Abstract
Members of the sea anemone genus Anthopleura are familiar constituents of rocky intertidal communities. Despite its familiarity and the number of studies that use its members to understand ecological or biological phenomena, the diversity and phylogeny of this group are poorly understood. Many of the taxonomic and phylogenetic problems stem from problems with the documentation and interpretation of acrorhagi and verrucae, the two features that are used to recognize members of Anthopleura. These anatomical features have a broad distribution within the superfamily Actinioidea, and their occurrence and exclusivity are not clear. We use DNA sequences from the nucleus and mitochondrion and cladistic analysis of verrucae and acrorhagi to test the monophyly of Anthopleura and to evaluate the pattern of distribution of acrorhagi and verrucae. We find that Anthopleura is paraphyletic: although species of the genus cluster together, some groups also include members of genera like Bunodosoma, Aulactinia, Oulactis, and Actinia. This paraphyly is explained in part by the discovery that acrorhagi and verrucae are pleisiomorphic for the subset of Actinioidea studied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Anthopleura Duchassaing de Fonbressin and Michelotti, 1860 (Cnidaria: Anthozoa: Actiniaria: Actiniidae) is one of the most familiar and well-known genera of sea anemones. Its members are found in both temperate and tropical rocky intertidal habitats and are abundant and species-rich when present (e.g., Stephenson 1935; Stephenson and Stephenson 1972; England 1992; Pearse and Francis 2000). Due to their diversity and abundance in predictable and accessible places, Anthopleura is the subject of many field studies of rocky intertidal ecology and physiology, including studies of thermal stress and nutrient transfer (e.g., Jennison 1978; Kruger and Griffiths 1998; Richier et al. 2008; Hiebert and Bingham 2012; Morar et al. 2011; Bingham et al. 2011; Quesada et al. 2014), the impact of pollution (e.g., Wicksten 1984), disease vectoring (e.g., Hopper et al. 2008), and the effect of local changes in habitat (e.g., Pineda and Escofet 1989; Haag and Dyson 2014). Specimens of species in this genus serve as model organisms for studying toxins and genomes (e.g., Hauck et al. 2007; Zhang et al. 2007; Xiang et al. 2008; Kohno et al. 2009; Peigneur et al. 2011, 2012; Alvarado et al. 2014; Macrander et al. 2015; Zhang and Zhu 2016, Ayala-Sumuano et al. 2017). Numerous studies have used species of Anthopleura to explore the symbiosis between anemones and microorganisms (e.g., Pearse 1974; Saunders and Muller-Parker 1997; Verde and McCloskey 2002; Weis et al. 2002; Bergschneider and Muller-Parker 2008; Letsch et al. 2009; McBride et al. 2009; Sanders and Palumbi 2011; Hiebert and Bingham 2012; Levine and Muller-Parker 2012; Towanda and Thuesen 2012; Miura et al. 2014; Borbón et al. 2016; Dimond et al. 2017).
Anthopleura is of particular interest in part because its members have inducible structures called acrorhagi that are deployed as part of a complex intraspecific interaction (reviewed in Francis 1988). Acrorhagi are bulbous marginal structures densely packed with holotrichous nematocysts (Fig. 1d, g, h); they are inflated and applied to the column of a conspecific individual during aggressive interactions (reviewed in Daly 2003). Acrorhagi, which are characteristic of Anthopleura and several other genera within the family Actiniidae (Table 1), have been documented to play a role in intra- and interspecific interactions with other anemones (Francis 1973, 1976; Bigger 1988; Ayre and Grosberg 2005) and in allorecognition (Bigger 1980; Grosberg 1988). This last capacity is clearly tied to the initiation and precision of the intraspecific behaviors mediated by acrorhagi (Foster and Briffa 2014). Because acrorhagial interactions are critical to fitness (Rudin and Briffa 2011), it is plausible that these structures have played a role in their diversification. However, as acrorhagi-bearing anemones are most species rich in temperate regions with hard substrate habitats (Fautin 2013), historical and ecological contingency cannot easily be disentangled as explanations: this habitat might favor the retention or re-evolution of acrorhagi, or this group may have undergone a radiation in this habitat because they have these structures (or both).
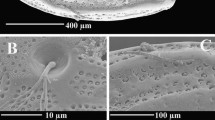
Anatomy and morphology of acrorhagi, verrucae, and vesicles. a External morphology of living, contracted specimen of Bunodactis verrucosa. Rows of verrucae on column extend to limbus and project over fosse. These marginal projections lack holotrichs and so are pseudoacrorhagi. b Longitudinal histological section through a verruca of Anthopleura thallia.c Longitudinal section through a vesicle of Bunodosoma californicum. Note that the vesicle lacks the dense glandular cells of the verruca in b. d External morphology of a living specimen of Anthopleura ballii. The marginal projections contain dense holotrichs on the surface facing the tentacles and fosse and so are acrorhagi. e External anatomy of vesicles on distal column of Bunodosoma cavernatum. Small white spherules visible between distal vesicles and tentacles are acrorhagi. f External morphology of the column of Anthopleura biscayensis, showing verrucae; some verrucae are holding gravel and are thus not visible. g Cross section through an acrorhagus of Anthopleura pallida. The holotrich-dense pad (H) in the center of the acrorhagus faces into the fosse in life. h Longitudinal section of an acrorhagus of A. pallida
Beyond their critical role in the ecology and biology of the anemones that bear them, acrorhagi are key diagnostic and taxonomic features (Stephenson 1935; Carlgren 1949). Nonetheless, practical and conceptual problems plague their application as a taxonomic feature: most critically, in at least some groups, acrorhagi do not manifest under all ecological circumstances or in all individuals, being induced through contact with conspecifics (reviewed by Daly 2003). Furthermore, because identification of an acrorhagus relies on relatively high-powered microscopy to differentiate between the types of nematocysts contained within the tissue, acrorhagi may not be correctly documented in species described before the middle of the twentieth century. Acrorhagi are sometimes confused with “pseudoacrorhagi,” bulbous structures that protrude from the distal column margin but that lack the nematocysts and behaviors associated with acrorhagi (Fig. 1a; see Daly 2003). Confusion happens because acrorhagi and pseudoacrorhagi are similar in external appearance and because the two structures may co-occur, mistakenly leading to the inference that only one is present.
The problems associated with acrorhagi as a diagnostic feature weigh heavily on systematic studies of Anthopleura because this feature is one of two that define the genus (Stephenson 1935; Carlgren 1949; Daly and den Hartog 2004). Anthopleura is differentiated from other members of Actiniidae in having both acrorhagi and protrusive, adhesive column structures called verrucae (Fig. 1b, f; see also Daly 2004a; Daly and den Hartog 2004). As for acrorhagi, some functions have been identified for verrucae, including reducing water loss at low tide (Hart and Crowe 1977) and retarding UV exposure (Dykens and Shick 1984). Verrucae are poorly defined in terms of anatomy and can be difficult to identify in preserved animals (reviewed in Häussermann 2004). The distinction between verrucae and so-called vesicles—non-adhesive, hollow outgrowths of the column (Fig. 1c, e)—is unclear (Stephenson 1928; Häussermann 2004), and the term vesicle may be overly general, referring to multiple structures. These inconsistencies make vesicles a problematic taxonomic feature: for example, both Phymactis and Bunodosoma are defined as having acrorhagi and non-adhesive vesicles (Table 1; see Gomes et al. 2012); as currently defined, these genera cannot be differentiated.
The taxonomic problems caused by the imprecise way in which acrorhagi and verrucae are defined extend beyond Anthopleura because acrorhagi and verrucae are also characteristic of groups other than Anthopleura (Table 1). The other genera are characterized either by the occurrence of only one of these features (e.g., verrucae but no acrorhagi in Aulactinia; acrorhagi but no verrucae in Actinia) or by the additional presence of additional anatomical structures (e.g., verrucae, acrorhagi, and marginal frills in Oulactis). In addition to taxonomic confusion, the evolutionary relationship between acrorhagi and pseudoacrorhagi (or between verrucae and vesicles) remains unknown: it is possible that acrorhagi and pseudoacrorhagi (or verrucae and vesicles) represent alternate manifestations of a homologous structure or that they are distinct and have no direct historical relationship (Daly 2003; Daly and den Hartog 2004).
We evaluate phylogenetic relationships among actinioidean sea anemones, with the goal of interpreting the phylogenetic, evolutionary, and taxonomic value of acrorhagi and verrucae. We include representatives of all genera characterized as having one or both of these features (Table 1), with especially dense sampling of Anthopleura (Table 2). Previous molecular phylogenies (Geller and Walton 2001; Daly et al. 2008; Rodríguez et al. 2014) and a morphological phylogeny (Daly 2004a) have demonstrated that Anthopleura is polyphyletic with respect to allied genera like Bunodosoma, Aulactinia, and Gyractis. Our more densely sampled analysis concurs in finding a non-monophyletic Anthopleura, although our trees indicate that the genus is paraphyletic with respect to other genera (and groups of genera) within Actiniidae rather than polyphyletic. Furthermore, we find that Actiniidae is paraphyletic with respect to other actinioidean lineages. Regardless of how we code or optimize acrorhagi or verrucae, we find both to be primitively present in our sample of taxa. Verrucae and vesicles are more labile than acrorhagi, evolving multiple times. Although our analyses support the interpretation of acrorhagi as homologous in the animals that bear them, they are inappropriate for recognizing subgroups within Actiniidae because they appear to be a shared primitive feature, rather than uniquely derived in any group. Our trees include the broadest taxonomic sample of the superfamily Actinioidea to date and so provide perspective on broader patterns of evolution in this major lineage of Actiniaria.
