Abstract
Agricultural, fruit and vegetable wastes were investigated as cost effective source for pectinase production. Orange peel proved as the best inducer of pectinase in a very simple culture medium used. Optimization of various cultural parameters was carried out by statistical method using response surface methodology and central composite design. The basic aim was to find out the best among local agri-wastes for maximum production of pectinase keeping in mind the agro-based economy of the country. Also the aim was to find the best pectinase of alkaline and thermophilic nature having maximum activity in minimum time, to be used in various industrial processes mainly textile industry which is the key industry in a country like Pakistan. The Bacillus licheniformis used in the present study was of special worth as it gave maximum pectinase activity of 219 U/ml in submerged fermentation, in a very simple medium with very few components (NaNO3, KH2PO4, KCl, MgSO4, Tryptone and Orange peel) under optimum conditions of pH 9.5, when incubated at 37 °C for 120 h in an alkaline culture medium of pH 9.5 supplementing with 0.3 % inoculum, 2.5 % orange peel and 0.5 % tryptone. The enzyme exhibited thermophilic nature by showing maximum activity when incubated with substrate (0.5 % citrus pectin solution pH 8.0) at 70 °C for 10 min. The study proved that orange peel has nutrients that enabled the microorganism understudy to produce high quantity (219 U/ml) of pectinase in very simple medium and can be better option for pectinase production on large scale for commercial use in industries.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Pectinase as a general term is used for pectic enzymes able to break down pectins. Pectins are negatively charged acidic glycosidic molecule found in all plants and form the major part of the plant cell walls. Pectinase have a share of 25 % in the global sale of food enzymes [1]. The global market has high demand for industrial enzymes [2]. The global market for industrial enzymes was worth nearly $4.5 billion in 2012 and nearly $4.8 billion in 2013. The market is expected to reach around $7.1 billion by 2018; a compound annual growth rate (CAGR) of 8.2 % from 2013 to 2018 is expected [3].
Pectic enzymes mainly occur in plants, bacteria, fungi, yeasts, insects, nematodes and protozoa. But microorganisms are generally used for industrial production of enzymes. Pectinases are produced from a wide variety of microorganisms such as bacteria [4–6], yeast [7, 8] and fungi [9, 10]. Out of more than 30 different genera of microorganism studied for pectinolytic activity, Erwinia, Bacillus, Saccharomyces, Kluyveromyces, Aspergillus, Penicillium, Fusarium and Rhizopus have been the genera most frequently studied in the last 15 years. They are used to produce many enzymes of industrial interest in processes relatively inexpensive and environment friendly. Enzymatic catalysis is always preferred over chemical methods due to its less aggressive and more specific nature.
The microorganism used for pectinase production depends on the type of culture required for pectinase production, (SSF or SmF), pH and thermal stability of the enzyme, and genotypic characteristic of the strain whether it is wild type or mutagenized [11–14]. Bacterial strains producing commercial enzymes are always preferred over fungal strains because of ease of fermentation process (for production) and implementation of strain improvement techniques or any modern technique to increase the yield of production.
As cell degrading enzymes, pectinases were long been used in the food and wine making industry [15, 16], in clarification of fruit juices [17–19], in retting of natural fibers i.e. ramie, hemp etc. [20, 21], in treatment of pectic waste water [22], in coffee and tea leaf fermentation [23], in oil extraction [24–26], in Starch extraction [27], in solid waste fermentation of citrus peel into bioethanol [28] and so.
Fruit processing industries produce huge amounts of orange peel which constitute almost 23 % of the fresh fruit weight. The average fruit weight for kinow is approximately 192 g and the average peel weight is 45 g [29]. Disposal of peels from industries is in itself a serious problem. As an alternative to the disposal of peels, it can be utilised as a substrate for production of pectinase enzyme [30, 31]. Traditionally, commercial source of pectin have been citrus peel and apple pomace.
Optimization of process conditions is one of the most critical stages in the development of an efficient and economic bioprocess. The classical method of studying one-variable-at-a-time (OVAT) is effective in some cases but for better optimization, studying the combined effect of all the factors is useful to consider. RSM is a model with a collection of statistical techniques wherein interactions between several variables can be identified with fewer experimental trials [32]. There are various advantages in using statistical methodologies in terms of rapid and reliable short listing of process conditions.
The goal of the present study was to investigate the best agricultural waste for maximum production of pectinase with an aim of cleaning environment by consuming solid waste in a fruitful way. The aim was also the application of RSM in the optimization process in order to achieve best results in minimum possible time and studying the interactions of different variables. The Bacillus licheniformis used in this study is alkalophilic (produce highest enzyme production at pH 9.5) although can grow in wide range of temperature from 4 to 60 °C and at pH from 4 to 10. It can better grow in LB medium as well its OD goes out of range at 630 nm after 18 h of incubation at 37 °C. It is also able to produce high enzyme activity in a medium with no need of addition of vitamins or minerals. The pectinase it produces is active in wide range of temperatures from 40 to 100 °C and wide pH range from 4 to 10.
