/10.1271/bbb.60672
Odelson DA, Breznal JA (1983) Volatile fatty acid production by the hindgut microbiota of xylophagous termites. Appl Environ Micobiol 45:1602–1613
Ohkuma M (2003) Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl Microbiol Biotechnol 61(1):1–9. https://doi.org/10.1007/s00253-002-1189-z
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30(21):3123–3124. https://doi.org/10.1093/bioinformatics/btu494
Pasti MB, Pometto AL 3rd, Nuti MP, Crawford DL (1990) Lignin-solubilizing ability of actinomycetes isolated from termite (Termitidae) gut. Appl Environ Microbiol 56(7):2213–2218
Schultz JE, Brezbak JA (1979) Cross-feeding of lactate between Streptococcus lactis and Bacteroides sp. isolated from termite hinguts. Appl Environ Microbiol 37(6):1206–1210
Tholen A, Brune A (2000) Impact of oxygen on metabolic fluxes and in situ rates of reductive acetogenesis in the hindgut of the wood-feeding termite Reticulitermes flavipes. Environ Microbiol 2(4):436–449. https://doi.org/10.1046/j.1462-2920.2000.00127.x
Thongaram T, Hongoh Y, Kosono S, Ohkuma M, Trakulnaleamsai S, Noparatnaraporn N, Kudo T (2005) Comparison of bacterial communities in the alkaline gut segment among various species of higher termites. Extremophiles 9(3):229–238. https://doi.org/10.1007/s00792-005-0440-9
Tokuda G, Yamaoka I, Noda H (2000) Localization of symbiotic clostridia in the mixed segment of the termite Nasutitermes takasagoensis (Shiraki). Appl Environ Microbiol 66(5):2199–2207. https://doi.org/10.1128/AEM.66.5.2199-2207.2000
Tuomela M, Hatakka A, Raiskila S, Vikman M, Itävaara M (2001) Biodegradation of radiolabelled synthetic lignin (14C-DHP) and mechanical pulp in a compost environment. Appl Microbiol Biotechnol 55(4):492–499. https://doi.org/10.1007/s002530000513
Vartoukian SR, Palmer RM, Wade WG (2007) The division “Synergistes”. Anaerobe 13(3-4):99–106. https://doi.org/10.1016/j.anaerobe.2007.05.004
Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernandez M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450(7169):560–565. https://doi.org/10.1038/nature06269
Yang H, Schmitt-Wagner D, Stingl U, Brune A (2005) Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ Microbiol 7(7):916–932. https://doi.org/10.1111/j.1462-2920.2005.00760.x
Acknowledgments
The authors would like to thank Maricel Cuevas David from UniversitiTun Hussein Onn Malaysia (UTHM), for the identification of the termite’s genus. This work was financed by Research University Grant (Q.J130000.2545.07H38) Universiti Teknologi Malaysia (UTM), Malaysia.
Author information
Authors and Affiliations
Contributions
Y.M.C. collected samples, extracted metagenome, analyses results and wrote the paper, S.F.L. performed bioinformatic analyses, M.M.S. and A.Y. designed the study and discussed the results.
Corresponding author
Rights and permissions
About this article
Cite this article
Chew, Y.M., Lye, S., Md. Salleh, M. et al. 16S rRNA metagenomic analysis of the symbiotic community structures of bacteria in foregut, midgut, and hindgut of the wood-feeding termite Bulbitermes sp.. Symbiosis 76, 187–197 (2018). https://doi.org/10.1007/s13199-018-0544-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-018-0544-5