Abstract
Three recent advances in immunology, genetics, and microbiology have ushered in a new era in the continued efforts to better understand and treat oral diseases, moving ever closer to the three Ps of modern healthcare: personalized, predictive, and preventive medicine (PPPM). The discovery of now 15 subtypes of innate lymphoid cells, the refinement of DNA sequencing, and culture-independent characterization of the entire microbial community begin to reveal this complex adaptive network. All these advances warrant a systematic review as they have changed and will continue to change dental medicine. We will update dental professionals on these advances as related to oral diseases and associated pathologies in other organ systems such as premature labor, arthrosclerosis, and cancer. The five objectives are:
-
1.
Introduce the concept of microbiota and microbiome
-
2.
Explain how we study microbiota and microbiome
-
3.
Describe the types and functions of innate lymphoid cells
-
4.
Inventory the unique demands of the oral cavity
-
5.
Provide a heuristic model to integrate the above
-
6.
Conclusions and expert recommendations
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to its frequent communication with the external environment and an extremely rich variety of microbial niches, the oral cavity has the most complex ecological systems of the human body. Within this average surface area of only 0.22 m2 reside 1010 to 1012 bacteria belonging to 1179 taxa [1]. The mouth is the proximal aperture of the alimentary canal and functions as the sole natural entrance for food, it controls the intake and initial interaction of what would become the gut microbiota, the total microbial community of the gastrointestinal tract. The sum total of their genetic materials make up the microbiome, thus these two terms describe different aspects of the gut microbial organisms. Microbiome is not microbiota even though they are certainly related; they are not interchangeable.
From the biomaterials and biomechanical perspectives, the mouth has the widest spectrum of tissue types. The hardest mineralized structure, enamel, has a compressive elastic modulus of 96 GPa (~ 1011 N/m2), while the pliable oral mucosa has only 100 kPa (105 N/m2) in compression, representing a difference of 1 million times [2]. Such variation contributes to the richness of the ecological niches. The masticatory system is also unique in that rigid living hard tissues, the teeth, firmly anchored by the supporting alveolar process must penetrate the surrounding soft tissue into a perpetually contaminated, microbe-laden, hostile, moist, environment—a condition faced by no other skeletal elements. Because of all these factors, the immune defense of the oral cavity is equally complex and critically important to maintain the proper microbiological and functional homeostasis. In this brief review, we highlight the latest development in innate immunity: innate lymphoid cells, pattern recognition receptors and tolerance, and most importantly, outline what must occur to prevent dysbiosis. Inappropriate host response to nominal tissue turnover and commensal microbial presence will cause tissue damage, while insufficient immune response will permit microbial overgrowth and invasion. These two pathological processes compounded by functional demands of the masticatory system underpin all oral diseases.
Until recently, microbial cultures are the principle means of investigating oral flora. One must culture the microbial species in order to identify and study them. However, in general, only less than 0.1% of the bacteria can grow in culture, many require conditions not yet recognized or that which we cannot duplicate in the laboratory. Using culture-independent characterization, such as 16s RNA gene sequencing, clinicians can now investigate the other 99+% of the bacterial species [3]. These new, eco-genetic, tools allow us to answer three important questions: how many bacteria species are present in a specimen, the quantity of each species, and what the imputed functions are of the gene expressions detected. This means that we now have the ability to identify any patient’s individual microbiome. Coupled with improved molecular techniques such as single cell-based RNA sequencing, the resolution of the differences among various subtypes of innate lymphoid cells is at the level that we can characterize the unique immunological “fingerprint” of each person in each location of the body. The interactions between the innate immune system and oral microbiota not only determine the health of the masticatory system, they also greatly influence the patient’s wellbeing at a systemic level. It is therefore crucial that healthcare professionals have a working knowledge of these new developments.
The key role of oral microbiome in PPPM-related research and medical services
There are four key characteristics common to all microbial organisms, including those inhabiting the human mouth. The first feature is that they are very small, as the word “microbe” so indicates. The most abundant oral microbial group belongs to the kingdom of bacteria, which are 10−6 m in size, 10- to 50-fold larger than viruses and 10- to 50-fold smaller than fungi. Such small size gives bacteria very large mass to surface area ratio—and all the physical and chemical properties inevitably associated with that. The second feature is that they reproduce extremely rapidly, 30 min to an hour on the average for bacteria. The third key feature about microbes, because of the above two characteristics, is that there are many, many of them. A single Streptococcus mutans under permissive conditions will become 16.8 million to 281 trillion (224 to 248) S. mutans in just 1 day [4]. The fourth characteristic is that they are promiscuous in exchanging their genetic materials. Together, these four features make bacteria ideal in a Darwinian world [5]. The fact that we carry only 22,000 Homo sapiens genes, whereas our microbiome has 8,000,000 genes attests to microbial success, and poses great physiological and therapeutic implications.