Materials and Methods
Raw Material and Chemicals Used
Agricultural raw materials used as carbon source were citrus peel, apple pomace, carrot pomace, wheat bran, peanut husk and gram bran. The substrates were obtained from local market. These raw materials were dried in oven at 60 °C and ground to fine granules to be used in experiment. The chemicals used were different nitrogen sources such as Ammonium Chloride, Ammonium Sulphate, Ammonium biPhosphate, Sodium Nitrate, Urea, Yeast Extract, Peptone and Tryptone. Other chemicals used as medium were Sodium chloride, potassium phosphate, potassium chloride, magnesium sulphate, Cacium chloride, Iron sulphate, glycine, citrus pectin and Dinitro Salicylic Acid. All the chemicals were of analytical grade.
Inoculums Preparation
50 ml LB media was prepared in 250 ml flask and sterilized at 121 °C for 20 min in autoclave. The prepared medium was then cooled to room temperature and Bacillus licheniformis colony grown on petri plate was shifted to flask by sterilized loop in contamination free laminar flow cabinet. The flask was incubated at 37 °C for 24 h in shaking incubator to get a homogenous dense growth of bacteria. This inoculum was used to inoculate the experimental flasks. For each experiment, fresh inoculum was prepared every time to exclude the chances of contamination.
Submerged Fermentation and Optimization
Media prepared has the following basic W/V composition: NaNO3; 0.2 %, KH2PO4; 0.1 %, KCl; 0.05 %, MgSO4; 0.05 % in addition to variable quantity of citrus peel and trypton. 100 ml media was prepared in 250 ml flask and sterilized at 121 °C for 20 min in autoclave. The prepared medium was cooled to room temperature and inoculated by 0.3 mg (on dry cell mass basis) concentration of Bacillus licheniformis inoculums, transferred to 100 ml medium. The inoculum was prepared fresh as above. For every experiment, after completion of required incubation period, medium was centrifuged at 4 °C and 5432×g for 10 min to obtain clear supernatant. Supernatant was used for enzyme activity assay. The Carbon and Nitrogen sources were optimized by one-variable-at-a-time (OVAT) method while pH, Temperature, Inoculum size, Nitrogen content and Incubation time were optimized by RSM using Central Compost Design (CCD).
Pectinase Assay
One unit of pectinase activity is defined as the number of reducing sugar (galacturonic acid) obtained from 1 ml of pectinase in 1 min. The resulted galacturonic acid was analyzed using modified Miller [33] method. Measurement of pectinase activity was carried out using 0.7 ml of 0.5 % pectin dissolved in 0.1 M Glycine buffer pH 8 and 0.3 ml of crude enzyme. This reaction mixture was incubated at 70 °C for 10 min in shaking water bath. 1 ml of Dinitro Salicylic Acid (DNS) was added to the mixture and boiled in water bath 100 °C for 10 min. The OD of the mixture was measured at 575 nm absorbance using UV–Vis spectrophotometer.
Experimental
Optimization experiment was designed by using software Design Expert (DE) version 9. Five factors full factorial design was created comprising of 50 experimental units. Each factor to be optimized had low and high values. Design was analyzed statistically and results were presented in form of graphs and tables.
The following equation describes the regression model utilized in the factorial planning, including the interaction terms:
where Y is the predicted response, \( \beta_{ \circ } \) is the intercept term, β i , β ij , and β ii is the measure of the effect of variables χ i , χ i χ j and χ 2 i respectively.
Results and Discussion
Owing to increasing demand of pectinase in industry, the demand for its production is increasing day by day but the high cost of production is a limiting factor so it is necessary to improve its yield without increasing the cost of production. One way to achieve high yield is to use agricultural waste and fruit and vegetable waste which are ideal commodities that can increase the enzyme production without increasing cost of production. The other way is to optimize the fermentation conditions for better growth and enzyme production by the organism. Growth of organism and enzyme production by it is strongly influenced by medium composition so optimization of the medium composition and various cultural parameters is a key to open a way for low cost, and high yield of pectinase. In this study both ways were used to achieve our target in a better way.
