Abstract
MicroRNAs (miRNAs) are noncoding RNAs involved in the regulation of the diverse biological processes such as metabolism, proliferation, and cell cycle, in addition to regulation of differentiation. So far, some miRNAs have been recognized to have important role in regulating hepatic functions. Statistically, let-7f has been revealed as a negative regulator of hepatic differentiation. In the present study, we investigated the effect of let-7f on hepatic differentiation of human adipose tissue-derived stem cells (hADSCs). hADSCs were transduced with recombinant lentivirus containing human inhibitor let-7 f. The expression of hepatocyte nuclear factors alpha (HNF4a), albumin (ALB), alpha fetoprotein (AFP), cytokeratin 18 (CK18), and cytokeratin 19 (CK19) was evaluated using quantitative real-time PCR (qRT-PCR). Immunocytochemistry was used to investigate the expression levels of the hepatocyte markers including ALB, AFP, and HNF4a, and biochemical analysis was implemented for hepatic function, glycogen deposition, and urea secretion. qRT-PCR showed significant upregulation in HNF4a, ALB, AFP, CK18, and CK19 expression in cells transduced with let-7f inhibitor lentiviruses. Moreover, positive staining was detected for ALB, AFP, and HNF4a using immunocytochemistry. Urea production and glycogen deposits were also found in the treated cells, the two specific features of the hepatic cells. Therefore, let-7f silencing led to the increased expression of the hepatocyte-specific factors and the accelerated hADSCs hepatic differentiation. Summing all these finding together, our present report has provided evidences that inhibition of let-7f would facilitate induction of hADSCs into hepatocyte-like cells and possibly in regenerative therapy of the liver disease in a wider spectrum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stem cell therapy is among the exceptional candidates that have been suggested for liver disorders [29]. Many types of stem cells have already been used for generation of hepatocyte-like cells in regenerative medicine. Some studies have indicated mesenchymal stem cells (MSCs) as a preferred cell source. MSCs are a group of multipotent cells characterized by high self-renewal potential and differentiation [22]. They can be isolated from different tissues including bone marrow [25], adipose tissue [34], umbilical cord blood [6], and the circulatory blood [24]. Adipose tissue contains an abundant and accessible number of MSCs with faster growth and higher proliferation capacity compared to MSCs from other sources [15, 16]. Adipose tissue-derived stem cells (ADSCs) hold the capacity to differentiate into osteocyte [35], neural cells [3], muscular cells [10], and hepatocyte [4]. Also, ADSCs can easily adapt hepatocyte-like function. These features make them an interesting source for cell therapy of the end-stage liver diseases in regenerative medicine [32].
MicroRNAs (miRNAs) are a group of posttranscriptional regulators with the key role in the diverse functions including cell proliferation, apoptosis, and organogenesis [2]. They are also well-known regulators of the differentiation, self-renewal, and division of stem cells [13, 14]. So far, specific and distinct miRNAs have been identified that mediate differentiation of MSCs into various cell types; for example, miR-1 [7] and miR-24 [27] play a role in differentiation of MSCs into myocardial cells, whereas miR-9 [20] is important during neural differentiation of MSCs. On the other hand, only few studies have characterized miRNA expression during differentiation of stem cells into the hepatocyte-like cells. Recently, the role of miR-122 was investigated during hepatocyte differentiation of embryonic stem cells and liver-derived progenitor cells [8, 17].
To our knowledge, there is only one study that has suggested the involvement of miRNAs in the differentiation of MSCs into hepatocyte-like cells [18]. Applying statistical analysis, Koh et al. have demonstrated that let-7 family miRNAs could act as negative regulator for hepatic differentiation by suppressing hepatocyte nuclear factor 4 alpha (HNF4a). Additionally, they have shown that among let-7 family members, let-7f expressed strongly both in the intracellular and extracellular of MSCs. HNF4a is a nuclear transcriptional factor which plays an important role in liver development in addition to functional differentiation of hepatocytes [26]. Furthermore, it functions as an essential factor in differentiation of hepatoblasts into the mature state in mouse [23]. Moreover, it has been shown that elevation in the HNF4a expression is in parallel with the upregulation of miR-122 (the liver-specific miRNA) during liver development in the mouse embryos [31].