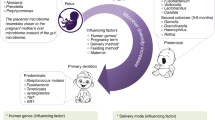
Even though the zygote is sterile, the fetus is not. The development of the infant oral microbiota, as a continuation of the fetal microbial community, begins in utero. Maternal transfer of microbes to the fetus, though at very low levels, occurs via uterine cervix and the placenta. Umbilical cord blood samples obtained under sterile condition following C-section have shown the presence of Enterococcus, Streptococcus, Staphylococcus, and Propionibacterium [6]. Amniotic fluids, previously considered sterile, also have maternal oral bacteria such as Porphyromonas gingivalis. Maternal oral health greatly influences the health of the fetus, adverse pregnancy outcomes correlate tightly with severity of periodontal infection [7]. Following birth, maternal microbial transfer contributes immensely to the formation of infant microbiome, and the maturation of its developing immune system. Within the human milk, four groups were present: staphylococci, streptococci, lactic acid bacteria, and bifidobacteria. The origin of these microbes is very likely the maternal gastrointestinal track, reaching the breast through enteric-mammary translocation. Passive transfer of human milk lactobacilli reduces infants’ gastrointestinal infections by an impressive 46% [8].
For bacteria to colonize, they must first attach to a surface. The mechanical, chemical, and microbial characteristics of the surface influence greatly which microbial species can attach. Neonates and infants have only soft surfaces of oral mucosa and thus do not have microbes that require or prefer hard surfaces. The primary colonizers of neonatal oral mucosa are maternal skin or genitourinary track in origin, depending on mode of delivery with C-sections for the former and eutocic births for the latter [9]. Streptococcus sanguis and Streptococcus mitis can cleave IgA and bind to sialylated mucin and fragments of dead bacteria. They are the principle components of first oral microbiome, producing matrices and cell surface anchors to allow the next microbial group to take hold. As primary tooth eruptions occur, there are also changes in dietary intake and oral hygiene practices. Dental plaque is thus a complex microbial ecology in dynamic equilibrium (Fig. 1). The oral microbiota becomes more diverse, organized, and co-develops with the host immune system, forming reciprocal dependences of varying degrees as the deciduous dentition gives way to mixed and then adult dentition. All three subtypes of innate lymphoid cells are present in the oral mucosa of human infants as early as 4 months of age, with ILC2 predominating [11].
The complex and highly organized microbial community on tooth surface. (Adapted after [10]) The colored lines represent adhesion-cognate receptors responsible for coaggregation. For the bacterial genus, starting from the bottom: S Streptococcus, P Propionibacterium (acnes), C Capnocytophaga, H Hemophilus, P Prevotella (loescheii, denticola, and intermedia), V Veillonella, E Eikenella, A Actinomyces (isralii and naslundii), T Treponema, P Porphyromonas (gingivalis)
Detailed survey of 5 normal adult human intra-oral samples from 9 different sites using culture-independent method based on 16S rRNA gene sequencing identified 2589 clones with 141 species being predominant. Most (60%) of these species could not grow in culture. Common to all sites were Gemella, Granulicatella, Streptococcus, and Veillonella, with S. mitis as the most common species. The microbiota changed with the sites, reflecting the importance of surface texture, hardness, oxygen tension, and presence of other microbes. Neisseria, a Gram-negative obligatory aerobe, for example, does not populate gingival crevices where oxygen level is low [12]. Poor oral hygiene causes microbial overgrowth. This quantitative change ushers in qualitative changes such as quorum sensing and group slide. Bacteria that demonstrate virulent behavior start to appear, producing organic acids, protease, phospholipases, tissue invasion, resisting opsonization, and evading phagocytosis. With the escalation in pathogenicity by the oral microbes, both innate and adaptive immunity also increase in their responses. In an attempt to eliminate the microbial invasion, immune responses may cause more extensive, collateral, tissue damage. The junctions between endothelial cells become more permeable and neutrophils translocate into interstitium, creating clinical findings of gingivitis. The crevicular fluid changes from transudate to exudate. If allowed to progress, this deteriorates into a vicious, self-perpetuating cycle via damage-associated molecular pathway (DAMP) and pathogen-associated molecular pathway (PAMP).