Material and methods
Taxonomic sampling and data collection
We include 27 representatives of Anthopleura: 23 nominal species, with two representatives of the widely distributed Anthopleura kurogane and Anthopleura nigrescens and two undescribed species (Anthopleura sp. “Green” and Anthopleura sp. South Africa, from Oman and South Africa, respectively). We broadly sample allied groups within Actinioidea, including representatives of 32 genera spanning eight families (Table 2) and the majority of species in Actinioidea that have verrucae or acrorhagi (Table 1). Guided by Rodríguez et al. (2014), we use the metridioideans Metridium senile and Diadumene leucolena as our chosen outgroups. Sequences from GenBank were included as appropriate (Table 2). We have generally only included those taxa from which we were able to amplify at least three of the five markers from a single specimen, and thus have analyzed a total of 328 sequences for 78 terminal taxa.
Specimens were collected by hand intertidally or during scuba dives. All specimens were identified using polyp anatomy and the distribution and size of cnidae in various regions of the polyp. Voucher specimens in formalin have been deposited at the American Museum of Natural History (AMNH), the Bavarian State Collection of Zoology (ZSM), the collection of Biodiversidad y Ecología de Invertebrados Marinos (BEIM) at the University of Seville, the California Academy of Sciences (CAS), the University of Kansas Natural History Museum (KUNHM), and the US National Museum of Natural History (USNM).
Genomic DNA was isolated from tentacle or column tissue using the Qiagen DNAeasy® Kit or by standard CTAB extraction. Template DNA was amplified from genomic samples using published primers and standard techniques. We sequenced three mitochondrial markers (partial 12S ribosomal RNA (rRNA) (12S), 16S rRNA (16S), and cytochrome c oxidase III (COIII)) and two nuclear markers (internal transcribed spacer (ITS) and partial 28S rRNA (28S)). We used primers published previously for 12S, 16S, 28S, and COIII (Geller and Walton 2001; Daly et al. 2008; Gusmão and Daly 2010). ITS was amplified using 5′-GGT TTC CGT AGG TGA ACC TGC GGA A-3′ as the forward primer and 5′-GTT CCC GCT TCA TTC GCC ATT AC-3′ as the reverse primer. Samples that could not be readily amplified using standard protocols were amplified with the high-fidelity enzyme Herculase® (Stratagene, La Jolla, CA), using manufacturer supplied protocols. All PCR products were cleaned using AMPure® magnetic bead solution (Agencourt, Beverly, MA) and re-hydrated with deionized, double-distilled water. Sequencing reactions used a total of 10 μL of cleaned PCR product, at a concentration of 25 ng/μL of product for every 200 bp of marker length. Cleaned PCR products were sequenced using amplification primers on an ABI 3730XL by staff at the sequencing facilities of Genaissance (New Haven, CT), Cogenics (Houston, TX) or Beckman Coulter (Beverley, MA). Forward and reverse sequences were assembled with Sequencher ver. 4.8 (Gene Codes Corporation, Ann Arbor, MI) or Geneious ver. 7.1.8 (Kearse et al. 2012). Once assembled, the contigs were queried against the nucleotide database of NCBI using BLAST to identify possible contaminants (from a symbiont or other contaminant) or cross contaminants. All sequences have been deposited in GenBank (Table 2).
Phylogenetic analyses
Sequences for each marker were aligned using the default settings of MUSCLE (Edgar 2004) implemented through Geneious ver. 7.1.8 (Kearse et al. 2012). Sequences were concatenated into mitochondrial (12S, 16S, and COIII) and nuclear (ITS and 28S) data sets; the mitochondrial and nuclear data sets were themselves combined into a single (“combined”) data set. All matrices were submitted to a test of the partitioning scheme using PartitionFinder ver. 1.1.1 (Lanfear et al. 2012) under the corrected Akaike Information Criterion (Akaike 1974). Analyses, described below, were conducted on all three data sets: the mitochondrial data set, the nuclear data set, and the combined data set of all markers. Matrix partition files generally separated each marker in each matrix into independent partitions (COIII further subpartitioned into codon positions) but failed to separate 12S and 16S in the mitochondrial matrix.
Twenty maximum likelihood runs were performed on each data matrix in RAxML ver. 8.1.16 (Stamatakis 2014). For all matrices, we used the GTR + G + I substitution model, per the results of jModelTest (Posada 2008). For each analysis, 1000 bootstrap replicates were performed on the best-scoring tree of the 20 maximum likelihood runs.
Character coding analysis
To understand acrorhagi, pseudoacrorhagi, verrucae, vesicles, and the evolutionary implications of their co-variation, we built a morphological matrix in which these features were coded as two multistate characters: marginal structures [absent; pseudoacrorhagi; acrorhagi] and column structures [absent (= smooth), verrucae, vesicles, solid papillae, or adhesive spots]. This multistate coding strategy approximates the conventional interpretation of the features, with acrorhagi and pseudoacrorhagi as equivalent alternatives to having no marginal swellings and papillae, verrucae, and vesicles as alternatives to a smooth column (Carlgren 1949). Although coding these as alternative states of a single feature implies homology at some level (Freudenstein 2005), the multistate characters propose no relationship among various manifestations of each feature, and we consider the states unordered in the analyses described below.
Although verrucae and vesicles are generally not treated as having underlying similarity beyond being attributes of the column, they are conceptually linked because both verrucae and vesicles are hollow outgrowths of the column (Stephenson 1928; Häussermann 2004), and thus verrucae and vesicles are more alike one another than either is to a smooth column or solid adhesive spots. To recognize this similarity, we also coded the column structures character as a pair of characters: column structures [smooth, solid growths, hollow outgrowths] and hollow outgrowths [cup-like, rounded]. For those taxa having a smooth column or solid outgrowths, the secondary character that describes the nature of the column outgrowth is scored as inapplicable.
To evaluate the pattern of character change, we optimize these characters on the tree of highest likelihood from the combined data set. For each coding strategy, we consider acctran, deltran, and unambiguous optimization strategies using the parsimony criterion as implemented in the “trace characters” function in Mesquite (Maddison and Maddison 2011) and conducted a simple likelihood optimization using Mesquite’s embedded stochastic model for character change.
Results
Interpretations of relationship among Actinioidea
This analysis concurs with broader-scale studies (e.g., Rodríguez et al. 2014) in finding a monophyletic Actinioidea (Figs. 2 and 3). The tree of highest likelihood for the combined data set has generally short internal branches with generally low support (Fig. 2). At the base of the ingroup is a comb-like series of sister group pairs and singleton taxa: Anthopleura sp. “pacifica”, Anthopleura fuscoviridis + Anthopleura midori, Anthopleura xanthogrammica + Anthopleura artemisia, A. kurogane Korea, and Phymactis clematis. These basal branches in the ingroup are not well supported and are generally resolved in other ways in the subset analyses (see Supplemental Figs. 1 and 2), suggesting that the arrangement of species in this part of the tree is unstable. Recognizing these limitations, we focus on the relationships among the major clades, emphasizing those groups that appear in the combined tree.
Optimization of focal characters onto combined data maximum likelihood tree. Colors correspond to the features discussed in the text: acrorhagi are indicated in pink, pseudoacrorhagi are indicated in blue, verrucae are indicated in yellow, vesicles are indicated in green, solid spots on the column are indicated in orange. The absence of marginal structures is indicated in purple and a smooth column is indicated in red. Pseudoacrorhagi and vesicles arise either de novo (from the state “absent” or “smooth”) or via a transformation of acrorhagi or verrucae, respectively. Transformed versus de novo originations of pseudoacrorhagi and vesicles are differentiated by the shade of blue (for pseudoacrorhagi) or green (for vesicles). Branch lengths not to scale (color figure online)
In the combined analysis, the large ingroup clade has three subclades whose interrelationship is not resolved. The smallest of these is the SA clade (Figs. 2 and 3), which consists of three South African species, Anemonia erythraea, Anemonia natalensis, and Pseudactinia varia. This clade is present in the mitochondrial tree, but is disassociated in the nuclear tree, perhaps because P. varia is not included in that analysis. The subclade described below and hereafter as ABEPP (Figs. 2 and 3) includes several species of Anthopleura, the sampled members of Bunodosoma, and the ptychodactiarians Dactylanthus and Preactis. The third clade, hereafter JSTEAL (Figs. 2 and 3), includes other species of Anthopleura and assorted members of Actiniidae, Stichodactylidae, and Liponematidae. The relationships among the SA, ABEPP, and JSTEAL clades are unresolved in the combined analysis and differ in the trees based on either nuclear or mitochondrial sequences alone. For example, the SA clade merges with the ABEPP clade in the tree based on mitochondrial sequences (Supplemental Fig. 1) and its constituents associate with members of this group in the nuclear tree (Supplemental Fig. 2), and in the nuclear tree, the taxa of the JSTEAL clade are paraphyletic with respect to the ABEPP clade (Supplemental Fig. 2), whereas in the mitochondrial tree (Supplemental Fig. 1), the taxa of the ABEPP clade nest within the JSTEAL clade.