Identification of factors involved in hepatic differentiation is likely to facilitate the development of novel therapeutic strategies for hepatic disorders in which, so far, only organ replacement therapy was implemented. Wide varieties of strategies have been employed for in vitro stem cells differentiation toward hepatocyte-like cells. However, currently, protocols mostly rely on using of expensive cytokines and growth factors. Therefore, regarding the importance of HNF4a in hepatic differentiation, in the present study, attempts were made to evaluate whether silencing of let-7f upregulates the expression of HNF4a in hADSCs and if it promotes hepatic differentiation without requirement for additional extrinsic factors, mainly due to the high manufacturing cost of recombinant proteins.
Material and methods
Isolation of hADSCs and cell culture
Consistent with the Institutional Medical Ethics Committee guideline, human adipose tissue was obtained from lipoaspirate, and isolation of hADSCs was carried out according to Zuk et al. [34]. Briefly, following to mincing the adipose tissue, fragments were digested with collagenase type I (Sigma, USA) at 37 °C for 1 h with vigorous shaking. Afterward, hADSCs were isolated using centrifugation at 1,200 × g for 10 min and were resuspended in the control medium containing Dulbecco’s modified Eagle’s medium (DMEM), 10 % fetal bovine serum (FBS), and 1 % penicillin and streptomycin (Sigma). Cells were plated in tissue culture flask (Nunc, Denmark) and were incubated at 37 °C supplied with 5 % CO2 incubator. The growing cell culture medium was changed twice a week. Confluent cells at 70 to 80 % were harvested and used for the following analyses and differentiation assays.
Hepatic differentiation of hADSCs
hADSCs were differentiated into hepatocyte-like cells as previously described [16]. Briefly, hADSCs were plated onto 24-well plastic cell culture plate (Nunc, Denmark) at a concentration of 1 × 103 cells/well. Hepatic induction was performed using two-step differentiation procedure. In the first step, cells were cultured in medium containing DMEM, 10 % FBS, 20 ng/ml hepatocyte growth factor (HGF), and 10−7 mol/l dexamethasone for 1 week. For the following 2 weeks, oncostatin M (OSM) was added to the medium at a concentration of 10 ng/ml. Culture medium was refreshed twice a week, and after 21 days, cells were used for hepatic differentiation assays.
Virus production and transduction of hADSCs
Production of lentiviral particles was performed according to Zuffery et al. [33]. Briefly, the inhibitor hsa-let-7f lentivector (abm, USA), psPAX2 (encoding the gag and Pol proteins), and pMD2.G (encoding VSV G envelop protein) were cotransfected in 293 T cells by calcium chloride/DNA precipitation method. Cell culture media was collected 24 and 48 h after transfection, and viral particles were concentrated using ultracentrifuge for 90 min at 72 × 103 × g. The resultant viral particles were stored at −70 °C. Moreover, 15 μl of the resultant viruses was taken for titration. 293 T cells were used as target cells, and the percentage of green fluorescent protein (GFP)-positive cells was determined by FACS.
Following to seeding hADSCs at density of 1 × 104 cells/well onto 24-well cell culture plate (Nunc, Denmark), cells were transduced with the concentrated virus particles at MOI of 50. Culture medium was refreshed a day after transduction. Additionally, hADSCs were transduced with the scramble lentiviral vector as a negative control for monitoring nonspecific changes in genes as a result of lentiviral transduction. Transduction efficiency of the both lentiviral vectors (encoding inhibitor let-7f and scramble) was evaluated using inverted fluorescence microscope.
miRNA extraction and quantitative RT-PCR
Total RNA was isolated using TRI reagent (Sigma, USA), and complementary DNA (cDNA) was synthesized by universal cDNA synthesis kit (Exiqon, USA), followed by amplification using SYBR green master mix (Exiqon, USA) and let-7f LNA PCR primer set (Exiqon, USA) according to the manufacturer’s instruction. The U6 snRNA PCR primer set (Exiqon, USA) was used to amplify U6 as a reference gene for normalization of target transcript levels. Amplification and detection were carried out by Applied Biosystems (StepOne Real-time PCR system, USA).