Innate immunity and microbiome
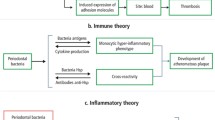
The immune system is a complex, diverse, and dynamic biologic network. Based on the rapidity of onset and the need for recombination of genes, it has two distinct though overlapping branches: innate immunity (congenital, rapid onset, without recombination, and less specific) and adaptive immunity (acquired, less rapid in onset, more specific due to genetic recombination). Both arms of the immune system provide host protection and are major contributors to the integrity and viability of host tissues. That said, however, innate immunity, as the first line of defense, plays a unique role by triggering a crucial systemic response to protect the host and maintain homeostasis. Further, innate defense is pivotal in activation and regulation of adaptive immunity. As stated, innate immunity reacts faster and in a less specific, or semi-specific, manner compared to the adaptive immunity. Importantly, in addition to the immediate responses to the first time challenges, innate immunity is able to elicit amplified responses to second challenges with increased intensity compared to the initial challenges. In fact, these enhanced responses to the second challenges represents what have been currently termed as “Primed Responses,” “Trained Immunity,” or more recently “Innate Immune Memory.” This is important because not only does it challenge the traditional concept that only adaptive immunity is able to develop immunologic memory but it also supports the notion that an immunologic memory can be established by innate immunity that is semi-specific, and without dependent on any specific antigen or stimulator. Interestingly, while innate immunity and concept of potential “innate memory” have attracted intense studies, very few investigations seek to determine whether and how innate immunity develop any form of tolerance. One of the most fundamental tenants in any host defense is the ability to discriminate self from all that are not, and more importantly, those that are safe to keep from ones that can cause harm. Last but certainly not least, during and after the deployment of effector arms to destroy these dangerous or harmful targets, how to minimize the collateral damages. Recent reports reveal that innate immune-induced tolerance is crucial in constraining hyperinflammatory responses and protecting the host against harmful inflammatory complications. The interactions between macrophages and relatively newly discovered innate lymphoid cells (ILCs), particularly group 3 of ILCs, ILC3s, may restore tissue homeostasis and promoting the tolerogenic microenvironment by induction of regulatory T cells (Tregs), as well as dendritic cells and macrophages with counter-inflammatory phenotype, M2.
The complex communities of microbes with over 100 trillion cells and their co-existence within the human body have advanced our understanding of immune system and its reciprocal relationship with the microbiota. In fact, several reports go further to support the notion that the evolution of such elaborated distinct immune network in mammals is to maintain a balance and tuned relationship with microbial flora. As a consequent of this basic symbiotic way of life, while immune system is affecting and preserving the microenvironment for the microbiota, the host microbial communities are re-adjusting and promoting the immune system to be tolerant toward the commensal and useful members of microbiota, and still elicit tuned, necessary responses against the pathologic and harmful microorganisms. This reciprocal interaction between host immune system and the microbiota is an important part of a continuous training scheme by which the immune system is educated to be an ingenious, or more accurately, intelligent system, capable of maintaining both local and systemic homeostasis while preserving host biological integrity. To achieve such, a very finely regulated and highly coordinated interaction between both innate and adaptive arms of immune system on one side and microbiota on the other side is required. It is clear and well documented that the co-existence and development of microbiota is possible only if an immune-tolerogenic mechanism is actively regulating the inflammation and containing the microbiota in its designated physiological niche, achieving tissue integrity at the local level as well as global homeostasis. Ironically, most studies to understand the interaction between host immune system and microbiota have focused on adaptive immunity and its mechanisms (e.g., Tregs induction, PD-1, antibody production, Pleiotropic effects, etc.) by which an “Immune Equilibrium” have guaranteed the symbiosis between host and microbiota (Fig. 2). In reality, considering the complexity of microorganisms in the oral cavity with frequent episodes of food and allergens entering the mouth, and innate immunity as the first line of defense, it is highly reasonable to suggest that a very potent innate tolerogenic mechanism exists to actively monitor and curb the interactions between host innate defense and microbiota. This mutual understanding between host and microbial communities has enabled both sides to co-exist in a symbiotic fashion. Interestingly, the repetitive nature pathogenic and non-pathogenic stimulation within the oral cavity make it plausible to propose, for the first time, that an active “innate tolerogenic memory” maybe responsible for regulating the immune responses in oral cavity, particularly in relation to the commensal microbiota. As such, an orchestrated exertion by components of innate immunity (e.g., macrophages, innate lymphoid cells, ILCs) may establish an immune-homeostasis in oral cavity which in turn provides an optimal condition for microbiota to develop and function. Importantly, this innate tolerogenic memory can be a potential immunotherapeutic target to not only predict but also prevent and treat a variety of inflammatory diseases as well as to help to resolve major problems of healthcare system such as antibiotic resistance.
