Abstract
Natural environmental gradients provide important information about the ecological constraints on plant and microbial community structure. In a tropical peatland of Panama, we investigated community structure (forest canopy and soil bacteria) and microbial community function (soil enzyme activities and respiration) along an ecosystem development gradient that coincided with a natural P gradient. Highly structured plant and bacterial communities that correlated with gradients in phosphorus status and soil organic matter content characterized the peatland. A secondary gradient in soil porewater NH4 described significant variance in soil microbial respiration and β-1-4-glucosidase activity. Covariation of canopy and soil bacteria taxa contributed to a better understanding of ecological classifications for biotic communities with applicability for tropical peatland ecosystems of Central America. Moreover, plants and soils, linked primarily through increasing P deficiency, influenced strong patterning of plant and bacterial community structure related to the development of this tropical peatland ecosystem.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Patterns of plant and soil microbial community structure across environmental gradients and relationships with ecosystem function are of primary interest in ecology, especially for developing predictions of environmental change (van der Heijden et al. 2008). This is a challenging goal because the growth and activity of plants and microbes operate on different temporal and spatial scales and vary in their response to resource availability and other environmental drivers (Hacker and Bertness 1999; Harrison et al. 2007). Studies that describe how plant and soil microbial communities correlate with ecosystem structure and function are needed in a multitude of ecosystems and, where possible, in the context of natural environmental variation. Natural gradients of ecosystem development provide an important framework for investigating patterns structuring ecosystems.
Several theoretical and empirical studies, along with some comprehensive reviews, suggest the importance of nutrient status in modulating plant-soil relationships and thus the potential for coupling between plant and microbial community composition (Aerts et al. 1999; Hooper et al. 2000; Ehrenfeld et al. 2005). These patterns are often explained by plant source effects via litter and root exudates that modulate the availability of a variety of labile organic constituents (i.e. Meier and Bowman 2008; Badri and Vivanco 2009). In peatland ecosystems, plant-soil feedbacks suggest a strong linkage between plant and microbial communities that covary with soil properties (Fisk et al. 2003; Peltoniemi et al. 2009; Dimitriu and Grayston 2010). In tropical peatlands, interdependence between soil and plant community structure has also been cited (Page et al. 1999; Troxler 2007) however relationships between plant and microbial community structure along environmental gradients have not been well studied.
The proximate control in ombrogenous peatland environments is hydrology, especially water source, which in turn influences soil chemistry (Bridgham and Richardson 1993). Development of ombrogenous peatlands in high latitudes is thought to have initiated with shifts in early Holocene climate (Tarnocai and Stolbovoy 2006) and has been used to explain bog initiation in some tropical areas (Anderson 1983; Phillips et al. 1997; Domaine et al. 2011). In temperate peatlands, limiting gradients of nutrient availability, hydrology and alkalinity along with low temperatures have been shown to be the principle drivers of peatland ecosystem structure (Bragazza et al. 2005). Chemical characteristics of peatland vegetation have also been recognized as limiting decomposition (Yavitt et al. 1997; Rydin and Jeglum 2006). Coincidentally, peatlands often have plant communities which are highly structured around nutrient status, oftentimes phosphorus (P) (Bridgham and Richardson 1993; Feller et al. 2002; Sjogersten et al. 2011). Notably, peatlands exhibit a special case of ecosystem development that runs contrary to classic soil development theory as related to phosphorus (Walker and Syers 1976). As opposed to soil weathering that reduces mineral phosphorus over time, peatland development results in increasing peat depth and nutrient deficiency resulting from increasing influence of atmospheric deposition as the sole nutrient source and as the peat surface simultaneously “raises” over mineral soil (Rydin and Jeglum 2006). Thus, feedbacks between plants and soils responding to peatland development and increasing P depletion over time suggest that plant and bacterial community structure would covary with P status (Aerts et al. 1999; Ehrenfeld et al 2005).
In this study, our goal was to determine how plant and bacterial communities covaried with environmental gradients structuring the peatland. We also sought to examine how soil microbial function (measured as enzyme activities and respiration) correlated with environmental gradients structuring the peatland, focusing on the natural P gradient. We hypothesized that plant and bacterial community structure and function would covary with phosphorus status along the ecosystem development gradient. Our work builds on previous studies describing biogeochemical characteristics of a tropical peatland (Troxler 2007; Sjogersten et al. 2011) by explicitly linking variation in community structure of canopy species and soil bacteria to environmental gradients that developed with peatland succession.
Materials and Methods
Study Site
In the province of Bocas del Toro, Panama, the Changuinola peat deposit developed on the western bank of Almirante Bay and is 90 km2 (Phillips et al. 1997). A large portion of this wetland, San San-Pond Sak, is a designated Wetland of International Importance (CWII 1971). Initiated by early Holocene climatic changes in the Caribbean region, this peatland is characterized by a P gradient that has been shown to be associated with the sequential development of a tropical ombrotrophic peat bog leading to progressive P depletion over the last 10,000 years (Phillips et al. 1997; Troxler 2007). Average maximum monthly temperature is 31.4°C. Average monthly rainfall is 288 mm with annual precipitation of about 3.5 m (World Meteorological Association; derived from 1971–2000 monthly averages). While the peatland is proximal to the coast, surface water from a canal excavated at its coastward edge shows salinity values less than 0.1 ppt (Davis, unpublished data). High annual precipitation, evenly distributed throughout the year, influences the peatland’s freshwater character (Anderson 1983; Phillips and Bustin 1996).