Quantitative RT-PCR analysis
In order to investigate the expression of hepatocyte-specific markers, quantitative real-time PCR (qRT-PCR) was carried out for alpha fetoprotein (AFP), albumin (ALB), cytokeratin 18 (CK18), cytokeratin 19 (CK19), and HNF4a, in addition to β-actin as a reference gene. The primer sets are listed in Table 1. RNA was extracted on days 1, 7, 14, and 21 using RNeasy mini kit (Qiagen) and was reverse transcribed to cDNA using power cDNA synthesis kit (Intron, Korea) according to the manufacturer’s guidelines. Three-step procedure PCR was carried out as follows: denaturation at 95 °C, 30 s, annealing at 56 °C, 30 s, and extension at 72 °C for 60 s were performed up to 40 cycles using Applied Biosystems, and the comparative CT method (ΔΔCt) served as the analysis method for the relative quantification of gene expression.
Immunocytochemistry
Cells were fixed with 4 % paraformaldehyde and permeablized in 2 % Triton-X100 for 30 min. Nonspecific bindings was blocked with 1 % BSA and 0.1 % Triton X-100; cells were then incubated overnight at 4 °C with 1:200 monoclonal anti-human albumin (Abcam, USA), 1:200 monoclonal anti-human alpha 1 fetoprotein (Abcam, USA), and 1:100 monoclonal anti-human HNF4a (Abcam, USA) separately. Subsequently, cells were incubated with the Alexa flour 594 donkey anti-mouse IgG for 1 h. Nuclear staining was carried out using 4,6-diamidino-2-phenylindole, and cells were visualized applying inverted fluorescence microscope (Nikon 200, Japan).
Glycogen staining
After fixation with 4 % paraformaldehyde, cells were oxidized with 1 % periodic acid for 5 min, treated with Schiff’s reagent (Sigma–Aldrich, USA), stained with hematoxylin and visualized under inverted microscope.
Urea detection
The presence of urea was determined in cell culture media using colorimetric assay kit (Zistchem, Iran) according to the manufacturer’s instruction.
Statistical analysis
Results were expressed as mean ± SEM, and statistical analysis was assessed using the Student’s t test; P values below 0.05 were considered significant.
Results
Identification of hepatocyte-specific markers during hepatic differentiation
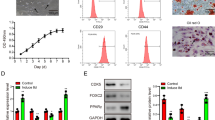
Hepatic differentiation of hADSCs was induced using cytokines and growth factors such as HGF, dexamethasone, and OSM for 21 days. Differentiation was assessed by morphological characterization in addition to functional evaluation of hepatocyte-like cells. During hepatic differentiation, cell morphology was changed from the fibroblast-like to polygonal shape remarkably. In addition, accumulation of granules was observed in the cytoplasm (Fig. 1). Production of glycogen and urea synthesis are two common features of the liver and specific to hepatocytes. Thus, we characterized these two features for assessing the success in the differentiation of hADSCs into the hepatocyte-like cells. Glycogen accumulation was evaluated using glycogen staining assay. On day 21, cells were strongly positive for glycogen deposition, indicating the acquisition of hepatocyte phenotype, while glycogen staining was negative for undifferentiated hADSCs (Fig. 2a). Consequently, urea synthesis was evaluated by measuring urea level in the cell culture media at different time intervals (days 1, 7, 14, and 21). Undifferentiated hADSCs did not produce detectable levels of urea, whereas urea production was significant in the hepatocyte-like cell culture media on days 14 and 21 (Fig. 2e).