In the combined analysis, the ABEPP clade has four subclades: a clade containing Anthopleura biscayensis plus a clade containing Anthopleura dowii, Anthopleura elegantissima, and Anthopleura sola (= EP clade; Fig. 2); a clade that joins the ptychodactiarians and the actiniids Korsaranthus and Anthostella (= P clade; Fig. 2); a clade containing members of Bunodosoma (= B clade; Figs. 2 and 3); and a clade that includes Oulactis, plus a clade of several South African species of Anthopleura and the type species of the genus Anthopleura, Anthopleura krebsi (= A clade; Figs. 2 and 3). Within this A clade, there is support for a group consisting of A. krebsi, Anthopleura pallida, and Anthopleura waridi, but other relationships are unresolved. Branches within this clade are very short but are resolved consistently: A. krebsi, A. pallida, and A. waridi are sister taxa to an assemblage of species from the South Atlantic. This South Atlantic assemblage includes Anthopleura annae, Anthopleura insignis, Aulactinia reynaudi, and Bunodosoma capense. Although Oulactis muscosa is the sister taxon to the rest of the A clade in the combined tree (Figs. 2 and 3) and is part of the ABP clade in all analyses, its position varies across data sets. The B clade (Figs. 2 and 3) includes the new-world Bunodosoma (Bunodosoma californicum, Bunodosoma cavernatum, Bunodosoma grande, and Bunodosoma granuliferum). In the combined (and nuclear) data sets, the B clade also includes Bunodactis verrucosa. The B clade never includes B. capense, the only old-world species of the genus included in this analysis, which is instead interpreted as part of the A clade. The B clade is always the sister taxon to the P clade, a finding congruent with that of Rodríguez et al. (2014). The EP clade groups well-known sibling species A. elegantissima and A. sola (see McFadden et al. 1997; Pearse and Francis 2000; Geller and Walton 2001) with the geographically close A. dowii; this novel grouping is consistent across data sets. In contrast, the association of A. dowii, A. elegantissima, and A. sola with A. biscayensis is seen only in the combined tree. Relationships of members of the EP clade are sensitive to data set. The P clade (Figs. 2 and 3) includes the two ptychodactiarians (Dactylanthus antarcticus, Preactis millardae) and the actiniids Anthostella stephensoni and Korsaranthus natalensis. All of these species are reported only from the Southern Ocean (Fautin 2013).
In the combined analysis, the B, P, and A clades form a clade to the exclusion of the EP clade; within this clade, the B and P clades are sister taxa to the exclusion of the A clade (Figs. 2 and 3). Other than the ptychodactiarians, all members of the ABEPP clade have historically been assigned to Actiniidae. The nuclear tree is consistent with the combined analysis in recovering a clade that encompasses the A, B, and P clades (Supplemental Fig. 2) and in resolving the B and P clades as sister taxa to the exclusion of the A clade. The A, B, and P clades are also present in the mitochondrial tree, and the B and P clades are sister taxa, but the B clade of the mitochondrial analysis does not include B. verrucosa and the relationship between the B + P clade and A clade is unlike that in the combined or nuclear tree (Supplemental Fig. 1).
The JSTEAL clade (Figs. 2 and 3) contains several members traditionally assigned to families other than Actiniidae: Actinostephanus (Actinodendridae); Capnea georgiana (Capneidae); Harenactis and Stephanthus (Haloclavidae); Liponema (Liponematidae) and Stichodactyla gigantea and Heteractis magnifica (Stichodactylidae). C. georgiana (Capneidae) is among the members of the larger JSTEAL clade that lack clear affiliation to any named clade. In the results of the combined analysis, the JSTEAL clade contains two subclades at the base: a clade that includes Phylctenactis tuberculosa and two species belonging to the genus Actinia and a clade that includes specimens of Gyractis sesere and an undescribed species of Anthopleura (Anthopleura sp. Green). The relative position of these clades varies across analyses, with weak support in all cases. These (unnamed) groups are sister taxa to a series of named clades that have more consistency in their membership and resolution.
The J clade (Figs. 2 and 3) includes species of Anthopleura from the Pacific; it is sister to a clade that includes Anemonia viridis, Anthopleura ballii, and Anthopleura dixoniana. The J clade has two further subgroups: a clade composed of the Japanese species Anthopleura atodai, Anthopleura inornata, A. kurogane (Japan), and Anthopleura japonica and a clade composed of the central to south Pacific species Anthopleura buddemeieri, Anthopleura handi, A. nigrescens, and Phymanthus loligo.
The ST clade (Figs. 2 and 3) includes members of Stichodactylidae plus the actiniids Anthopleura thallia and Macrodactyla doreensis. Although not currently classified among the Stichodactylidae, Macrodactyla has historically been considered among the Stichodactylidae (see Dunn 1981), and, as is the case with Stichodactylidae, some of its members harbor clownfish (Dunn 1981). Although we consider the resolution of M. doreensis within the ST clade credible (Figs. 2 and 3), in the nuclear analysis (Supplemental Fig. 2), M. doreensis is interpreted as the sister to the A.dowii + A. elegantissima clade rather than as part of the ST clade. The inclusion of A. thallia within the ST clade is surprising but consistent across data sets. Despite this consistency, we regard it as suspicious, because A. thallia differs from other members of the ST clade in size, ecology, and cnidom (compare e.g., Dunn 1981; Fautin et al. 2008; Daly and Picton 2012).
The EA clade (Figs. 2 and 3) includes the Northern hemisphere members of Epiactis (Epiactis japonica, Epiactis lisbethae, Epiactis prolifera), species of Aulactinia (Aulactinia incubans, Aulactinia stella), the sole included representative of Capnea, and Urticina grebelnyi. Aulactinia veratra is the sister to the union of the EA + L clade rather than part of the EA clade. U. grebelnyi groups with the EA clade in the combined and nuclear trees but is part of a polytomy that includes the EA clade in the mitochondrial trees. C. georgiana is within the EA clade in the combined analysis but is reconstructed near the base of the ingroup in the mitochondrial tree (Figs. 2 and 3; Supplementary Fig. 1). This taxon is interpreted differently yet in the analyses of Rodríguez et al. (2014): C. georgiana lies outside of Actinioidea in that analysis.
The L clade (Figs. 2 and 3) includes more family-level diversity than any of the other well-supported groups, comprising members of Actiniidae (Bolocera kerguelensis, Epiactis thompsoni, Glyphoperidium bursa, Isosicyonis alba, Isotealia antarctica, Urticina coriacea), Haloclavidae (Peachia cylindrica, Harenactis argentina, Stephanthus antarcticus), and Liponematidae (Liponema brevicornis, Liponema multiporum). Actinostephanus sp., a member of Actinodendridae, associates with this clade based on mitochondrial sequences (no nuclear loci sampled for this species). Haloclavids and actinodendrids are burrowers in soft sediments and share aspects of their cnidom, most notably the absence of microbasic p-mastigophores (Riemann-Zürneck and Griffiths 1999; Rodríguez and López-González 2003). In the nuclear tree (Supplemental Fig. 2), monophyly of the L clade is disrupted by the association of G. bursa, H. argentina, and S. antarcticus with the basal node of the ingroup. Furthermore, A. biscayensis is reconstructed within the L clade, as the sister of E. thompsoni (Supplemental Fig. 2), rather than with A. dowii and A. elegantissima in the EP clade.
We find several taxa that are resolved in dramatically different ways in our analyses. The inferred position of Haloclava producta is inconsistent across data sets. These differences in topology have consequences for interpreting evolution, because the different positions for H. producta force differences in its inferred branch length: the total branch length (root to tip) is relatively long when H. producta is associated with the ST clade (Figs. 2 and 3) and relatively short when it is at the base of the actinioidean clade (Supplemental Fig. 1). In the analysis of Rodríguez et al. (2014), H. producta associates with the species that are here within the L clade; that interpretation uses nearly the same genes but a different taxon sample, including more diversity at the ordinal level but less diversity at the subfamilial level. All other haloclavids are part of the L clade in the combined analysis (Figs. 2 and 3), suggesting a relationship between the haloclavids and the other members of the L clade that may be obscured by the peculiarities of sequences for H. producta. However, the inclusion of P. cylindrica within the L clade is also marker-dependent: although S. antarcticus and H. argentina remain within the L clade in all analyses, P. cylindrica (and H. producta) fall to the base of the ingroup in the mitochondrial tree.
A. dixoniana is interpreted as the sister to the clade that includes A. ballii and A. viridis in the combined analysis, but is sister to the clade consisting of Isosicyonis striata and L. brevicornis in the nuclear tree and associates with G. sesere in the mitochondrial tree. A. biscayensis is sister to the A. elegantissima/sola/dowii clade in the combined analysis but groups with H. producta, A. nigrescens Galapagos, and E. thompsoni in the nuclear tree and with the Anthopleura clade within the ABP clade in the mitochondrial tree.