Different agricultural wastes were used in this study as carbon sources to investigate among them the better producer and inducer of pectinase by Bacillus licheniformis. Fruit and vegetable peels and pomace as well as bran of several cereals were used as raw carbon source. Agricultural wastes previously used by many other investigators as raw carbon sources for pectinase production include citrus peel [9, 34–36], wheat bran [37], Carrot waste [38], Banana peel powder [39], date syrup [5], sunflower heads [40], corn cob [41] etc. In this study, several raw materials used as sole carbon source were wheat bran (WB), gram bran (GB), corn cob (CC), peanut shell (Pn.S), apple pomace (AP), carrot pomace (Cpom), banana peel (BP) orange peel (OP), citrus pectin (CP) and glucose (G) (Fig. 1). Orange peel at the rate of 2.5 % gave highest production of pectinase (34.9 U/ml) followed by wheat bran (29.67 U/ml), corn cob (28.88 U/ml), gram bran (26.26 U/ml), apple pomace (24.63 U/ml) and carrot pomace (24.45 U/ml). These results are in concordance with Kumari et al. [39] and Irshad et al. [42]. All of them used citrus peel as carbon source and obtained high pectinase activity as compared to other agro-wastes. Neagu et al. [43] observed wheat bran as best pectinase producer which is also in accordance to our results as in this study wheat bran was second best pectinase producer.
Various Nitrogen sources were also investigated to conclude their impact on pectinase activity. The nitrogen containing compounds used in this study are Ammonium Chloride, Ammonium Sulphate, Ammonium biPhosphate, Sodium Nitrate, Urea, Yeast Extract, Peptone and Tryptone and some of their combinations as shown in Fig. 2. Both organic and inorganic nitrogen sources can improve cell growth and synthesis of enzymes. Dinarvand et al. [44] observed that high productivity of enzymes was obtained under low C/N ratio condition. Among various nitrogen sources investigated for best production of pectinase, Tryptone (Tryp) 0.5 % alone when added in basic culture medium gave 38.86 U/ml of pectinase which is almost equal to a combination of Tryptone 0.25 % + Yeast Extract (YE) 0.25 % + salts (CaCl2 0.001 % + FeSO4 0.001 %) together capable of producing 38.06 U/ml (Fig. 2). Similar results were obtained by Embaby et al. [45], where trypton (0.5 %) gave maximum units.
Other physical parameters of medium optimized for pectinase production were pH, temperature, incubation time, inoculums size and nitrogen content. The combined effect of these parameters were studied and optimized by response surface methodology (RSM). Combination of various statistical methods useful for designing an experiment, optimizing a procedure or analyzing some data is actually RSM [46]. RSM includes the experimental designs that explores independent variables, as well as used to optimize values of independent variables that create desirable responses of variables [47]. Till now Pectinase and various other enzymes have been optimized by RSM [48–51]. The designed model was evaluated by analyzing the predicted versus actual values in order to quantitatively determine their relevance using RMSE analysis, analysis of variance (ANOVA) and p- and F values. The adequacy of the model was determined by lack of fit and the coefficient of determination, R2.
In order to actualize the predicted results, the experimental conditions optimized by Design Expert software were performed in 250 ml shake flasks to obtain the actual results. Using CCD for above five factors, 50 experimental combinations were generated. The Design of Experiment (DE) version 9.0 was used to set up a statistical model. The final equation in terms of significant coded factors is as follows:
where Y is predicted response and χ1, χ2, χ3, χ4 and χ5 are the coded values of independent variables indicating initial pH, Temperature, Inoculum size, Nitrogen content and Incubation time respectively.
The upper and lower limits of each variable as shown in Table 1 were chosen in the range found in literature servey of pectinase production. From data in Table 2, it is obvious that highest production of Pectinase was observed at 37 °C, pH 9.5, Inoculum size 0.3 ml, Nitrogen Content 0.5 % and Incubation time of 120 h. In these conditions, the production of pectinase was 219 U/ml. In repeated experiments, the above set of combinations gave the values of pectinase activity as 215 and 217 U/ml which were highest among other combinations and shows the validity of the experiment. The predicted values given in Table 2 closely match the actual values which is also an indication of the validity of the model.
The model/variables having larger F-ratio and smaller the p value are considered more significant (p < 0.05) [46]. It is evident from Table 3 that model F value of 128.08 implies the model is significant. There is only a 0.01 % chance that an F value this large could occur due to noise. Values of “Prob > F” <0.05 indicate model terms are significant. In this case pH (A), Temperature (B), Inoculum size (C), Nitrogen content (D) has p value <0.0001 while Incubation time (E) had p value of 0.0110 so all these variables are important. Interactions like pH and Inoculum size (AC), pH and Nitrogen content (AD), pH and Incubation time (AE), Temperature and Inoculum size (BC), Temperature and Nitrogen content (BD), Temperature and Incubation time (BE), Inoculum size and Nitrogen content (CD), Inoculum size and Incubation time (CE) and Nitrogen content and Incubation time (DE) are significant model terms because they all have p values < 0.05. Values >0.1000 indicate the model terms are insignificant so in this model the interaction AB (Interaction between pH and Temperature) has p value of 0.5610 means that this interaction is not important and will have very limited contribution in enhancing enzyme production. The “Lack of Fit p value” is also >0.1000. The “Lack of Fit F value” of 2.36 implies, the Lack of Fit is not significant relative to the pure error. There is a 10.38 % chance that a “Lack of Fit F value” this large could occur due to noise. Non-significant lack of fit is good showing the model is fit.