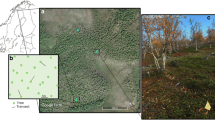
In 2005 and 2006, we characterized bacterial community structure, bulk soil properties, porewater dissolved constituents, conductivity, soil enzyme activities and soil microbial respiration at nine sites that paralleled canopy plots established at five sites along the peatland development gradient. Although the first site along the coastal end-member is referred to as “near-coast”, this site is 300 m from a nearby canal used to access the peatland and >500 m from the Caribbean sea (Fig. 1a). Previous work demonstrated that this strong soil P gradient corresponded with observations of changing canopy species composition and height of aboveground biomass with a vegetation community that graded into a Sphagnum bog (Fig. 1; Phillips and Bustin 1996; Phillips et al. 1997; Troxler 2007). This pattern has also been reported for some tropical peat swamps of Malesia (Anderson 1983).
Soil phosphorus (P) resource gradient with approximate distribution of plant successional communities in the coastal peatland study area near Changuinola, Panama. A) The white triangle outline corresponds to decreasing soil P concentration s with distance into the interior peatland from the coast. Black boxes mark approximate locations of 0.25 ha canopy plots. White stars mark soil sampling locations and the distal ends of the transect. Aerial image made available by ANAM (2004). B) Pictorial description of general plant community types occurring along the soil P gradient
Plant Community Structure and Nutrient Status
Along a 2.7 km transect, soil samples were collected from each of nine stratified, randomly selected 1 m2 plots. Along this same transect, five stratified, randomly selected 0.25 ha plots were established with five of the nine 1 m2 soil sampling plots co-located within each of the 0.25 ha plots (Fig. 1a). The other four soil sample locations were conducted in 1 m2 plots intervening these 0.25 ha plots along the transect. Each 0.25 ha plot was subdivided into four 625m2 quadrants, and individual trees ≥10 cm in diameter at breast height (DBH) were measured, identified to species, and tagged (Condit 1998). Raphia taedigera (Mart.) is a clumping palm, and stem density was determined from number of individuals found in each plot. Given the morphology of R. taedigera, DBH was difficult to determine for this species. We thus used number of stems per individual and approximate diameter per stem to estimate average DBH for the plot where R. taedigera was found (0.3 km). Abundance for each species was determined relative to the total number of individuals in all canopy plots. Canopy diversity and richness were calculated using the Shannon-Wiener (H′) index. Nutrient status and 15N values of canopy vegetation [C, nitrogen (N) and P content of live leaves] from the nine sites was drawn from previous work (Troxler 2007).
Bacterial Community Structure
Bacterial community structure of surface soils was characterized in each of the nine soil sampling sites along the transect. Five samples within a 1 m2 subplot at each site were collected from a depth of two cm using a 2 cm diameter PVC tube. Sub-samples were composited, mixed and preserved with RNAlater (Quiagen, Valencia, CA, USA; used for preservation of both RNA and DNA, Gorokhova 2005), stored frozen overnight, transported to Florida International University in an iced cooler within 24 h of collection, and frozen at −80°C until processed. Because of the remote location of the study site and impossibility of immediate preservation under −80°C conditions, RNAlater was used as an additional method of preservation. Bacterial structure was then determined with Polymerase Chain Reaction amplification of 16S rRNA gene sequences combined with Denaturing Gradient Gel Electrophoresis (PCR-DGGE). PCR-DGGE characterizes the rDNA fragments from those species that dominate the community and is very useful for identifying changes in community structure (Muyzer and Smalla 1998; Ikenaga et al. 2010). See “Electronic Supplementary Materials” for further details.
Using information about sequenced and cloned bands, we characterized the relative dominance of ribotypes for each site. We calculated bacterial diversity using the Shannon-Wiener (H′) index. While this is not a true measure of bacterial species diversity, we employed it as a useful index to compare the community structure of dominant ribotypes across sites (Kowalchuk et al. 2002). All of the DGGE bands were used in diversity estimates.
Soil Enzyme Activities, Microbial Respiration and Chemical Properties
Soil samples were collected at these same nine sites with a 2.5 cm diameter serrated plastic coring tube. A subsample was first dried to a constant weight at 70°C for soil moisture determination. A second subsample was slurried by adding a volume of water equal to the mass of wet soil after removing bulk roots. Slurries were used in the determination of enzyme activities and microbial respiration (CO2).
We determined soil enzyme activities using 4-methylumbelliferone (MUF) substrates (Chróst and Krambeck 1986; Hoppe 1993) for the determination of four enzyme activities: phosphatase (AP), β-1-4-glucosidase (BGC), β-N-acetylglucosaminidase (BAN) and sulfatase (SUF). The substrates used for each of these enzyme assays were: 4-methylumbelliferyl-phosphate, MUF-β-D-glucoside, MUF–N-acetyl-β-D-glucosaminide, and MUF-sulfate respectively. Enzyme activity was determined as the difference between fluorescent substrate liberated during incubation time (tf) and time zero (t0). Accumulation of fluorescence was monitored over time to ensure linearity. Fluorescence was measured using a Cytofluor 4000 multiwell plate reader (PerSeptive Biosystems, Inc., Carlsbad, CA, USA).