Although A. biscayensis is a relatively long branch (Fig. 2), long branches are not the likely explanation for the differences in resolution. First, not all of the unstable taxa are relatively long branches: P. clematis and H. producta both vary in position from data set to data set, but neither is associated with a long branch. Second, many of the longer-branched taxa are stable: A. atodai is always the sister taxon to A. kurogane (Japan) and part of a clade that includes A. inornata and A. japonica; the resolution of M. doreensis is similarly stable within the ST clade despite the length of the branch; D. antarcticus is always the sister to P. millardae and part of the P clade; and L. brevicornis and I. alba are both long but stable within the L clade.
Character analyses
All means of coding and optimizing the focal features on the combined data tree agree that acrorhagi and verrucae are shared primitive features of the ingroup. Both of these characters change across the tree, with the number of transformations depending on the tree used, optimization method, and the coding strategy.
On the combined tree, the most parsimonious interpretation is for acrorhagi to be present ancestrally; the likelihood interpretation of acrorhagi as present in the common ancestor of the focal taxa is 0.975 (probability of pseudoacrorhagi being the ancestral state = 0.01, probability of no spherules = 0.016). Acrorhagi are inferred to have been transformed nine times (Fig. 3): three losses (P clade; Phlyctenactis; STEAL clade), three transformations to holotrich-less pseudoacrorhagi (B. verrucosa; P. loligo, Gyractis), and three independent originations (A. nigrescens Galapagos; A. thallia; A. stephensoni). In this scenario, pseudoacrorhagi arise four times de novo: in A. stephensoni (which has acrorhagi and pseudoacrorhagi), in A. veratra, in the clade composed of A. incubans and A. stella, and in I. antarctica.
When verrucae are as part of a single multistate character (Fig. 3), they are inferred to be primitively present, with verrucae transforming into vesicles five times (P. clematis; BP clade; Bunodosoma sp. South Africa; B. capense; H. producta) and being wholly lost five times (SA clade; Korsaranthus, Actinia + Phylctenactis; A. viridis; within EAL clade). In this coding scheme, vesicles are lost twice (Korsaranthus, I. striata), transform into solid papillae once (Anthostella), and arise de novo (Phylctenactis, U. grebelnyi) from a smooth column or transform from verrucae (see above), but never give rise to verrucae.
When verrucae are broken into two independent characters, hollow outgrowths on the column are interpreted to have been lost seven times (Fig. 3: in the SA clade; Korsaranthus + Anthostella; A. viridis; Actinia + Phylctenactis; within the L clade; within the EA clade) and gained once (Phylctenactis). In the coding scheme in which verrucae and vesicles are alternate states of the character “hollow outgrowth,” outgrowths are present at the base of the tree (Fig. 3) and verrucae (adhesive, cup-like) are the initial state, with transformations to vesicle (rounded, non adhesive) in P. clematis, the BP clade, Bunodosoma sp. South Africa, B. capense, and H. producta (Fig. 3). These are all branches in which verrucae are inferred to transform into vesicles in the single character coding scheme.
The probability of a particular ancestral state as inferred by likelihood reconstruction differs slightly depending on the atomization of the column features. The likelihood value for verrucae at the ingroup node when verrucae are part of a single multistate character is >0.9 (probability of vesicle being the ancestral state >0.01, probability of smooth column >0.01). When the column feature is atomized into two characters, the probability increases slightly: at the ingroup node, hollow column outgrowths have a probability of 0.988 (probability of smooth column >0.01). However, this increase in support obscures ambiguity at another level, as the probability of these hollow outgrowths being cup-like (and thus equivalent to verrucae) is 0.82, a value slightly lower than the inferred probability of verrucae being present at this node in the single-character analysis.
Discussion
Phylogeny of Actinioidea and Actiniidae
Our trees concur with most broad-scale analyses of Actiniaria (e.g., Daly et al. 2008; Rodríguez et al. 2012; but see Rodríguez et al. 2014) in finding a monophyletic Actinioidea. One notable difference is that E. lisbethae was not recovered within Actinioidea by Rodríguez et al. (2014); our denser sampling of Epiactis specifically and Actinioidea effectively resolves what Rodríguez et al. (2014) considered a spurious placement. Despite consistent and relatively strong support for ingroup monophyly, our analyses do not effectively resolve relationships among the major groups within Actinioidea. The internal branches at the deepest nodes are short, their resolution is not well supported, and the differences among the data sets largely reflect alternative interpretations of the basal branching order.
We find that Actiniidae is polyphyletic with respect to other families of Actinioidea: the node at which all taxa currently within Actiniidae are monophyletic is the node that includes all members of Actinioidea. Despite failing to recover a monophyletic Actiniidae, the combined, mitochondrial, and nuclear trees agree on many clades and recover several groups consistently or with high support, and thus provide a starting point for circumscribing groups within Actinioidea. Our taxon sampling precludes taxonomic revision for most groups because of its incompleteness at the genus- and family-level and because almost all of the clades we identify include members of multiple genera, if not multiple families. Furthermore, at present, none of these groups are clearly definable based on features typically used for classification. Defining the boundaries and membership of these and identifying the features by which these can be recognized is beyond the scope of the present study, but will be facilitated by the delimitation of these broad groupings.
The combined tree and the single-data set analyses differ in their interpretation of the primary split within Actinioidea. In both the mitochondrial and nuclear trees, species that are part of the EA and L clades form a grade sister to the majority of ingroup species (Supplemental Figs. 1 and 2), whereas the combined data tree recovers the EA and L as sister clades within the large ingroup clade (Fig. 2). Although Larson and Daly (2016) studied relatively fewer lineages in Actiniidae, their analyses included relatively denser sampling of what here constitutes the EA group, and they found support for EA and L as a grade outside of a clade that included species of e.g., Actinia, Anthopleura, Bunodactis, Bunodosoma, Gyractis, and Oulactis. The ecology, bathymetric distribution, and natural history of species in the EA and L groups are more varied than that of the species in the rest of the ingroup. The different interpretations of the relative position of the taxa in the EA and L groups are important for interpreting the evolution of features like acrorhagi (discussed below) and for understanding the evolution of photosymbiosis or asexual reproduction. Especially in light of their diversity in biology, the EA and L lineages are probably under sampled relative to the rest of the ingroup in our analyses, but this is necessary, given our primary interest in Anthopleura and its monophyly. We anticipate that denser sampling of the groups represented in the EA and L clades will resolve this instability.
All of our analyses find a sister-group relationship between the ptychodactiarians and the South African species K. natalensis and A. stephensoni. In their description of Korsaranthus, Riemann-Zürneck and Griffiths (1999) note the similarity of K. natalensis to Dactylanthus and other ptychodactiarians with respect to the anatomy of the actinopharynx and cnidom. However, they interpret these as convergent similarities based on similar diet and mode of locomotion, an interpretation accepted by Cappola and Fautin (2000). Although several attributes that have been interpreted to support their separation as a higher taxon (reviewed by Cappola and Fautin 2000), the affiliation between ptychodactiarians and actiniids has been recovered in several molecular phylogenetic analyses (Daly et al. 2003, 2008; Rodríguez et al. 2012, 2014) and the sister group relationship between the clade containing the ptychodactiarians Dactylanthus and Preactis and members of the B clade is consistent across data sets.
Our results have clear implications for a handful of relatively minor taxonomic issues. We never find monophyly of the included species of Anemonia, Aulactinia, Epiactis, or Urticina (Fig. 2). Members of Bunodosoma are divided between the B and A clades within the ABEPP clade in groupings that are both consistent across analyses and well supported in the combined analysis. The placement of A. biscayensis outside of Bunodosoma (and sister to other nominal Anthopleura) supports the re-assignment of this species proposed by Daly (2004b). A. viridis and A. ballii are recovered as sister taxa in all analyses. Although assigned to different genera and distinct in the anatomy of the column (smooth in A. viridis, verrucose in A. ballii), these northern hemisphere species are similar in having weakly muscled columns and non-retractile tentacles and are also similar in coloration (see Stephenson 1935). The other species of Anemonia included in this analysis, A. erythraea and A. natalensis, are southern hemisphere species that group with P. varia in the highly supported SA clade. The close association we find between Aulactinia and the Northern hemisphere species of Epiactis was also found in a more narrowly focused analysis (Larson and Daly 2016) but is not obvious in terms of anatomy: the included species of Aulactinia have a verrucose column whereas that of Epiactis is smooth (Tables 2 and 3). All of these polyphyletic genera (Anemonia, Aulactinia, Bunodosoma, Epiactis, Urticina) require revision. In addition, we find that Actinia, Isosicyonis, and Liponema are paraphyletic, but acknowledge that support for their paraphyly is weak and these lineages have not been the focus of our sampling.
Dunn et al. (1980) conditionally proposed a synonymy between Bunodactis and Aulactinia. Although this proposal was limited in the scope of the synonymy and advocated further study, it has been generally adopted (without the recommended specimen-level studies), and most authors now apply Aulactinia as the valid name for the species originally described as Actinia verrucosa (see Fautin 2016), although this is not uniformly the case (Spano et al. 2013, Garese et al. 2014). Because we never find a close relationship between B. verrucosa and species of Aulactinia, we reject the proposition that Bunodactis is wholly in synonymy with Aulactinia and consider Bunodactis the appropriate generic epithet for the species originally described as A. verrucosa. The limits of our taxon sampling for Aulactinia and the non-monophyly of its included species (which remains non-monophyletic even if we ignore B. verrucosa) prevent us from making broader taxonomic recommendations for Aulactinia or Bunodactis.