Symbiosis and dysbiosis are two extremes of the complex relationships between the oral microbial community and the immune responses to its presence. Long-term healthy homeostasis is not possible with excessive immune permissiveness or reactivity to oral microbes as the former fosters microbial overgrowth and virulence and the latter produces destruction through humeral and cellular factors
Periodontal health and diseases
The periodontium is the mechanical and physiological interphase between the alveolar bone and the teeth. Humans, like other mammals, have two sets, a deciduous dentition with 20 teeth followed by permanent dentition with 32 teeth. The principle attachment from bone to cementum (outer calcified tissue over the root surfaces) is the periodontal ligament. The elements bearing tensile stress are Sharpe’s fibers, originating from alveolar bone at a more coronal position (closer to the crown of the tooth) to insert in cementum at a more apical location (closer to the root apex). The intra oral soft tissue lining consists of pink stippled gingiva and red, smooth oral mucosa, meeting at the muco-gingival junction, or the MGJ. The gingiva has two parts: attached gingiva and free gingiva. The space between the tooth surface and free gingiva is the gingival crevice or sulcus, normally less than 3 mm and not inflamed. It contains a liquid transudate called crevicular fluid. When inflamed, this fluid changes to an exudate. The attached gingiva contains many hemidesmosomes to adhere very firmly to the buccal and labial surfaces of the alveolar bone, creating the surface stippling. The mastication of food generates significant shear stress and attached gingiva, under normal conditions, can withstand such mechanical challenges. To masticate, the lower teeth close against the upper teeth by the coordinated action of the temporalis, masseter, and medial pterygoid, with molars bearing most of the compressive forces. These muscles can generate a maximum bite forces between 100 to 120 N in the molar regions in human. [13] The morphology of the incisors and canines allows them to disarticulate the molars as the lower jaw glides forward or sideways. Too much force can cause destruction of the periodontal support, known as primary occlusal traumatism. With bacterial invasion, eroding the normal support, even physiologic chewing forces can cause tissue destruction, known as secondary occlusal traumatism. While both conditions are pathological, oral pathobionts play a much larger role in the latter, especially if the host happens to have susceptible tissue types, such as HLA DR4 [14].
The microbial species colonizing the pliable soft tissue are different from those that form over the rigid surfaces. S. mutans for example, cannot establish themselves without hard surfaces. Young infants before tooth eruption have low to no S. mutans. Recent investigations of infants receiving acrylic naso-alveolar molding plates as initial managements of cleft lip and palate before dental eruption demonstrated significant increase in S. mutans. This supports the concept that, at least in this case, it is not the chemical composition but the mechanical properties that determine the microbial species. As plaque builds up, calcium deposits form, firmly attached to the surface of enamel or cementum, providing mechanical hindrance for cleansing and more rigid surfaces for bacterial to anchor and grow. The niches within the plaque increases as the external part of the microbial communities become progressively more different from the internal milieu. Many other variables participate in this bacterial growth determination, including oxygen tension, nutrient type and availability, hydrogen ion concentration, other microbial species present, and importantly, host immune responses. The microbes show cooperative as well as competitive behaviors: some are high producers of key substrates for the colony while others make adhesive matrices to provide physical supports. The emergent effect is increase in colony fitness [15]. In health, a state of sustained homeostasis exists with the microbial population and composition, remaining in cyclic dynamic equilibrium: elimination balances accumulation and immune activation balanced by suppression [16]. Like most mixed competitive-cooperative conditions, first occupants have the advantage. Since the total available surface area and space are limited and only one physical entity can occupy any given area or space, allowing commensal microbes to form the inaugural microbiota is very important. If a mutualistic existence between the host and microbes is present first, the pathobionts must compete for the same area and space—similar to the new entrants attempting to penetrate a mature, established market. This occupation by “good microbes” resisting colonization by “bad microbes” is colonization resistance. Unfortunately, the microenvironment, the behaviors and compositions of microbes, host immune defenses change over time. “Good” or “bad” can be ephemeral, depending on the perspective and context. In labor economics, the threshold for change