Microbial respiration rates were determined employing the modified method of Amador and Jones (1993). Rates were measured on known mass of replicate soil slurries (approximately 4 g slurry) that were incubated for 40 h in 20 mL gas headspace vials that were purged with CO2-free air prior to incubation. Headspace gas was measured on a gas chromatograph HP 5890 Series II equipped with a flame ionization detector (Hewlett Packard, Palo Alto, CA, USA) after conversion to methane in a Shimadzu MTN-1 Methanizer.
Soil total P, nitrogen (TN) and carbon (TC) analyses were performed using the modified acid-digestion method of Solorzano and Sharp (1980) for TP, and with a Carlo Erba elemental analyzer (ThermoFisher Scientific, Waltham, MA, USA) for TN and TC. Organic matter content was determined by ashing 5 mg oven dried soil at 400°C. Conductivity values were determined on soil slurries using a YSI handheld conductivity meter (YSI Inc., Yellow Springs, OH, USA). Soil porewater was analyzed for concentrations of soluble reactive phosphorus (SRP), nitrate + nitrite (NO3 + NO2), ammonium (NH4) and dissolved organic carbon (DOC) on samples collected with a porewater sipper (aquarium filter stone inserted to 10 cm depth, attached to fine Tygon® tubing and extracted with a 120 mL syringe). Filtered (Whatman GF/F) water samples were analyzed for SRP, NH4, NO3 + NO2, and NO2 concentrations using a four-channel auto-analyzer (Alpkem model RFA 300; Technicon Instruments, Tarrytown, NY) at the Florida International University Southeast Environmental Research Water Quality Laboratory.
Data Analysis
We used the data reduction technique of Principal Components Analysis (PCA) using a correlation matrix to examine the variance associated with soil abiotic structure (n = 9; excluding salinity, pH and peat depth). We applied a varimax rotation to the eigenvectors to improve the structure for the components. We then used the soil parameters that explained the variance of each axis to determine how plant and bacterial community structure (including canopy taxa and soil bacteria ribotypes) and soil microbial function (enzyme activities and soil respiration) varied along environmental gradients described using best fit regression models. PCA and regression analyses were performed in JMP 7.0 (SAS Institute, Cary, NC, USA). Cluster analyses were performed for all DGGE bands of the soil bacterial community using the Ward method in R (R Development Core Team 2008) with box plots describing the average variation in soil TP (μg P g−1 soil) among four clusters. We used an arcsine-square root transformation for relative abundances of canopy species and performed statistical tests with log-transformed data where appropriate.
Results
Summary of Plant and Bacterial Community Structure and Soil Parameters
Canopy species identified at the five 0.25 ha plots with DBH 10 cm or greater were Campnosperma panamensis, Symphonia globulifera, Euterpe precatoria, Ilex guiensis, Ardisia sp. Cassipourea elliptica, Alchornea latifolia, Raphia taedigera and Cyrilla racemiflora. R. taedigera was found in one site only (0.3 km). At the 0.3 km site, average DBH was 53.7 cm ± 0.3 and 23 cm ± 1.0 with and without (i.e. hardwoods and solitary palms only) R. taedigera DBH estimates, respectively (Table 1). In further analyses, we did not include R. taedigera DBH values because of the uncertainty associated with how DBH was estimated for this species. Overall, average DBH declined with distance toward the interior. Stem density was fairly similar along the transect but was lowest at 2.7 km into the mire interior. In both near-coast (0.3 km) and most developed bog (2.7 km) sites, a single tree species dominated species composition (R. taedigera and C. racemiflora, respectively). Canopy species richness and diversity were lowest in the 0.3 and 2.7 km sites. Canopy tissue P and N concentrations were included from Troxler (2007; Table 1).
A greater number of bands was identified with the primer set 357F-GC and 517R (first primer set; n = 72) as compared with 968F-GC and 1401r (second primer set; n = 51; Tables 1 and 2). In general, diversity of dominant bands was higher for the second primer set, but were more variable across the transect using the first primer set. Sequencing the dominant bands, five bacterial phyla were identified with the first primer set identifying more bands of unique but undetermined phylogenetic classification. Acidobacteria were the most abundant operational taxonomic units (OTU) or ribotypes identified, regardless of primer set used (Table 1). Using the second primer group, dominant bands within the proteobacteria phylum were identified to class, including α-, β-, δ- and γ-proteobacteria.
With respect to general soil characteristics, inland sites had lower total nutrients, porewater SRP and NO3 + NO2, generally higher phosphatase activity and soil moisture and greater peat depths (Table 1). In soil porewater, the near-coast sites (0.3 km and 0.6 km) had higher SRP, NO3 + NO2 and conductivity but lower DOC. Overall, there was low variability in pH (3.26 ± 0.22) with all values below 4.0. Variability in organic matter content and enzyme activities (except AP) generally did not appear to covary with peatland development (i.e. distance along the transect).
Principal Components Analysis
Incorporating supplementary data sources, we used the dataset of soil parameters (Table 1; salinity, pH and peat depth were excluded because of fewer data points, n = 5) to reduce dimensionality in the dataset so that broad-scale environmental gradients could be identified. The PCA reduced the dataset to four principal axes that described 92 % of the variance (Fig. 2). The four factors had eigenvalues greater than 1 and were more useful in parsing out environmental gradients within the dataset as compared with the unrotated factors.