Phylogeny of Anthopleura
We find no evidence for monophyly of Anthopleura: its members are scattered across the tree, associated with members of other actiniid genera and with members of other actinioidean families (Fig. 2; Supplemental Figs. 1 and 2). Genera that have been proposed as closely related to Anthopleura, such as Anthostella, Aulactinia, Bunodactis, Bunodosoma, and Gyractis, are closely related to it in the sense that they group with nominal species of Anthopleura. The type of Anthopleura, A. krebsi, lies within the A clade. The interrelationships among nominal Anthopleura are difficult to determine given the paraphyly of the genus and the relatively low support for internal branches. Of the well-supported and consistent clades, only the A and ST clades contain members of Anthopleura; the majority of included species of Anthopleura are part of poorly supported clades. Because we have over-sampled Anthopleura relative to other genera, this may reflect sampling depth rather than something more significant about Anthopleura relative to other actinioideans: for example, the support for the L, EA, or P clades may be inflated because many members of those clades have not been included and so the support is not divided among the same number of nodes or subjected to the same number of possible resolutions as clades of Anthopleura.
Previous studies of relationships among species of Anthopleura (Geller and Walton 2001; Daly 2004a) have not included as many nominal species or as many taxa from outside of Anthopleura. In light of the broad polyphyly of Anthopleura, this difference makes comparing trees difficult. However, we see points of congruence with the results of Geller and Walton (2001), including finding a close relationship between A. handi and A. nigrescens and between these and a clade of Anthopleura from Japan. Geller and Walton (2001) and Daly (2004a) both find that Anthopleura is not monophyletic with respect to Bunodosoma; the present results extend this finding to include several other genera, but concur in finding a close relationship between Bunodosoma and several species of Anthopleura (Figs. 2 and 3; Supplemental Figs. 1 and 2). Similarity between the present results and those of Geller and Walton (2001) is not surprising given that their focal markers are among those we analyze here. The tree in Daly (2004a) is based on morphological data and is generally less congruent with these new results than is the tree of Geller and Walton (2001); other than the close association between Bunodosoma and Anthopleura, only the proposed relationship between A. dowii and A. elegantissima is seen in our trees and in those of Daly (2004a).
Although the frequency with which species of Anthopleura occur in sympatry is explained in part by the broad polyphyly of the genus, geography provides a means of interpreting and synthesizing the diversity of Anthopleura and its actinioidean relatives, if only because several of the well-supported or consistently recovered subunits have some geographic signature. Of course, because we have not sampled all members of Anthopleura and because the relationships of a few species of Anthopleura are highly labile across data sets, these groups and inferences about them are provisional.
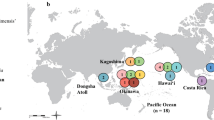
In terms of the biogeography of the North Pacific, based on our results, there are at least two colonization events each in the East and West Pacific (Fig. 4). Species of Anthopleura from the North East Pacific span two groups: the clade that includes A. dowii, A. elegantissima, and A. sola and the clade containing A. xanthogrammica and A. artemisia. These two groups are never siblings. A. xanthogrammica and A. artemisia are generally associated with one another but are not siblings in all analyses. The included species from the Southern and Central West Pacific (A. handi, A. buddemeieri, A. nigrescens) are sister to a clade from the North West Pacific (A. atodai, A. inornata, A. kurogane (Japan), A. japonica), together forming the J clade (Fig. 2). Of these, there seem to be two in situ radiations: the dowii/elegantissima/sola group has undergone an in situ radiation on the North American coast and the atodai/inornata/kurogane/japonica group has undergone and in situ radiation in Asia. Our finding of multiple origins for the East and West Pacific Anthopleura was previously predicted by Geller and Walton (2001).
Recorded occurrences of focal species mapped onto combined data maximum likelihood tree. Distributional data from Fautin (2013). Branch lengths are proportional to change
Geography is more predictive of relationship than taxonomy, for species from the Atlantic (Fig. 4). Anthopleura from the Atlantic belong to three clades: A. balli is sister to A. viridis (also an Atlantic species); A. thallia is part of the ST clade; and the remaining species (A. annae, A. insignis, A. krebsi, A. pallida, Anthopleura sp. South Africa) are part of the A clade, which also contains the Red Sea-Northern Indian Ocean species A. waridi. A. waridi is reported from the Red Sea, Persian Gulf, and the coasts of East Africa and India (Fautin 2013) and is most closely related to two species from the North Atlantic (A. krebsi, A. pallida). The taxa within the B and P clades, which are sister to the A clade, are also primarily Atlantic in their distribution. The distribution of the members of the ABEPP clade suggests an Atlantic (or possibly Tethyan) origin for the group, with B. californicum dispersing into the Pacific and A. waridi dispersing into the Indian Ocean.
The North American and Northern European costs were under ice during the Quaternary glaciation, as were parts of Argentina and Chile. In light of their current restriction to the shallow subtidal and intertidal zones, it is probable that Anthopleura species currently inhabiting these regions are migrants from warmer waters rather than from deepwater refugia.
Evolution of acrorhagi and verrucae
Our results suggest that acrorhagi are a shared, inherited feature in the majority of taxa that bear them. The combined analysis interprets them as ancestral for the Actinioidea (Fig. 3). The nuclear tree moves the optimization of acrorhagi higher into the tree: because the members of this assemblage at the base of the ingroup (the clade that includes Stephanthus, Peachia, Glyphoperidium, and A. viridis, among other taxa) and of the EA clade generally lack acrorhagi, acrorhagi are interpreted as originating in parallel in A. viridis + A. ballii and in the clade that contains the ABP, ST, and EP clades (see asterisks, Supplemental Fig. 2). The mitochondrial tree also posits that acrorhagi arose relatively later in actinioidean history (see asterisks, Supplemental Fig. 1) and are convergent; the purported instance of convergence is in Anthopleura sp. pacifica, rather than A. viridis + A. ballii as in the nuclear tree. These differences between the mitochondrial, nuclear, and combined trees relate to the relative position of the EA and L clades because members of these groups lack acrorhagi.
Holotrich-bearing acrorhagi are interpreted to have been lost several times. In B. verrucosa, P. loligo, and Gyractis, this loss of acrorhagi creates pseudoacrorhagi; in all other cases, pseudoacrorhagi either arise in the absence of acrorhagi (A. veratra, A. stella + A. incubans, I. antarctica) or arise concomitant with the de novo origination of acrorhagi (Anthostella) (Fig. 3). This pattern of character evolution means that acrorhagi and pseudoacrorhagi are homologous only in the case of B. verrucosa, P. loligo, and Gyractis (and possibly in the case of Anthostella). However, the inducibility of acrorhagi (reviewed by Daly 2003) and the relatively poor state of anatomical descriptions for some of the species inferred to have lost acrorhagi means that some of these instances of loss may be mistaken. Careful re-evaluation of pseudoacrorhagi and of species inferred to have lost acrorhagi are needed to verify the absence of acrorhagi and to document the structure of pseudoacrorhagi, which, because they are convergent in many of the taxa that bear them, are likely more diverse in structure than currently appreciated.
Although we interpret acrorhagi as primitive for the majority of taxa that bear them, the phylogenetic tree we reconstruct here also supports the inference of convergence for acrorhagi. The inferred instances of re-evolution of acrorhagi within the ingroup in the combined tree are generally also required by the single-locus analyses. Acrorhagi are inferred to re-evolve in Anthostella; this interpretation is compelling because the resolution of this species is well supported and consistent across data sets, and thus unlikely to change. In contrast, the inferred re-evolution of acrorhagi in A. nigrescens and in A. thallia is less clear, as these species are each weakly supported within or associated with of the ST clade (in all analyses), and denser sampling of stichodactyline taxa might resolve these two convergences as a single event or align A. nigrescens and A. thallia with other lineages in ways that eliminate the convergence. In the combined tree, optimization at the base of the node that includes Anthopleura sp. Green is ambiguous.
In all of the cases in which acrorhagi are interpreted as re-evolving, holotrichs have been reported from the sister species (see Carlgren 1938; Dunn 1981; Cappola and Fautin 2000; González-Muñoz et al. 2012; Laird 2013; Rodríguez and López-González 2013), which makes the re-acquisition of acrorhagi more plausible. Although acrorhagi are inferred to have been lost at the base of the P clade, A. stephensoni, D. antarcticus, and P. millardae all have holotrichs in their cnidom (see Dunn 1983; England and Robson 1984). Although holotrichs have not been reported in Korsaranthus (see Riemann-Zürneck and Griffiths 1999), previous accounts of cnidae are cursory in terms of the tissues examined and number of capsules reported; we have found a few capsules in the specimens sequenced here. Holotrichs are absent from most members of the EA and L clades (e.g., Larson and Daly 2016), although they have been reported from E. prolifera and E. lisbethae (see Fautin and Chia 1986), E. japonica (see Sanamyan and Sanamyan 1998), and A. incubans (Dunn et al. 1980). This pattern of character distribution suggests that the loss of the nematocyst is distinct from the loss of the structure or behavior in which it is deployed and argues for a more atomized and finer-scale study of the constituent elements of acrorhagi in the clades in which they show convergent evolution or loss.