Morphology of hADSCs at days 1 and 21 after induction of hepatic differentiation. Cells were treated with HGF (20 ng/ml) and dexamethasone (10−7 mol/l) for 1 week and with OSM (10 ng/ml), HGF (20 ng/ml), and dexamethasone (10−7 mol/l) for the subsequent 2 weeks. hADSCs represented the spindle shape and fibroblast-like morphology (a); hepatic induction of these cells resulted in significant changes toward a round shape at 21 days after hepatic induction (b). hADSCs human adipose-derived stem cells, HGF hepatocyte growth factor, OSM oncostatin M
Differentiation of hADSCs into hepatocyte-like cells using growth factors. Glycogen staining of hepatocyte-like cells derived from hADSCs and undifferentiated hADSCs (a). Immunocytochemistry assay for AFP, ALB, and HNF4a in hepatocyte-like cells, respectively (b, c, d). Urea production was evaluated in the cell culture medium at different time points of the culture in the hepatic differentiation media (e). qRT-PCR analysis of the selected hepatocyte-specific markers during hepatic differentiation. The level of expression for AFP, ALB, CK18, CK19, and HNF4a was as measured with qRT-PCR, and data were normalized using β-actin as an internal expression control and were compared with the undifferentiated hADSCs (day 0). Results represent mean ± SEM (f). ALB albumin, AFP alpha fetoprotein, CK18 cytokeratin 18, CK19 cytokeratin 19, HNF4a hepatocyte nuclear factor, hADSCs human adipose-derived stem cells, qRT-PCR quantitative real-time PCR
In addition, hepatocyte-like cells were tested for expression of hepatocyte-specific genes, such as AFP, ALB, CK18, CK19, and HNF4a by qRT-PCR at different time intervals. Undifferentiated hADSCs and HepG2 were used as a negative and positive control, respectively. qRT-PCR showed no significant change in the expression of CK19, whereas the expression of CK18 and HNF4a was increased significantly on day 21 in the cells treated with growth factors compared to undifferentiated cells (Fig. 2f). Also, the expression level of ALB and AFP was evaluated and compared with the undifferentiated hADSCs. The expression of AFP was significantly increased 7 days after culturing and continued until the day 21 (Fig. 2f). Changes in the ALB levels were noticed later on day 14 after induction of the differentiation and reached up to 2-fold on day 21 (Fig. 2f).
We also examined expression levels of the AFP, ALB, and HNF4a at protein level through immunofluorescence assay. In agreement with the gene expression, staining of cells showed positive staining for AFP, ALB, and HNF4a on day 21 of differentiation, while undifferentiated cells were negative for all the above three hepatocyte markers (Fig. 2b, c, d).
Silencing of let-7f in human hADSCs
Analyzing the level of let-7f, we noticed that it became subject of gradual downregulation in expression during hepatic differentiation of hADSCs through the process of growth factor application (Fig. 3). This finding encouraged us to evaluate whether hepatic differentiation of hADSCs would be affected by silencing of let-7 f. In order to examine this hypothesis, we investigated the effect of let-7f on hepatic differentiation of hADSCs by decreasing the level of let-7f via lentiviruses containing inhibitor let-7 f. Also, hADSCs were transduced with lentiviruses containing scramble as a negative control. The calculated titer for both lentiviral particles was 109 viral particles/ml (data not shown). After 48 h of transduction, the percentage of GFP-positive cells was assessed using fluorescence microscope. Cells with 80 % or more of the transduction efficiency were used for further analysis.
Let-7f expression in hADSCs through the course of hepatic differentiation induced by growth factors. qRT-PCR was used to assess let-7f expression level on the days 1, 7, 14, and 21. Data were normalized using U6 snRNA and expressed relative to undifferentiated hADSCs (day 0). Results represent mean ± SEM. hADSCs human adipose derived stem cells, qRT-PCR quantitative real-time PCR
The level of let-7f expression in the transduced hADSCs either with inhibitor let-7f or scramble lentivectors was measured by qRT-PCR on the days 7, 14, and 21. Results showed a significant (P < 0.05) decline in let-7f expression in cells transduced with the inhibitor let-7f compared to negative control (Fig. 4).