Factor 1 described 33.5 % of the variance and was strongly correlated with soil P (r = 0.85, p = 0.0080) and soil N:P (r = −0.97, p < 0.0001). Sites that loaded positively on this axis were at the near-shore end member of the transect (0.3 km, 0.6 km and 0.9 km sites). Factor 2 described an additional 26 % of the variance and was positively correlated with percent organic matter (r = 0.90, p = 0.0025) and negatively correlated with porewater DOC (r = −0.85, p = 0.0075). This axis was differentiated by 0.3 km and 0.6 km sites with higher %OM and lower DOC that contrasted 0.9 km, 1.5 km and 1.8 km sites. Factor 3 was positively correlated with soil N (r = 0.92, p = 0.0012) and negatively correlated with soil C:N (r = −0.94, p = 0.0005) and described 20.9 % of the variance. Patterns of soil N and soil C:N in the peatland grouped 1.8 km and 2.4 km sites with lower soil N (and higher C:N) in contrast to 0.6 km and 2.1 km sites. Finally, factor 4 was positively correlated with porewater NH4 (r = 0.95, p = 0.0003) and partitioned an additional 11.7 % variance in the dataset. This factor grouped sites that were highly differentiated by other components, especially soil P (0.3 km, 1.5 km, and 2.7 km).
Variation in Community Structure of Canopy Species
Canopy biomass (DBH) was related to soil P in a positive, linear relationship (r2 = 0.86, F = 17.78, p = 0.024; Fig. 3a). Soil P explained 95 % of the variance in canopy diversity in a polynomial relationship (F = 21.48, p = 0.044; Fig. 3b). There was a corresponding pattern in richness, but this relationship was not significant (r2 = 0.89, F = 7.98, p = 0.1113; Fig. 3b). Soil N:P was negatively related to DBH (r2 = 0.85, F = 16.60, p = 0.0267). Otherwise, soil parameters did not determine strong variation in canopy community structure.
We determined how soil gradients described variance in the relative abundance of canopy species in the peatland. We plotted relative abundance with soil P to describe general trends (Fig. 4a). R. taedigera and C. racemiflora were confined to sites with high and low P soils, respectively. C. racemiflora increased significantly with decreasing soil P within low P sites in a polynomial relationship (r2 = 0.97, F = 30.15, p = 0.032) whereas R. taedigera showed an insignificant trend (i.e. abundant at one site only). We found C. panamensis to increase in relative abundance with decreasing soil P until it dropped to low abundance at lowest soil P in a non-significant, polynomial trend (p > 0.2). Both E. precatoria and C. elliptica increased in abundance with increasing soil P until decreasing to lower abundance or absent at highest soil P (non-significant, polynomial trends). Furthermore, several canopy species increased in abundance with moderate soil P including S. globulifera (p = 0.070) and Ardisia sp. (p = 0.090) in non-significant polynomial trends. Ilex guiensis (p > 0.2) and an unidentified species (p > 0.2) were in low abundance or only present at one site with moderate soil P. Along a secondary soil gradient, C. elliptica, E. precatoria, and Ardisia sp. were negatively related to organic matter content (CE: linear, r2 = 0.82, F = 13.44, p = 0.035; EP: polynomial, r2 = 0.99, F = 691.02, p = 0.001; and ARD: polynomial, r2 = 0.88, F = 49.85, p = 0.020). S. globulifera followed this trend (p = 0.162) in a non-significant relationship. There was also a strong relationship between an unidentified species and organic matter content with high relative abundance at moderate %OM (r2 = 0.96, F = 27.40, p = 0.035). Relationships with soil porewater DOC illustrated positive and negative polynomial relationships with R. taedigera and I. guinensis (r2 = 0.99, F = 226.80, p = 0.004 and r2 = 0.94, F = 17.45, p = 0.054, respectively), but these relationships were based on presence in a single site.
Variation in Bacterial Community Structure
Bacterial community composition clustered strongly along the soil P gradient (Fig. 5). Bacterial communities showed similar changes regardless of primer set used. Cluster analyses of soil bacteria displayed taxon assemblages (OTU) that grouped according to the range in soil P level: 1) low: ~400, 2) low-moderate: 600–800, 3) moderate: 900–1000, and 4) high: 1100–1250 μg P g−1 soil, respectively (Fig. 5). Applying our index of bacterial diversity using the first primer group illustrated a positive, linear relationship with soil P (r2 = 0.54, F = 7.19, p = 0.036; Fig. 6a). Further, soil N:P described 76 % of the variance in bacterial diversity (F = 18.93, p = 0.005; Fig. 6b). Patterns in bacterial diversity were similar using the second primer group with the exception of 2.4 km and 2.7 km sites which exhibited among the higher diversity values than the sites with moderate soil P concentration. Other soil gradients were not important in describing variation in bacterial diversity.