in established mature market facing new entrants is the compensating differential: sufficient additional advantages imparted by and to the new entrants to tip the existing balance [17]. Without proper behavioral intervention known collectively as oral hygiene, oral microbes increase with time. This increase comes with changes in composition, often progressing to pathological states if continued. The innate immune responses result in inflammatory mediators such as IL-1 and TNF-α causing destruction of periodontal attachments and bone loss [18]. This type of obvious weakness is uncommon in biology, as evolution would have not permitted the continued existence of organisms with this kind of fault. However, two relevant observations, both valid, may provide the explanation: periodontal disease is very new, appearing only 10,000 years ago, and we humans live much longer than other mammals similar in size to us. Culture evolution is Lamarckian: fast, directed, and learnable, far outpacing Darwinian evolution, which is slow and random. As Neolithic humans transitioned from hunter-gatherer to an agrarian existence, their diet changed. Four key changes came with this transition. First, increase in carbohydrate intake in the form of starch. Second, more frequent and regular food intake rather than episodic feeding with prolonged fasting. Third, reduction in varieties of both animal proteins and plant types. Fourth, closer living conditions and with larger populations. Each of the four contributes to the development of oral diseases including periodontitis. Oral hygiene practices such as chewing plant roots, branches, or leaves appeared contemporaneously with this transition. This is a good example of the Baldwin effect. A learned behavior, transmitted across generations, can overcome environmental challenges without genetic solutions. This underpins the critical dependence of oral hygiene to prevent oral dysbiosis. Another, more novel, adjunct is to engineer the microbiota by introducing directly commensal bacteria such as bifidobacteria and lactobacilli to benefit the host [19].
Systemic implications
There is little doubt that oral microbiota has significant systemic effects, from the early days after conception to aging and senescence. Our microbiome has co-evolved with us, with clear evidence of both vertical and horizontal transfers [20]. At the individual level, the initial transfers of microbes are maternal in origin, either directly through the cervical plug or indirectly by blood across the placenta. These early microbial transfers are permissible partly due to the immune suppressive nature of the fetal stem cells and NK cells within the umbilical cord. If microbial population exceeds a certain threshold, spontaneous abortion will occur with expulsion of the fetus and all related extra-embryonic structures. Prospective randomized control trial has clearly shown that reducing abnormal bacterial population in the mother reduces both spontaneous abortion and preterm labor [21]. Increase in amniotic fluid inflammatory mediators such as TNF-α and IFN-γ due to microbial presence is part of the complex mechanism that initiates labor. Importantly, maternal infection and immune activation during pregnancy can also alter the neurobehavioral development of the infant such as autism and schizophrenia [22]. Recent studies have highlighted the importance of the MHC loci of chromosome 6 in schizophrenia and may lead to possible specific intervention [23].
The method of delivery greatly affect the initial oral microbiota; vaginal delivery tends to load the baby with genitourinary flora like lactobacilli while babies delivered by C-section have more skin flora such as staphylococci and streptococci. Mothers continue to influence the babies’ microbiota, especially if they breastfeed. There is a bi-directional traffic: infants’ oral flora will reflux into mothers’ milk ducts while milk leukocytes and maternal bacteria enter the babies’ mouth and GI tract. These maternal transfers help set up and shape the infants’ developing immune system and their oral microbiome. The recent discovery of milk ILC strengthens this important concept [24].
Throughout childhood and adolescence, the oral microbiota continues to evolve, depending on dental eruption, which provides hard surface for bacterial attachment, and multitude of other factors such as diet, oral hygiene habits, exercise, body mass index, host HLA, and the socioeconomic status [25]. Obviously, some of these variables can affect one another in complex cause-effect loops and do not lend themselves easily to traditional reductive study designs. Still, recent evidence clearly shows the presence of oral bacteria in distant systemic sites [26]. Aspiration of oral content occurs and causes well-described anaerobic pulmonary infection and abscesses [27]. Atherosclerosis can result from bacterial overgrowth and identification of microbes in the atheromatous plague of major arteries first appeared in the literature in the mid-1990s, with early links between bacterial eicosanoids and arthrosclerosis reported in 1988 [28].