Coding verrucae and vesicles as a set of characters rather than a single multistate character reduces the number of inferred character transformations, but does not change the overall inference that verrucae are not necessarily homologous across those Actiniidae that have them. The phylogenetic results suggest that the verrucae of A. annae, for example, and the vesicles of B. capense are homologous (albeit modified in B. capense) but that the verrucae of A. annae and e.g., A. veratra are not. Detailed functional and anatomical studies are necessary to determine if verrucae show previously unrecognized diversity as a result of their independent origins or whether they represent a convergence that can only be recognized through phylogenetic investigation. Although there are small differences in the interpretation of verrucae depending on the way in which they are coded, at least one major point is consistent across coding strategies (Fig. 3): cup-like verrucae are never interpreted to have arisen from rounded vesicles, although verrucae give rise to vesicles in several instances. The apparent irreversibility of the cup-like morphology, which is inferred to make the structures adhesive (Häussermann 2003), is interesting and suggests that the microanatomy that differentiates these structures may be more complex than previously appreciated.
Although they frequently co-occur, are symplesiomorphic for Actinioidea in this analysis, and both represent modifications of column tissue, verrucae and acrorhagi are functionally, structurally, and logically distinct. Other than their co-occurrence at the base of the ingroup and in a few clades, we see no evidence for correlation in the evolution of verrucae and acrorhagi. Verrucae are lost in taxa that retain acrorhagi (and vice versa). However, we do see a link between verrucae and pseudoacrorhagi: all species that have been described as bearing pseudoacrorhagi (Tables 1 and 2) have verrucae. This raises the possibility that in some cases, what have been described as pseudoacrorhagi are distal marginal projections bearing verrucae, caused either by differential growth of the column or by distortion of the column by the distension of the verrucae.
In light of their frequency of loss or modification to a functionally distinct state, the adaptive value of acrorhagi and verrucae may be over interpreted. We see no ecological differences between those species with verrucae and those with vesicles or a smooth column. For example, A. annae, A. natalensis, and B. capense co-occur despite differing in their column morphology and B. verrucosa is abundant and successful in the same localities as A. thallia, despite lacking acrorhagi. Although verrucae have been shown to retard desiccation and UV exposure (e.g., Hart and Crowe 1977; Dykens and Shick 1984) by holding debris to the column, they may serve additional functions. For example, because they are hollow, verrucae increase the surface area of the polyp, which might be valuable for animals that rely on diffusion for many of their physiological processes (Shick 1991); this function would be conserved in vesicles.
Although all major lineages of Actiniaria have shallow water members, Actinioidea is the lineage with the greatest proportion of shallow-water members. The interpretation of verrucae and acrorhagi as symplesiomorphic for Actinioidea could be construed as support for a shallow-water origin for Actinioidea in light of the association of both of these traits with functions that confer advantages in shallow water. However, because Anthopleura is a genus whose members are restricted to waters shallower than about 60 m, our sampling strategy emphasized shallow-water species, and this emphasis may contribute to both the inference of symplesiomorphy and to the inference of shallow water as the ancestral habitat for Actinioidea.
References
Akaike, H. (1974). A new look at the statistical model identification. IEEE Transactions on Automatic Control, 19(6), 716–723.
Alvarado, J., Álvarez, Y., Pedrera, L., Ros, U., Lanio, M. E., Valle, A., & Álvarez, C. (2014). Isolation and partial purification of a hemolytic sphingomyelin-inhibitable fraction from the sea anemone Anthopleura nigrescens. Biotecnología Aplicada, 31(1), 53–56.
Ayala-Sumuano, J.-T., Licea-Navarro, A., Rudiño-Piñera, E., Rodríguez, E., & Rodríguez-Almazán, C. (2017). Sequencing and de novo transcriptome assembly of Anthopleura dowii Verrill (1869), from Mexico. Genomics Data, 11, 92–94.
Ayre, D. J., & Grosberg, R. K. (2005). Behind anemone lines: factors affecting division of labour in the social cnidarian Anthopleura elegantissima. Animal Behavior, 70, 97–110.
Belém, M. J. D., Herrera Moreno, A., & Schlenz, E. (1996). On Isoaulactinia stelloides (McMurrich, 1889), n. gen., n. comb. (Cnidaria; Actiniaria; Actiniidae). Biociencias, 4, 77–88.
Bergschneider, H., & Muller-Parker, G. (2008). Nutritional role of two algal symbionts in the temperate sea anemone Anthopleura elegantissima Brandt. The Biological Bulletin, 215, 73–88.
Bigger, C. H. (1980). Interspecific and intraspecific acrorhagial aggressive behavior among sea anemones, a recognition of self and not-self. The Biological Bulletin, 159, 117–134.
Bigger, C. H. (1988). The role of nematocysts in anthozoan aggression. In D. A. Hessinger & H. M. Lenhoff (Eds.), The biology of nematocysts (pp. 295–308). San Diego: Academic.
Bingham, B. L., Freytes, I., Emery, M., Dimond, J., & Muller-Parker, G. (2011). Aerial exposure and body temperature of the intertidal sea anemone Anthopleura elegantissima. Invertebrate Biology, 130(4), 291–301.
Borbón, H., Váldes, S., Alvarado-Mesén, J., Soto, R., & Vega, I. (2016). Antimicrobial properties of sea anemone Anthopleura nigrescens from Pacific coast of Costa Rica. Asian Pacific Journal of Tropical Biomedicine, 6(5), 418–421.
Cairns, S. D., den Hartog, J. C., & Arenson, C. (1986). Anthozoa. In W. Sterrer & C. Schoepfer-Sterrer (Eds.), Marine flora and fauna of Bermuda (pp. 159–194). New York: Wiley.
Cappola, V. A., & Fautin, D. G. (2000). All three species of Ptychodactiaria belong to order Actiniaria (Cnidaria: Anthozoa). Journal of the Marine Biological Association of the UK, 80(06), 995–1005.
Carlgren, O. (1938). South African Actiniaria and Zoantharia. Kungliga Svenska Vetenskaps Akademiens Handlingar 3, 17(3), 1–148.
Carlgren, O. (1949). A survey of the Ptychodactiaria, Actiniaria and Corallimorpharia. Kungliga Svenska Vetenskapsakademiens Handlingar (4th series), 1, 1–121.
Daly, M. (2003). The anatomy, terminology, and homology of acorhagi and pseudoacrorhagi in sea anemones. Zoologische Verhandelingen, 345, 89–101.
Daly, M. (2004a). Phylogeny and biogeography of Anthopleura in the North Atlantic Ocean. Hydrobiologia, 530–531(1), 241–248.
Daly, M. (2004b). A redescription of three sea anemones from Baja California, including Isoaulactinia hespervolita. Pacific Science, 58, 377–390.
Daly, M., & den Hartog, J. C. (2004). Taxonomy, circumscription, and usage in Anthopleura (Cnidaria: Anthozoa: Actiniaria) from the Gulf of Mexico and Caribbean. Bulletin of Marine Science, 74(2), 400–421.
Daly, M., & Picton, B. (2012). Description of the sea anemone Anthopleura thallia (Gosse 1854). Biology and Environment: Proceedings of the Royal Irish Academy, 112B(2), 235–240.
Daly, M., Fautin, D. G., & Cappola, V. A. (2003). Systematics of the Hexacorallia (Cnidaria: Anthozoa). Zoological Journal of the Linnean Society, 139(3), 419–437.
Daly, M., Chaudhuri, A., Gusmão, L. C., & Rodriguez, E. (2008). Phylogenetic relationships among sea anemones (Cnidaria: Anthozoa: Actiniaria). Molecular Phylogenetics and Evolution, 48(1), 292–301.
Dimond, J. L., Orechovesky, S., Oppenheimer, J., Rodríguez-Ramos, J., & Bingham, B. L. (2017). Photophysiology and hydrogen peroxide generation of the dinoflagellate and chlorophyte symbionts of the sea anemone Anthopleura elegantissima. Journal of Experimental Marine Biology and Ecology, 489, 43–47.
Duchassaing de Fonbressin, P., & Michelotti, G. (1860). Mémoir sur les corralliares des Antilles. Turin: Imprimerie Royale.
Dunn, D. F. (1981). The clownfish sea anemones: Stichodactylidae (Coelenterata: Actiniaria) and other sea anemones symbiotic with pomacentrid fishes. Transactions of the American Philosophical Society, 71(1), 3–115.
Dunn, D. F. (1983). Some Antarctic and sub-Antarctic sea anemones (Coelenterata: Ptychodactiaria and Actiniaria). In L. S. Kornicker (Ed.), Biology of the Antarctic seas XVI, Antarctic research series, 39 (pp. 1–67). Washington D.C.: American Geophysical Union.
Dunn, D. F., Chia, F.-S., & Levine, R. (1980). Nomenclature of Aulactinia (= Bunodactis), with a description of Aulactinia incubans n. sp. (Coelenterata: Actiniaria), an internally brooding sea anemone from Puget Sound. Canadian Journal of Zoology, 58, 2071–2080.