We used results from the first primer group to determine how ribotypes varied along the soil gradients structuring the peatland (Fig. 4b). We found positive, polynomial relationships with soil P for several bacterial ribotypes including two unidentified taxa w (r2 = 0.88, F = 19.40, p = 0.004) and 2x (2x: r2 = 0.95, F = 49.99, p = 0.001), Holophaga sp. (3b: r2 = 0.91, F = 26.94, p = 0.002), an Hyphomicrobiaceae bacterium (3i: r2 = 0.93, F = 33.21, p = 0.001), an Acidobacteria bacterium (3n: r2 = 0.82, F = 11.71, p = 0.013) and Alpha-proteobacterium (3q: r2 = 0.97, F = 87.17, p = 0.0001). For another Acidobacteria bacterium (2v), there was a unimodal relationship where abundance increased with moderate soil P (r2 = 0.87, F = 17.21, p = 0.006). We also found negative polynomial and linear relationships for two taxa of Acidobacteria bacterium (2 s: r2 = 0.75, F = 7.59, p = 0.031 and 3c: r2 = 0.62, F = 9.83, p = 0.020, respectively) and a linear relationship with an Actinobacteria bacterium (2z: r2 = 0.65, F = 11.09, p = 0.016). Relationships with soil N covaried with soil P for ribotype w and with soil N:P for ribotypes w, 2s, 2z, 3b, 3i, 3m, and 3q. The abundance of two additional unidentified ribotypes (2l and 2x) decreased and another Acidobacteria bacterium increased with increasing soil porewater DOC (2v: r2 = 0.53, F = 8.02, p = 0.025; r2 = 0.71, F = 17.59, p = 0.004; r2 = 0.62, F = 11.46, p = 0.0117, respectively). Otherwise, soil gradients describing variation in peatland ecosystem structure did not describe significant variance in the abundance of soil bacteria ribotypes.
Environmental Gradients and Soil Microbial Function
To determine how soil microbial function varied relative to the environmental gradients described by the PCA, we related parameters describing the soil gradients to enzyme activities and microbial respiration rates. Soil P explained 71 % of the variance in PA in a negative, polynomial relationship (F = 6.23, p = 0.044; Fig. 7a) while soil N:P was positively related to PA (r2 = 0.68, F = 12.95, p = 0.011; Fig. 7b). Neither soil N, C:N, %OM or DOC were important in describing variance in enzyme activities or soil microbial respiration. In contrast, soil porewater NH4 explained approximately 60 and 80 % of the variance in BGC and MR in negative, linear relationships (BGC: r2 = 0.61, F = 10.80, p = 0.013 and MR: r2 = 0.77, F = 19.66, p = 0.004; Fig. 8a and b).
Discussion
Soil Abiotic Gradients, Community Structure and Soil Microbial Function
The Changuinola peatland was characterized by highly structured plant and bacterial communities that correlated with gradients in phosphorus status and soil organic matter content. The P gradient followed a peatland development gradient that covaried with strong relationships in canopy P status, C:P and N:P molar ratios and nitrogen isotope composition indicating increasing P deficiency with peatland development (Troxler 2007). The P gradient (thus considered as a time for space substitution with the development of ombrogeneous conditions) influenced aspects of canopy community structure including aboveground biomass, the relative abundance of C. racemiflora, and to a lesser extent, other canopy taxa including S. globulifera and Ardisia sp. Soil P is often cited as a strong determinant of aboveground biomass in low P systems like peatlands (Bridgham and Richardson 1993; Feller et al. 2002) and C. racemiflora is an evergreen species associated with low nutrient environments. This species has been shown to exhibit high P resorption efficiency (nearly 90 %) in the Okeefenokee swamp of Georgia, USA (DeLucia and Schlesinger 1995) and in pocosin bogs of North Carolina, USA (Walbridge 1991).
The concomitant increase in soil phosphatase activity with decreased soil P is further evidence of increasing P deficiency toward the mire interior (Olander and Vitousek 2000). While other factors may coincide with changes associated with soil development, increasing AP activity is generally explained by increased inputs low in P that are reflected in plant nutrient status (Richardson et al. 2004, Parfitt et al. 2005, Allison et al. 2007). This is consistent with the patterns we found in this and other studies of the Changuinola peatland (Troxler 2007, Sjogersten et al. 2011). Moreover, as hydrology and soil chemistry are implicitly linked in peatland ecosystems, especially with the development of ombrogenous conditions and colonization of Sphagnum, the gradient in P status as an important driver of overall canopy community structure is well founded. The increase in abundance of canopy taxa S. globulifera and Ardisia sp. at moderate soil P coincided with a unimodal relationship between soil P and canopy diversity. We also found an increase in the abundance of two additional canopy taxa with lower soil organic matter, a secondary gradient coinciding with the abundance of rarer canopy species (and moderate soil P; Appendix 1) in this system. Variation in organic matter content often reflects wetland hydropattern (Mitsch and Gosselink 2000) and a drier condition may have resulted in the higher abundance of several species toward the center of the peatland. Phillips et al. (1997) suggested that the interior peatland was raised above the inland and near-coast vegetation communities corroborating this conclusion.