Sustained immune response with chronic inflammation is one of the principle causes of cancer. An important and inevitable consequence of sustained inflammation is perturbation in tissue dynamics. Disturbance in tissue dynamics increases the proliferation rate and leads to mucosal hyperplasia, which can progress to frank malignancy [29]. This chronic inflammation to carcinogenesis sequence is present in many sites from oral cancer to intestinal polyp-carcinoma spectrum, to Marjolin’s ulcer arising from non-healing wounds. Oral microbiota thus has great impact on general health [30].
Conclusions and expert recommendations
The masticatory system with all its soft and hard tissues components must perform two important functions: break up the food ingested and remain stable to carry this function for the lifetime of the individual. There is no apposition of enamel and external dentine after the initial formation of the tooth. The wear from chewing, on the other hand, removes these hard tissues relentlessly on the occlusal (incisal) and buccal (labial) surfaces. Taken together, this means there is an inevitable attrition of the dentition, even with dental restorations. This inevitability and the alterations in diet and systemic factors usher in the associated changes of oral microbiota. The oral microbes form complex ecologies within the mouth and require repeated periodic cleansing to prevent overgrowth and dysbiosis. No other tissue or organ in the human body has such behavioral dependence for its maintenance. These complex oral ecologies take various forms from biofilms on enamel and dentinal surfaces to anaerobic polymicrobial communities within gingival crevice and periodontal pockets. Without proper oral hygiene, microbial overgrowth and compositional change will develop over time, triggering immune response that may exacerbate the tissue damage and loss of supporting bone, especially for those with susceptible HLA types. This impairs the tooth stability and creates new niches for more pathobionts, setting up an escalating vicious spiral. Once initiated, the microbiota select out progressively more virulent and synergistic pathogens, evading ineffective or eliciting frankly destructive immune defenses. Such dysbiotic oral microbiota cause dental caries, gingivitis, periodontitis, dental abscess, and tooth loss. In addition, they can exert significant distant effects on the cardiovascular, gastrointestinal, hepato-biliary, pancreatic, neuroendocrine, and genitourinary systems [31]. The patient’s own immune system plays both suppressive and supportive roles in this complex pathogenesis, depending on the HLA genotype and epigenetic changes through histone acetylations and methylations. Going forward, patients with gingivitis and early periodontitis should have tissue typing to determine if the personal HLA subtypes are prone to excessive immune activations. They should also have complete characterization of the dental plague and calculus obtained from scaling, root planning, and curettage, using culture-independent assays such as 16S rRNA sequencing-based methods and megagenomics by pyrosequencing. Selective use of antibiotics and targeted alterations in innate immune profiles, tailored to each based on the above personal data, can permit the prevention in some and predicting disease progression in others, allowing for a personalized treatment to return to a more commensal microbiome with just the right immune, inflammatory response. Introduction of commensal bacteria (probiotics) has proven to be an important component of this immune restoration [32]. The individualized, personal, approaches described above follow the principles of EPMA [33]. When combined with evidence-based, data-driven, standardized multidisciplinary protocols following the ERAS principles [34], we can significantly improve global oral health.
References
Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–17.
Brunski JB, Puleo DA, Nanci A. Biomaterials and biomechanics of oral and maxillofacial implants: current status and future developments. Int J Oral Maxillofac Implants. 2000;15(1):15–46.
Sharpton TJ. An introduction to the analysis of shotgun metagenomic data. Front Plant Sci. 2014;5:209.
Beckers HJ, Van der Hoeven JS. Growth rates of Actinomyces viscosus and Streptococcus mutans during early colonization of tooth surfaces in gnotobiotic rats. Infect Immun. 1982;35(2):583–7.
Yu JC, Khodadadi H, Malik A, Davidson B, Salles ÉD, Bhatia J, et al. Innate immunity of neonates and infants. Front Immunol. 2018;9.
Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. 2005;51(4):270–4.
DiGiulio DB, Romero R, Kusanovic JP, Gómez R, Kim CJ, Seok KS, et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol. 2010;64(1):38–57.
Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69(1):1–0.
Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci. 2010;107(26):11971–5.
Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ Jr., Communication among oral bacteria., Microbiol Mol Biol Rev. 2002;66(3):486–505
Simmerman E, Qin X, Marshall B, Perry L, Cai L, Wang T, et al. Innate lymphoid cells: a paradigm for low SSI in cleft lip repair. J Surg Res. 2016;205(2):312–7.
Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–32.
Kikuchi M, Korioth TW, Hannam AG., The association among occlusal contacts, clenching effort, and bite force distribution in man., J Dent Res. 1997;76(6):1316–25
Marotte H, Farge P, Gaudin P, Alexandre C, Mougin B, Miossec P. The association between periodontal disease and joint destruction in rheumatoid arthritis extends the link between the HLA-DR shared epitope and severity of bone destruction. Ann Rheum Dis. 2006;65(7):905–9.
Palmer TM, Doak DF, Stanton ML, Bronstein JL, Kiers ET, Young TP, et al. Synergy of multiple partners, including freeloaders, increases host fitness in a multispecies mutualism. Proc Natl Acad Sci USA. 2010;107(40):17234–9.
Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8(7):481–90.
Rosen S. Hedonic prices and implicit markets: product differentiation in pure competition. J Polit Econ. 1974;82(1):34–55.
Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol. 2003;74(3):391–401.
Bubnov RV, Babenko LP, Lazarenko LM, Mokrozub VV, Spivak MY. Specific properties of probiotic strains: relevance and benefits for the host. EPMA J. 2018;9(2):205–23.
Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–4.
Ugwumadu A, Manyonda I, Reid F, Hay P. Effect of early oral clindamycin on late miscarriage and preterm delivery in asymptomatic women with abnormal vaginal flora and bacterial vaginosis: a randomised controlled trial. Lancet. 2003;361(9362):983–8.
Heijtz RD. Fetal, neonatal, and infant microbiome: perturbations and subsequent effects on brain development and behavior. In seminars in fetal and neonatal medicine 2016 Dec 1 (Vol. 21, no. 6, pp. 410-417). WB Saunders.
Amare AT, Schubert KO, Baune BT. Pharmacogenomics in the treatment of mood disorders: strategies and opportunities for personalized psychiatry. EPMA J. 2017;8(3):211–27.
Baban B, Malik A, Bhatia J, Yu J. Presence and profile of innate lymphoid cells in human breast Milk. JAMA Pediatr. 2018;172(6):594–6.
Sampaio-Maia B, Monteiro-Silva F. Acquisition and maturation of oral microbiome throughout childhood: an update. Dental Res J. 2014;11(3):291–301.
Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 2014;5(2):e01012–4.
Maddi AB, Scannapieco FA. Oral biofilms, oral and periodontal infections, and systemic disease. Am J Dent. 2013;26(5):249–54.
Yazawa K, Araki K, Okazaki N, Watanabe K, Ishikawa C, Inoue A, et al. Production of eicosapentaenoic acid by marine bacteria. J Biochem. 1988;103(1):5–7.
Yu JC, Cai L, Wang TH, Berdel HO, Lee JH, Lam PS, et al. Tissue dynamics: lessons learned from sutural morphogenesis and cancer growth. Ann Plast Surg. 2016;77:S87–91.
Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18(2):109–20.
Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582–8.
Bubnov RV, Spivak MY, Lazarenko LM, Bomba A, Boyko NV. Probiotics and immunity: provisional role for personalized diets and disease prevention. EPMA J. 2015;6(1):14.
Golubnitschaja O, Costigliola V. Dental health: EPMA recommendations for innovative strategies. EPMA J. 2014;5,S1:A119
Ljungqvist O, Scott M, Fearon KC. Enhanced recovery after surgery: a review. JAMA Surg. 2017;152(3):292–8.
Author information
Authors and Affiliations
Contributions
JCY contributed to the scientific conception, preparations, writing, and editing. HK participated in preparations and writing. BB contributed to the scientific conception, preparations, writing and editing as well as participated in the sequence alignment.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethical approval
This article is a mini-review and does not contain any studies with human or animal performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yu, J.C., Khodadadi, H. & Baban, B. Innate immunity and oral microbiome: a personalized, predictive, and preventive approach to the management of oral diseases. EPMA Journal 10, 43–50 (2019). https://doi.org/10.1007/s13167-019-00163-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-019-00163-4