Dykens, J. A., & Shick, J. M. (1984). Photobiology of the symbiotic sea anemone, Anthopleura elegantissima: defenses against photodynamic effects, and seasonal photoacclimatization. The Biological Bulletin, 167(3), 683–697.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797.
England, K. W. (1992). Certain Actiniaria (Cnidaria: Anthozoa) from Hong Kong with additional data on similar species from Aden, Bahrain and Singapore. In B. Morton (Ed.), The marine flora and fauna of Hong Kong and southern China III. Proceedings of the Fourth International Marine Biological Workshop: The Marine Flora and Fauna of Hong Kong and Southern China, Hong Kong, 11–29 April 1989 (pp. 49–95). Hong Kong: Hong Kong University Press.
England, K. W., & Robson, E. A. (1984). A new sea anemone from South Africa (Anthozoa, Ptychodactiaria). Annals of the South African Museum, 94, 305–329.
Fautin, D. G. (2005). Three species of intertidal sea anemones (Anthozoa: Actiniidae) from the tropical Pacific: description of Anthopleura buddemeieri, n. sp., with remarks on Anthopleura asiatica and Gyractis sesere. Pacific Science, 59(3), 379–391.
Fautin, D. G. (2013). Hexacorallians of the World. http://hercules.kgs.ku.edu/hexacoral/anemone2/index.cfm
Fautin, D. G. (2016). Catalog to families, genera, and species of orders Actiniaria and Corallimorpharia (Cnidaria: Anthozoa). Zootaxa, 4145(1), 1–449.
Fautin, D. G., & Chia, F.-S. (1986). Revision of the sea anemone genus Epiactis (Coelenterata: Actiniaria) on the Pacific coast of North America, with descriptions of two new brooding species. Canadian Journal of Zoology, 64(8), 1665–1674.
Fautin, D. G., Crowther, A. L. & Wallace, C. C. (2008). Sea anemones (Cnidaria: Anthozoa: Actiniaria) of Moreton Bay. In Davie, P.J.F. & Phillips, J.A. (Eds.), Proceedings of the Thirteenth International Marine Biological Workshop, The Marine Fauna and Flora of Moreton Bay, Queensland. Memoirs of the Queensland Museum — Nature 54(1): 35–64. Brisbane. ISSN 0079-8835.
Foster, N. L., & Briffa, M. (2014). Familial strife on the seashore: Aggression increases with relatedness in the sea anemone Actinia equina. Behavioural Processes, 103, 243–245.
Francis, L. (1973). Intraspecific aggression and its effect on the distribution of Anthopleura elegantissima and some related sea anemones. The Biological Bulletin, 144(1), 73–92.
Francis, L. (1976). Social organization within clones of the sea anemone Anthopleura elegantissima. The Biological Bulletin, 150(3), 361–376.
Francis, L. (1988). Cloning and aggression among sea anemones (Coelenterata: Actiniaria) of the rocky shore. The Biological Bulletin, 174(3), 241–253.
Freudenstein, J. V. (2005). Characters, states and homology. Systematic Biology, 54(6), 965–973.
Garese, A., Longo, M. V., Martin, J. P., & Acuña, F. H. (2014). The sea anemone Bunodactis octoradiata (Anthozoa: Actiniaria) from southern Patagonia: morphological study and new records. Zoologia, 31(5), 475–481.
Geller, J. B., & Walton, E. D. (2001). Breaking up and getting back together: evolution of symbiosis and cloning in sea anemones (genus Anthopleura) inferred from a molecular phylogeny. Evolution, 55, 1781–1794.
Gomes, P. B., Schama, R., & Solé-Cava, A. M. (2012). Molecular and morphological evidence that Phymactis papillosa from Argentina is, in fact, a new species of the genus Bunodosoma (Cnidaria: Actiniidae). Journal of the Marine Biological Association of the United Kingdom, 92(5), 895–910.
González-Muñoz, R., Simoes, N., Sánchez-Rodríguez, J., Rodríguez, E., & Segura-Puertas, L. (2012). First inventory of sea anemones (Cnidaria: Actiniaria) of the Mexican Caribbean. Zootaxa, 3556, 1–38.
González-Muñoz, R., Simões, N., Mascaró, M., Tello-Musi, J. L., Brugler, M. R., & Rodríguez, E. (2015). Morphological and molecular variability of the sea anemone Phymanthus crucifer (Cnidaria, Anthozoa, Actiniaria, Actinoidea). Marine Biological Association of the United Kingdom. Journal of the Marine Biological Association of the United Kingdom, 95(1), 69–79.
Grosberg, R. K. (1988). The evolution of allorecognition specificity in clonal invertebrates. QUARTERLY REVIEW OF BIOLOGY, 63(4), 377–412.
Gusmão, L. C., & Daly, M. (2010). Evolution of sea anemones (Cnidaria: Actiniaria: Hormathiidae) symbiotic with hermit crabs. Molecular Phylogenetics and Evolution, 56(3), 868–877.
Haag, E., & Dyson, K. (2014). Trade-off between safety and feeding in the sea anemone Anthopleura aureoradiata. New Zealand Journal of Marine and Freshwater Research, 48(4), 540–546.
Hand, C. (1955). The sea anemones of central California, part II. Wasmann Journal of Biology, 13, 37–99.
Hart, C. E., & Crowe, J. H. (1977). The effect of attached gravel on survival of intertidal anemones. Transactions of the American Microscopical Society, 96(1), 28–41.
Hauck, L. L., Phillips, W. S., & Weis, V. M. (2007). Characterization of a novel EF-hand homologue, CnidEF, in the sea anemone Anthopleura elegantissima. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 146(4), 551–559.
Häussermann, V. (2003). Redescription of Oulactis concinnata (Drayton in Dana, 1846) (Cnidaria: Anthozoa: Actiniidae), an actiniid sea anemone from Chile and Perú with special fighting tentacles; with a preliminary revision of the genera with a “frond-like” marginal ruff. Zoologische Verhandelingen, 345, 173–207.
Häussermann, V. (2004). Re-description of Phymactis papillosa (Lesson, 1830) and Phymanthea pluvia (Drayton in Dana, 1846) (Cnidaria: Anthozoa), two common actiniid sea anemones from the south east Pacific with a discussion of related genera. Zoologische Mededelingen, 78(23), 345–381.
Häussermann, V., & Försterra, G. (2001). A new species of sea anemone from Chile, Anemonia alicemartinae n. sp.(Cnidaria: Anthozoa). An invader or an indicator for environmental change in shallow water? Organisms Diversity & Evolution, 1, 211–224.
Hiebert, T. C., & Bingham, B. L. (2012). The effects of symbiotic state on heterotrophic feeding in the temperate sea anemone Anthopleura elegantissima. Marine Biology, 159, 939–950.
Hopper, J. V., Poulin, R., & Thieltges, D. W. (2008). Buffering role of the intertidal anemone Anthopleura aureoradiata in cercarial transmission from snails to crabs. Journal of Experimental Marine Biology and Ecology, 367, 153–156.
Jennison, B. L. (1978). Effects of thermal effluents on reproduction in a sea anemone. In Thorp, J. H., & Gibbons, J. W. (Eds.), Symposium on energy and environmental stress in aquatic systems: selected papers from a symposium held at Augusta, Georgia, November 2–4, 1977, pp. 470–483. Washington, Technical Information Center, U.S. Department of Energy, Florida Atlantic University, Harbor Branch Oceanographic Institute contribution, no. 91. [online version at http://www.iczn.org/iczn/index.jsp]
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649.
Kohno, Y., Satoh, H., Iguchi, A., & Nagai, H. (2009). Characterization of a new hemolytic protein toxin from the sea anemone Anthopleura asiatica. Fisheries Science, 75, 1049–1054.
Kruger, L. M., & Griffiths, C. L. (1998). Sea anemones as secondary consumers on rocky shores in the south-western Cape, South Africa. Journal of Natural History, 32(5), 629–644.
Laird, M. C. (2013). Taxonomy, systematics and biogeography of South African Actiniaria and Corallimorpharia (Doctoral dissertation, University of Cape Town).
Lanfear, R., Calcott, B., Ho, S. Y. W., & Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29(6), 1695–1701.
Larson, P. G., & Daly, M. (2016). Phylogenetic analysis reveals an evolutionary transition from internal to external brooding in Epiactis Verrill (Cnidaria: Anthozoa: Actiniaria) and rejects the validity of the genus Cnidopus Carlgren. Molecular Phylogenetics and Evolution, 94, 548–558.
Letsch, M. R., Muller-Parker, G., Friedl, T., & Lewis, L. A. (2009). Elliptochloris marina sp. nov. (Trebouxiophyceae, Chlorophyta), symbiotic green alga of the temperate pacific sea anemones Anthopleura xanthogrammica and A. elegantissima (Anthozoa, Cnidaria). Journal of Phycology, 45, 1127–1135.
Levine, M. R., & Muller-Parker, G. (2012). Distribution patterns and nutritional contributions of algal symbionts in the sea anemone Anthopleura xanthogrammica. Marine Ecology Progress Series, 453, 79–94.
Macrander, J., Brugler, M. R., & Daly, M. (2015). A RNA-seq approach to identify putative toxins from acrorhagi in aggressive and non-aggressive Anthopleura elegantissima polyps. BMC Genomics, 16(1), 1.