Silencing of let-7f in hADSCs. qRT-PCR result of let-7f expression level in hADSCs transduced with lentiviruses containing either inhibitor let-7f or negative control (scramble). Data were normalized using U6 snRNA and expressed relative to undifferentiated hADSCs (day 0). Mean ± SEM of the measurements are presented. hADSCs human adipose derived stem cells
Effect of let-7f inhibition on the expression of hepatocyte-specific factors in hADSCs
Likewise, to determine the effect of let-7f inhibition on hepatocyte-specific factors, we analyzed the expression levels of AFP, ALB, CK18, CK19, and HNF4a at different time intervals (day 1, day 7, day 14, and day 21) using qRT-PCR. Compared to the negative control, we observed a significant upregulation of ALB and AFP expression in transduced hADSCs with the inhibitor let-7f after 14 and 21 days of transduction (P < 0.05) (Fig. 5f). Interestingly, the same result was found for CK18, although the expression level of CK19 was increased significantly (2.5-fold) only on the day 21 (Fig. 5f). Noteworthy that we found significant upregulation of HNF4a 14 and 21 days following transduction with inhibitor let-7f by 2.5- and 4-fold in comparison with negative control (Fig. 5f). This finding supports suppression of HNF4a expression by let-7 f. In addition, positive staining was detected for AFP, ALB, and HNF4a in the cells transduced with lentivirses containing inhibitor let-7 f. In contrast, such staining was negative for negative control (Fig. 5b, c, d). Biochemically, this notion was supported by observing glycogen deposition (Fig. 5a) and urea secretion (Fig. 5e) in the presence of inhibitor let-7f on day 21. These findings indicate a negative regulatory function for let-7f in hADSCs in the path toward hepatic differentiation.
The effect of let-7f inhibition on the expression of hepatocyte-specific markers in hADSCs. Glycogen deposition was detected using Shiff’s reagent in hADSCs transduced with lentiviruses containing either inhibitor let-7f or negative control (scramble) (a). Immunocytochemistry assay for AFP, ALB, and HNF4a in hADSCs transduced with lentiviruses containing either inhibitor let-7f or negative (scrambled) control (b, c, d). Urea was detected in the cell culture medium on days 1, 7, 14, and 21 following to lentiviral transduction of hADSCs; either with inhibitor let-7f or negative (scramble) control (e). Hepatic differentiation was evaluated with qRT-PCR for selected factors including AFP, ALB, CK18, CK19, and HNF4a at different time intervals. Data were normalized with β-actin and expressed relative to undifferentiated hADSCs. The mean ± SEM of the results are presented (g). Abbreviations are the same as those in Fig. 3. Adobe Photoshop CS 8.0 was used to create the figures
Discussion
Liver is the central organ for a wide range of vital reactions among which detoxification, biochemical, and metabolic reactions. Currently, millions of patients worldwide suffer from end-stage liver disease, and the only available therapy since 1982 has been liver transplantation (LT) [1]. Due to the accompanying chronic complications in LT, cell-based therapy has been introduced as an alternative strategy for hepatic disease. In this regard, various strategies have already been employed for the in vitro stem cell differentiation toward hepatocyte-like cells [21]. hADSCs have a hepatogenic potential allowing them to easily differentiate into hepatocyte-specific phenotype [32]. Moreover, these cells can be obtained from patients with a minimally invasive procedure, compared to BMSCs. The abundance, easy access, and high proliferation rate of these cells, as well as the possibility to keep the cells in culture over a long time period, were features making hADSCs the ideal choice for this study.
Recent studies have indicated roles that are played by specific miRNAs during differentiation of MSCs into specific cell type, such as osteoblasts [12], chondrocytes [28], neurons [20], and adipocytes [11]. Among the miRNAs, only the let-7 family of miRNAs has been shown to be involved in the MSCs differentiation to hepatocyte-like cells [18]. Applying analysis of deep sequencing, Koh et al. have shown the possible role of let-7 miRNA family in the regulation of HNF4a. This transcriptional factor has shown to regulate expression of several hepatic genes [30] and functions as an essential transcription factor in the morphological and functional differentiation of hepatocytes [26]. In addition, a correlation between liver development and the expression of HNF4a has also been shown by previous works [30, 26]. In accordance with these results, we examined the effect of silencing let-7f which is the only known miRNAs during hepatic differentiation of MSCs on the differentiation of hADSCs toward hepatocyte-like cells in the absence of any extrinsic factors.