Bacterial community structure was also strongly clustered by P status and bacterial diversity positively covaried with P status. In general, there were fewer OTU present at the 2.7 km site as compared with other sites. Underlying shifts in dominance of several soil bacteria coincided with this diversity pattern. Soil bacteria identified as dominant in this tropical peatland have been found to be abundant in other low latitude peatlands, especially Acidobacteria and α-proteobacteria (Jackson et al. 2009). Generally, Acidobacteria are more frequently associated with soils that are low in labile C (Marilley and Aragno 1999) and considered to be among the oligotrophic bacteria (Fierer et al 2007). However, given the low and well-constrained soil pH values, environmental factors like nutrient status appear to be important in determining dominance of Acidobacteria ribotypes. For example, Dimitriu and Grayston (2010) found the greatest abundance of Acidobacteria to correlate with lowest pH (4–5). The authors suggest that within this narrow, range where growth of Acidobacteria is optimal, other abiotic factors may drive abundance of this group. A study by Clarkson et al. (2005) found that pH was less important than P-limitation because of the limited range of variability in pH values. As pH was constrained to values below 4.0 with little variation among sites, it is concluded to be a less important factor influencing patterns of plant and bacterial community structure and function within this peatland. However, abundance of Acidobacteria ribotypes did not always similarly covary with soil P. Given the limited number of ribotypes identified with each of the primers, assessing variation in bacterial community structure of ribotypes grouped by phylum or subphylum may be misleading without genomic analyses that survey all bacterial ribotypes (i.e. pyrosequencing). We could however assess important variation in community composition and individual ribotypes that was informative in describing covariation in plant and bacterial community structure with the soil P gradient and peatland ecosystem development.
A further key result of this study showed that neither soil microbial respiration nor BGC were correlated with P status, but to porewater NH4, contrary to our expectation. D’Angelo and Reddy (1999) found that microbial respiration correlated with inorganic N and indices of available C but not pH or soil P that was similar to our results. Hydrology is also an important control on microbial respiration rates, but this relationship has not been demonstrated consistently for tropical peatland soils (Chimner 2004; Hirano et al. 2009).
Moreover, a successional sere appears to develop in the peatland center toward a differentiated community type with structure that is more strongly correlated with canopy diversity, organic matter content and intermediate P status. Along the peatland development gradient, several strongly correlating factors suggest that peatland development influences water source and availability, soil P status and subsequently canopy biomass, canopy species abundances, and bacterial community composition. Following this rationale, the development of subsequent gradients that emerge within the maturing peatland may have a more important influence, albeit indirect, on soil C mineralization (Yavitt et al. 2005; Belyea and Baird 2006; Kramer and Holscher 2010).
Developing an Ecological Context for Community Structure in a Tropical Peatland
Studies that explicitly evaluate the influence of nutrient status on biotic communities show the strongest links between plant and microbial community structure and diversity. Recent studies have illustrated how nutrient status or nutrient availability regulate bacterial diversity (Langenheder and Prosser 2008), the effect of plant diversity on fungal diversity (Waldrop et al. 2006), and the interactions of aboveground and belowground communities (Haase et al. 2008). Differential resource supply (i.e. N:P) has also been shown to be well correlated with soil microbial composition (i.e. preferential colonization by fungi and bacteria; Güsewell and Gessner 2008) and functional diversity (Liu et al. 2010). Our study provided several lines of evidence supporting correlated plant and bacterial community structure that largely covaried with a P status gradient that emerged with peatland development.
While the distribution of plant species along environmental gradients is a well-founded area of ecology, insights into the vast diversity of microbial taxa using various molecular profiling techniques have led to significant advances in developing an ecologically useful context for bacterial phyla (i.e. Fierer et al 2007). Only recently has the community structure and species variation of both plants and soil bacteria been evaluated together. Characterizing shifts in community taxa along environmental gradients, especially successional gradients, can provide a meaningful framework for inferring correlated physiological and ecological constraints on plant and bacterial community composition as well as functional significance of bacterial taxa (Dimitriu and Grayston 2010; Nemergot et al. 2010; Philippot et al. 2011). Microbial succession processes are thought to be analogous to mechanisms that drive plant succession including dispersal, resource availability, disturbance and environmental stress (Aerts 1999; Fierer et al. 2010). Our study showed that plant and bacterial community composition were determined by soil P content and indicators of ecosystem nutrient retention and efficient P cycling (AP and canopy C:P) strongly covaried with peatland development. Interestingly, however, we found that plant communities, through changes in aboveground biomass allocation and species shifts, also responded strongly but not necessarily always in similar ways as did bacterial communities.
Considering shifts in canopy species and bacterial taxa along the peatland gradient, we found that community patterns often followed a copiotroph-oligotroph spectrum posed by Aerts (1999) and Aerts et al. (1999) for plants and Fierer et al. (2007) for soil bacteria. From this, we garnered important insights into the ecological tolerances of biotic communities and specific taxa relative to changes in P status. Moreover, our study suggested that strong plant-soil linkages with increasing P deficiency over the time course of tropical peatland ecosystem succession led to strong patterns in both plant and bacterial communities. We found evidence that community composition of the forest canopy and soil bacteria covaried relative to P status, but secondary environmental gradients suggested this was not the case for microbial function related to soil communities. Our study advances a general understanding of the potential for strong plant-soil feedback in peatland ecosystems to include systems of tropical America.