Maddison, W. P., & Maddison, D. R. (2011). Mesquite: a modular system for evolutionary analysis. Version 2.75. Available from: http://mesquiteproject.org.
McBride, B. B., Muller-Parker, G., & Jakobsen, H. H. (2009). Low thermal limit of growth rate of Symbiodinium californium (Dinophyta) in culture may restrict the symbiont to southern populations of its host anemones (Anthopleura spp.; Anthozoa, Cnidaria). Journal of Phycology, 45, 855–863.
McFadden, C. S., Grosberg, R. K., Cameron, B. B., Karlton, D. P., & Secord, D. (1997). Genetic relationships within and between clonal and solitary forms of the sea anemone Anthopleura elegantissima revisited: evidence for the existence of two species. Marine Biology, 128, 127–139.
Miura, O., Keawtawee, T., Sato, N., & Onodera, K.-I. (2014). Vertical zonation of endosymbiotic zooxanthellae within a population of the intertidal sea anemone, Anthopleura uchidai. Marine Biology, 161(8), 1745–1754.
Morar, S. R., Bury, S. J., Wilkinson, S. P., & Davy, S. K. (2011). Sedimentary nitrogen uptake and assimilation in the temperate zooxanthellate sea anemone Anthopleura aureoradiata. Journal of Experimental Marine Biology and Ecology, 399(2), 110–119.
Parry, G. (1951). The Actiniaria of New Zealand. A check-list of recorded and new species a review of the literature and a key to the commoner forms. Records of the Canterbury Museum, 6(1), 83–119.
Pearse, V. B. (1974). Modification of sea anemone behavior by symbiotic zooxanthellae: expansion and contraction. The Biological Bulletin, 147, 641–651.
Pearse, V., & Francis, L. (2000). Anthopleura sola, a new species, solitary sibling species to the aggregating sea anemone Anthopleura elegantissima. Proceedings of the Biological Society of Washington, 113, 596–608.
Peigneur, S., Billen, B., Derua, R., Waelkens, E., Debaveye, S., Béress, L., et al. (2011) A bifunctional sea anemone peptide with Kunitz type protease and potassium channel inhibiting properties. Biochemical Pharmacology, 82(1), 81–90.
Peigneur, S., Lescrinier, E., Moller, C., Marí, F., Béress, L., & Tytgat, J. (2012). A natural point mutation reveals target promiscuity of toxins isolated from the sea anemone Anthopleura elegantissima. Biophysical journal, (3, Supplement 1). doi:10.1016/j.bpj.2011.11.3582.
Pineda, J., & Escofet, A. (1989). Selective effects of disturbance populations of sea anemones from northern Baja California, Mexico. Marine Ecology Progress Series, 55, 55–62.
Posada, D. (2008). jModelTest: phylogenetic model averaging. Molecular Biology and Evolution, 25(7), 1253–1256.
Quesada, A. J., Acuña, F. H., & Cortés, J. (2014). Diet of the sea anemone Anthopleura nigrescens: composition and variation between daytime and nighttime high tides. Zoological Studies, 53(1), 26.
Richier, S., Rodriguez-Lanetty, M., Schnitzler, C. E., & Weis, V. M. (2008). Response of the symbiotic cnidarian Anthopleura elegantissima transcriptome to temperature and UV increase. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 3(4), 283–289.
Riemann-Zürneck, K. (1980). Actiniaria des Südwestatlantik V. Bolocera, Isotealia, Isosicyonis (Actiniidae). Mitteilungen aus dem Hamburgischen Zoologischen Museum und Institut, 77, 19–33.
Riemann-Zürneck, K., & Griffiths, C. L. (1999). Korsaranthus natalensis (Carlgren, 1938) nov. comb. (Cnidaria: Actiniaria) a mobile sea anemone attacking octocorals. South Afrrican Journal of Zoology, 34, 190–196.
Rodríguez, E., & López-González, P. J. (2003). Stephanthus antarcticus, a new genus and species of sea anemone (Actiniaria, Haloclavidae) from the South Shetland Islands, Antarctica. Helgoland Marine Research, 57(1), 54–62.
Rodríguez, E., & López-González, P. J. (2013). New records of Antarctic and sub-Antarctic sea anemones (Cnidaria, Anthozoa, Actiniaria and Corallimorpharia) from the Weddell Sea, Antarctic Peninsula, and Scotia Arc. Zootaxa, 3624(1), 001–100.
Rodríguez, E., Barbeitos, M., Daly, M., Gusmão, L. C., & Häussermann, V. (2012). Toward a natural classification: phylogeny of acontiate sea anemones (Cnidaria, Anthozoa, Actiniaria). Cladistics, 28, 375–392.
Rodríguez, E., Barbeitos, M., Brugler, M. R., Crowley, L. M., Grajales, A., Gusmão, L. C., et al. (2014). Hidden among sea anemones: The first comprehensive phylogenetic reconstruction of the order Actiniaria (Cnidaria, Anthozoa, Hexacorallia) reveals a novel group of hexacorals. PLoS one DOI. doi:10.1371/journal.pone.0096998.
Rudin, F. S., & Briffa, M. (2011). The logical polyp: assessments and decisions during contests in the beadlet anemone Actinia equina. Behavioral Ecology, 22(6), 1278–1285.
Sanamyan, N., & Sanamyan, K. (1998). Some Actiniaria from the Commander Islands (Cnidaria: Anthozoa). Zoosystematica Rossica, 7(1), 1–8.
Sanders, J. G., & Palumbi, S. R. (2011). Populations of Symbiodinium muscatinei show strong biogeographic structuring in the intertidal anemone Anthopleura elegantissima. The Biological Bulletin, 220(3), 199–208.
Saunders, B. K., & Muller-Parker, G. (1997). The effects of temperature and light on two algal populations in the temperate sea anemone Anthopleura elegantissima (Brandt, 1835). Journal of Experimental Marine Biology and Ecology, 211, 213–224.
Shick, J. M. (1991). A functional biology of sea anemones. New York: Chapman & Hall.
Spano, C., Rozbaczylo, N., Häussermann, V., & Bravo, R. (2013). Redescription of the sea anemones Anthopleura hermaphroditica and Bunodactis hermafroditica (Cnidaria: Anthozoa: Actiniaria) from Chile. Revista de Biología Marina y Oceanografía, 48, 521–534.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. doi:10.1093/bioinformatics/btu033.
Stephenson, T. A. (1928). The British sea anemones. Volume I. London: The Ray Society.
Stephenson, T. A. (1935). The British sea anemones. Volume II. London: The Ray Society.
Stephenson, T. A., & Stephenson, A. (1972). Life between tidemarks on rocky shores. San Francisco: W. H. Freeman.
Towanda, T., & Thuesen, E. V. (2012). Prolonged exposure to elevated CO2 promotes growth of the algal symbiont Symbiodinium muscatinei in the intertidal sea anemone Anthopleura elegantissima. Biology Open, 1(7), 615–621.
Verde, E. A., & McCloskey, L. R. (2002). A comparative analysis of the photobiology of zooxanthellae and zoochlorellae symbiotic with the temperate clonal anemone Anthopleura elegantissima (Brandt), II. Effect of light intensity. Marine Biology, 141, 225–239.
Weis, V. M., Verde, E. A., Pribyl, A., & Schwartz, J. A. (2002). Aspects of the larval biology of the sea anemones Anthopleura elegantissima and A. artemisia. Invertebrate Biology, 121(3), 190–201.
Wicksten, M. K. (1984). Survival of sea anemones in Bunker C fuel. Marine Pollution Bulletin, 15(1), 28–33.
Xiang, H., Tao, W., Wang, L., Wang, F., & Xu, A. (2008). The effect of recombinant neurotoxins from the sea anemone Anthopleura sp. on sodium currents of rat cerebral cortical neurons. Cellular and Molecular Neurobiology, 28(8), 1119–1128.
Zhang, L., & Zhu, Q. (2016). Complete mitochondrial genome of the sea anemone, Anthopleura midori (Actiniaria: Actiniidae). Mitochondrial DNA Part A, 1–2.
Zhang, M., Liu, X. S., Diochot, S., Lazdunski, M., & Tseng, G. N. (2007). APETx1 from sea anemone Anthopleura elegantissima is a gating modifier peptide toxin of the human ether-a-go-go- related potassium channel. Molecular Pharmacology, 72(2), 259–268.
Acknowledgements
This work would not have been possible without specimens and field support from A. Ardelean, P. Cartwright, H. Cha, A. D’ Orazio, L. Francis, R. Goodwill, L. Gusmão, V. Haüssermann, C. McFadden, B. Picton, A. Schulze, and K. Yanagi. Lab support was provided by A. Chaudhuri, A. Lindgren, and A. Reft. Funding for this project came from the American Museum of Natural History, NSF DEB-9978106 to DGF, and NSF EF-0531763 to MD.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Daly, M., Crowley, L.M., Larson, P. et al. Anthopleura and the phylogeny of Actinioidea (Cnidaria: Anthozoa: Actiniaria). Org Divers Evol 17, 545–564 (2017). https://doi.org/10.1007/s13127-017-0326-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-017-0326-6