Using growth factors and cytokines, we were able to induce hepatic differentiation of hADSCs. This differentiation was determined through elevation of hepatocyte-specific factors, such as AFP, ALB, CK18, CK19, and HNF4, and further was confirmed by positive immunostaining for AFP, ALB, and HNF4a. In accordance to hepatocyte phenotype, we also detected glycogen deposition and urea secretion in hepatocyte-like cells.
Applying qRT-PCR, our data indicate that the expression of let-7f subjects to the downregulation during hepatic differentiation of hADSCs using growth factors. Hence, to evaluate the impact of let-7f on hepatic differentiation, lentiviral transduction method was used to silence let-7f in hADSCs. Cells with at least 80 % efficiency were selected, and subsequent qRT-PCR displayed that expression of let-7f was downregulated in cells transduced by inhibitor let-7 f. Furthermore, our results showed that inhibition of let-7f in hADSCs could intensify hepatic differentiation without requirement for any extrinsic factors. This notion could be judged by significant upregulation of AFP, ALB, CK18, CK19, and HNF4a in cells transduced with inhibitor let-7 f. Protein production is considered as the ultimate reflection of gene expression. As a result, works were focused to evaluate the levels of proteins related to each of these hepatocyte-specific genes. Performing immunocytochemistry, we were able to show the expression of AFP, ALB, and HNF4a on day 21 and acquisition of hepatocyte-like phenotype in hADSCs transduced with inhibitor let-7 f. The observations of glycogen deposition and urea secretion are further evidences for the hepatic differentiation of hADSCs through let-7f silencing.
Based on our findings, the inhibition of let-7f miRNA would result in the expression of HNF4a in hADSCs; therefore, let-7f clearly mediates regulation of HNF4a. Indeed, HNF4a functions as a positive transcriptional regulator for many hepatocyte genes [5, 23] and plays a significant role in hepatic differentiation and liver morphogenesis as well [9, 19]. Based on the above reports, our results also validate the importance of HNF4a as an essential factor in hepatic differentiation of stem cells.
In conclusion, in the present study, we have shown that miRNAs such as let-7f act as a negative regulator for hepatic differentiation of hADSCs through suppression of HNF4a, as silencing of this factor accelerates hepatic differentiation of this type of cells. Such findings are likely to facilitate the development of novel therapeutic strategies for managing liver diseases.
References
Åberg F, Iisoniemi H, Höckerstedt K (2011) Long-term results of liver transplantation. Scand J Surg 100:14–21
Alvarez-Garcia I, Miska EA (2005) MicroRNA functions in animal development and human disease. Development 132:4653–4662
Ashjian PA, Elbarbary AS, Edmonds B, DeUgarte D, Zhu M, Zuk PA, Lorenz HP, Benhaim P, Hedrick HK (2003) In vitro differentiation of human processed lipoaspirate cells into early neural progenitors. Plast Reconstr Surg 111:1922–1931
Banas A, Teratani T, Yamamoto Y, Tokuhara M, Takeshita F, Osaki M, Kato T, Okochi T, Ochiya T (2009) Rapid hepatic fate specification of adipose-derived stem cells and their therapeutic potential for liver failure. J Gastroenterol Hepatol 24:70–77
Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA (2006) Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci 103:8419–8424
Bieback K, Kern S, Kluter H, Eichler H (2004) Critical parameters for the isolation of mesenchymal stem cells from umbilical cord blood. Stem Cells 22:625–634
Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38:228–233
Chen Y, Zhou HO, Sarver AL, Zeng Y, Roy-Chowdhury J, Steer CJ, Sahin MB (2010) Hepatic differentiation of liver-derived progenitor cells and their characterization by microRNA analysis. Liver Transolant 16:1086–1097
DeLaForest A, Nagaoka M, Tayeb KS, Noto FK, Konopka G, Battle MA, Duncan SA (2011) HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Dev Stem Cells 138:4143–4153