References
Aerts R (1999) Interspecific competition in natural plant communities: mechanisms, trade-offs and plant-soil feedbacks. J Exp Bot 50:29–37
Aerts R, Verhoeven JTA, Whigham DF (1999) Plant-mediated controls on nutrient cycling in temperate fens and bogs. Ecology 80:2170–2181
Allison VJ, Condron LM, Peltzer DA, Richardson SJ, Turner BL (2007) Changes in enzyme activities and soil microbial community composition along C and nutrient gradients at the Franz Josef chronosequence, New Zealand. Soil Biol Biochem 39:1770–1781
Amador JA, Jones RD (1993) Nutrient limitations on microbial respiration in peat soils with different total phosphorus content. Soil Biol Biochem 25:793–801
Anderson JAR (1983) The tropical peat swamps of western Malesia. In: Gore AJP (ed) Ecosystems of the world: mires: swamp, bog, fen and moor. 4B, Regional Studies. Elsevier, New York, pp. 181–199
Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32:666–681
Belyea LR, Baird AJ (2006) Beyond “the limits to peat bog growth”: cross-scale feedback in peatland development. Ecol Monogr 76:299–322
Bragazza L, Rydin H, Gerdol R (2005) Multiple gradients in mire vegetation: a comparison of a Swedish and an Italian bog. Plant Ecol 177:223–236
Bridgham SD, Richardson CJ (1993) Hydrology and nutrient gradients in North Carolina peatlands. Wetlands 13:207–218
Chimner RA (2004) Soil respiration rates of tropical peatlands in Micronesia and Hawaii. Wetlands 24:51–56
Chróst RJ, Krambeck HJ (1986) Fluorescence correction for measurements of enzyme activity in natural waters using methylumbelliferyl-substrates. Arch Hydrobiol 106:79–90
Clarkson BR, Louis AE, Schipper A, Moyersoen B, Silvester WB (2005) Foliar 15N natural abundance indicates phosphorus limitation of bog species. Oecologia 144:550–557
Condit R (1998) Tropical forest census plots: methods and results from Barro Colorado Island, Panama and a comparison with other plots. Springer-Verlag
Convention on Wetlands of International Importance especially as Waterfowl Habitat. Ramsar (Iran) (1971) UN Treaty Series No. 14583. As amended by the Paris Protocol, 3 December 1982, and Regina Amendments, 28 May 1987
D’Angelo EM, Reddy KR (1999) Regulators of heterotrophic microbial potentials in wetland soils. Soil Biol Biochem 31:815–830
DeLucia EH, Schlesinger WH (1995) Photosynthetic rates and nutrient-use efficiency among evergreen and deciduous shrubs in Okefenokee Swamp. Int J Plant Sci 156:19–28
Dimitriu PA, Grayston SJ (2010) Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microb Ecol 59:563–573
Domaine R, Couwenberg J, Joosten H (2011) Development and carbon sequestration of tropical peat domes in south-east Asia: links to post-glacial sea-level changes and Holocene climate variability. Quaternary Sci Rev 30:999–1010
Ehrenfeld JG, Ravit B, Elgersma K (2005) Feedback in the plant-soil system. Annu Rev Environ Resour 30:75–115
Feller IC, McKee KL, Whigham DF, O’Neill JP (2002) Nitrogen vs. phosphorus limitation across an ecotonal gradient in a mangrove forest. Biogeochemistry 62:145–175
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Fierer N, Nemergut D, Knight R, Craine JM (2010) Changes through time: integrating microorganisms into the study of succession. Research in Microbiology 161:635–642
Fisk MC, Ruether KF, Yavitt JB (2003) Microbial activity and functional composition among northern peatland ecosystems. Soil Biol Biochem 35:591–602
Gorokhova E (2005) Effects of preservation and storage of microcrustaceans in RNA later on RNA and DNA degradation. Limnol Oceanogr Meth 3:143–148
Güsewell S, Gessner MO (2008) N:P ratios influence litter decomposition and colonization by fungi and bacteria in microcosms. Funct Ecol 23:211–219
Haase J, Brandl R, Scheu S, Schädler M (2008) Above- and belowground interactions are mediated by nutrient availability. Ecology 89:3072–3081
Hacker SD, Bertness MD (1999) Experimental evidence for factors maintaining plant species diversity in a New England salt marsh. Ecology 80:2064–2073
Harrison KA, Bol R, Bardgett RD (2007) Preferences for different nitrogen forms by coexisting plant species and soil microbes. Ecology 88:989–999
Hirano T, Jauhiainen J, Inoue T, Takahashi H (2009) Controls on the carbon balance of tropical peatlands. Ecosystems 12:873–887
Hooper DH, Bignell DE, Brown VK, Brussaard L, Dangerfield JM, Wall DH, Wardle DA, Coleman DC, Giller KE, Lavelle P, van der Putten WH, DeRuiter PC, Rusek J, Silver WH, Tiedje JM, Wolters V (2000) Interactions between aboveground and belowground biodiversity in terrestrial ecosystems: patterns, mechanisms, and feedbacks. Bioscience 50:1049–1061
Hoppe HG (1993) Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurement of bacteria. In: Kemp PF et al (eds) Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, pp 423–431
Ikenaga M, Guevara R, Dean AL, Pisani C, Boyer JN (2010) Changes in community structure of sediment bacteria along the Florida Coastal Everglades Marsh–Mangrove–Seagrass Salinity Gradient. Microb Ecol 59:284–295
Jackson CR, Liew KC, Yule CM (2009) Structural and functional changes with depth in microbial communities in a tropical Malaysian peat swamp forest. Microb Ecol 57:402–412
Kowalchuk GA, Buma D, de Boer W, Klinkhamer PGL, van Veen JA (2002) Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Anton van Leeuw 81:509–520