Deslex S, Negrel R, VannierC EJ, Ailhaud G (2007) Differentiation of human adipocyte precursors in a chemically define d serum-free medium. Exp Cell Res 313:2875–2886
Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R (2004) MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279:52361–52365
Eskildsen T, Taipaleenma KH, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M (2011) MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A 108(15):6139–6144
Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H (2005) Stem cell division is regulated by the microRNA pathway. Nature 435:974–978
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Kern S, Eichler H, Stoeve J, Kluter H, Bieback K (2006) Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells 24:1294–1301
Khoshdel A, Lotfi AS, Soleimani M, Daliri M, Mota A, Adibi B (2012) Evaluation of biochemical markers in hepatocyte like cells differentiated from adipose derivation stem cells. WASJ 16(5):693–698
Kim N, Kim H, Jung I, Kim Y, Kim D, Han YM (2011) Expression profiles of miRNAs in human embryonic stem cells during hepatocyte differentiation. Hepatol Res 41:170–183
Koh W, Sheng CT, Tan B, Lee QY, Kuznetsov V, Kiang LS, Tanavde V (2010) Analysis of deep sequencing micro-RNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics 11:6
Li J, Ning G, Duncan SA (2000) Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev 14:464–474
Lim PK, Patel SA, Gregory LA, Rameshwar P (2010) Neurogenesis: role for microRNAs and mesenchymal stem cells in pathological states. Curr Med Chem 17:2159–2167
Lodi D, Iannitti TM, Palmieri (2011) Stem cells in clinical practice: applications and warnings. J Exp Clin Cancer Res 30:9
Otto RW, Wright NA (2011) Mesenchymal stem cells: from experiment to clinic. Fibrogenesis Tissue Repair 4:20
Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA (2003) Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34:292–296
Phinney DG, Isakova I (2005) Plasticity and therapeutic potential of mesenchymal stem cells in the nervous system. Curr Pharm 11:1255–1265
Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147
Spath GF, Weis MC (1998) Hepatocyte nuclear factor 4a provokes expression of epithelial marker genes, acting as a morphogen in dedifferentiated hepatoma Cells. J Cell Biol 140(4):935–946
Sun F, Wang J, Pan Q, Yu Y, Zhang Y, Wan Y, Wang J, Li X, Hong A (2009) Characterization of function and regulation of miR-24-1 and miR-31. Biochem Biophys Res Commun 380:660–665
Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T (2006) The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 580:4214–4217
Vosough M, Moslem M, Pournasr B, Baharvand H (2011) Cell-based therapeutics for liver disorders. Brit Med Bull 1–16
Watt AJ, Garrison WD, Duncan SA (2003) HNF4: a central regulator of hepatocyte differentiation and function. Hepatology 37(6):1249–1253
Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH (2010) Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1during liver development. Hepatology 52:1431–1442
Zemel R, Bachmetov L, Ad-El D, Abraham A, TurKaspa R (2009) Expression of liver specific markers in naive adipose derived mesenchymal stem cells. Liver Int 1326–1337
Zufferey R, Trono D (2000) Production of high-titer lentiviral vectors. Curr Protoc Human Genet 12(10):1–12
Zuk PA, Zhu M, Mizuno H (2001) Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 7:211–228
Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH (2002) Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell 13:4279–4295
Acknowledgements
This work was supported by a grant (91000780) from Iran National Science Foundation (INSF).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Davoodian, N., Lotfi, A.S., Soleimani, M. et al. Let-7f microRNA negatively regulates hepatic differentiation of human adipose tissue-derived stem cells. J Physiol Biochem 70, 781–789 (2014). https://doi.org/10.1007/s13105-014-0346-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13105-014-0346-z