Kramer I, Holscher D (2010) Soil water dynamics along a tree diversity gradient in a deciduous forest in Central Germany. Ecohydrology 3:262–271
Langenheder S, Prosser JI (2008) Resource availability influences the diversity of a functional group of heterotrophic bacteria. Environ Microbiol 10:2245–2256
Liu Z, Fu B, Zheng X, Liu G (2010) Plant biomass, soil water content and soil N:P ratio regulating soil microbial functional diversity in a temperate steppe: a regional scale study. Soil Biol Biochem 42:445–450
Marilley L, Aragno M (1999) Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol 13:127–136
Meier CL, Bowman WD (2008) Links between plant litter chemistry, species diversity, and below-ground ecosystem function. Proc Natl Acad Sci 105:19780–19785
Mitsch WJ, Gosselink JG (2000) Wetlands, 3rd edn. Wiley, New York
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Anton van Leeuw 73:127–141
Nemergot DR, Cleveland CC, Wieder WR, Washenberger CL, Townsend AR (2010) Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol Biochem 42:2153–2160
Olander LP, Vitousek PM (2000) Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochemistry 49:175–190
Page SE, Rieley JO, Shotyk OW, Weiss D (1999) Interdependence of peat and vegetation in a tropical peat swamp forest. Philos Trans R Soc Lond B 354:1885–1897
Parfitt RL, Ross DJ, Coomes DA, Richardson SJ, Smale MC, Dahlgren RA (2005) N and P in New Zealand soil chronosequences and relationships with foliar N and P. Biogeochemistry 75:305–328
Peltoniemi K, Fritze H, Laiho R (2009) Response of fungal and actinobacterial communities to water-level drawdown in boreal peatland sites. Soil Biol Biochem 41:1902–1914
Phillips S, Bustin R (1996) Sedimentology of the Changuinola peat deposit: organic and clastic sedimentary response to punctuated coastal subsidence. Geol Soc Am Bull 108:794–814
Phillips S, Rouse G, Bustin R (1997) Vegetation zones and diagnostic pollen profiles of a coastal peat swamp, Bocas del Toro, Panamá. Palaeogeogr Palaeocol Palynol 128:301–338
Philippot L, Tscherko D, Bru D, Kandeler E (2011) Distribution of high bacterial taxa across the chronosequence of two alpine glacier forelands. Microb Ecol 61:303–312
R Development Core Team (2008). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org
Richardson SJ, Peltzer DA, Allen RB, McGlone MS, Parfitt RL (2004) Rapid development of phosphorus limitation in temperate rainforest along the Franz Josef soil chronosequence. Oecologia 139:267–276
Rydin H, Jeglum J (2006) Chapter 7, Peatland succession and development. The Biology of Peatlands, Oxford University Press Inc, New York, USA, pp. 119–137
Sjogersten S, Cheesman AW, Lopez O, Turner BL (2011) Biogeochemical processes along a nutrient gradient in a tropical ombrotrophic peatland. Biogeochemistry 104:147–163
Solorzano L, Sharp J (1980) Determination of total dissolved P and particulate P in natural waters. Limnol Oceanogr 25:754–758
Tarnocai C, Stolbovoy V (2006) Northern peatlands: their characteristics, development and sensitivity. In: Martini IP, Martinez Conrtizas A, Chesworth W (eds) Peatlands: evolution and records of environmental and climate changes, Developments in Earth Surface Processes, 9. Elsevier, New York, pp. 181–199
Troxler TG (2007) Patterns of phosphorus, nitrogen and δ15N along a peat development gradient in a coastal mire, Panama. J Trop Ecol 23:683–691
Van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310
Walbridge MR (1991) Phosphorus availability in acid organic soils of the lower North Carolina coastal plain. Ecology 72:2083–2100
Waldrop MA, Zak DR, Blackwood CB, Curtis CD, Tilman D (2006) Resource availability controls fungal diversity across a plant diversity gradient. Ecol Lett 9:1127–1135
Walker TW, Syers JK (1976) The fate of phosphorus during pedogenesis. Geoderma 15:1–19
Yavitt JB, Williams CJ, Wieder RK (1997) Production of methane and carbon dioxide in peatland ecosystems across North America: effects of temperature, aeration, and organic chemistry of peat. Geomicrobiol J 14:299–316
Yavitt JB, Williams CJ, Wieder RK (2005) Soil chemistry versus environmental controls on production of CH4 and CO2 in northern peatlands. Eur J Soil Sci 56:169–178
Acknowledgments
The authors would like to acknowledge B. Giraldo, G. Jacome, P. Gondola, E. Brown, M. Barry, D. Johnson and I. Mendoza for field, lab and logistical assistance. Drs. S. Davis, D. Liptzin and J. Edmonds provided helpful comments on earlier drafts of this manuscript. We are grateful to the Smithsonian Tropical Research Institute and R. Collins for assistance with travel funding through a fellowship to T. Troxler. This material is based upon work supported by the National Science Foundation through the Florida Coastal Everglades Long-Term Ecological Research program under Grants No. DBI-0620409 and No. DEB-9910514. This is contribution #548 of the Southeast Environmental Research Center at Florida International University.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Electronic Supplementary Materials
(DOC 36 kb)
Rights and permissions
About this article
Cite this article
Troxler, T.G., Ikenaga, M., Scinto, L. et al. Patterns of Soil Bacteria and Canopy Community Structure Related to Tropical Peatland Development. Wetlands 32, 769–782 (2012). https://doi.org/10.1007/s13157-012-0310-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13157-012-0310-z