The fitness of the model is also judged by the R2 (coefficient of determination) value. R2 is the ratio of explained variation to the total variation as a measure of the degree of fit. As much as R2 value is closer to or equal to 1, means that the empirical data will better represent the actual data. Kutner et al. [52] suggested that a good fit model should possess a minimum R2 of 0.80 thus R2, in this model is 0.9826 (Table 4), suggesting a good fit. The predicted model seemed to better represent the observed values. Thus, the responses were sufficiently explained by the models.
The adjusted R2 is actually a corrected value for R2 after elimination of the unnecessary/non-significant model terms. If many non significant terms have been included in the model, the adjusted R2 would be remarkably smaller than the R2. In this study, the adjusted R2 for pectinase is 0.9749 which are very close to their corresponding R2 value in model. A high value of adjusted R2 also advocates significance of the models for all responses. The “Pred R-Squared” of 0.9585 is in reasonable agreement with the “Adj R-Squared” of 0.9749; i.e. the difference is <0.2. “Adeq Precision” measures the signal to noise ratio. A ratio >4 is desirable. In this model, ratio of 43.430 indicates an adequate signal. It means that the model can be used to navigate the design space. The C.V. (coefficient of variation) describes the extent to which the data are dispersed. The coefficient of variation is a measure of residual variation of the data relative to the size of the mean; the small values of CV give better reproducibility. The small CV value of 2.76 reveals that the experimental results are precise and reliable (Table 4). From all the above analysis, it is evident that model shows significant terms and is reliable to better explain the results.
Response Surface 3D Plots for Various Combinations of Variables
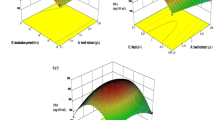
In order to examine the interaction between different variables and their impact on each other, 3D Response Surface plots were studied and their impact was evaluated. Response surface plots are generally aimed to describe the function of two factors at a time, keeping all other factors at fixed levels. So these plots are more helpful in understanding the interaction effects of these two factors. The 3D response surface graphs and contour plots are shown in Figs. 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12. Interactions among various variables can be easily explained by keenly observing the contour plots because, as a rule, the more elliptical the shape of the contour, the more good is the interaction between the two variables while circular shape indicates least interaction between the variables. Detail of various interactions between variables is described in forth coming lines below. From our results, shown in Fig. 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12, it is clear that there is a significant interaction between every two variables except one temperature versus pH interaction.
Temperature Verses pH Interaction
The pH and temperature (AB) interaction is not significant as is clear from Table 3 and from Fig. 3a, a′, it is obvious that low pH and low temperature has negative effect on pectinase production if other variables are kept at centeral level. The contour lines shown in Fig. 3a′, are almost parallal revealing least interaction among the variables. The corresponding variables are considered more significant (p < 0.05) as the F-ratio became larger and the p value became smaller [46]. But in our case it is vice versa and it is obvious from our results in Table 3 that pH and temperature interaction has smaller F value of 0.34 and larger p value of 0.5610.
Inoculum Size and pH Interaction
Inoculum size and pH interaction is significant in terms it has p value of 0.0011. Figure 4b, b′ illustrates clearly that maximum activity can be achieved as the pH increases but the activity declines with increase in inoculum size. At low pH and high inoculum size, there is a negative tendency in pectinase activity while at high pH and low inoculum the activity tendency is positive showing the alkalophilic nature of the organism understudy. This is in concordance with other studies illustrating that most Bacillus species have been reported to produce pectinase notably at an alkaline pH ranging from 7.0 to 9 [20, 53]. Decrease in activity at high Inoculumn size is probably due to limitations in getting adequate nutrients for metabolites production in limited and controlled environment of the experimental flasks.
Nitrogen Content and pH Interaction
Nitrogen content and pH interaction is important as it has p value of <0.0001 Fig. 5c, c′ shows the same interaction tendencies as were described for inoculum size and pH. The activity inceases with increase in pH level and decreases with increase in nitrogen content. The activity increase is from 100 U/ml to above 200 U/ml as the pH level rises from 6.5 to 10.5 which shows a 100 % increase in pectinase activity while increase in pectinase activity is from 120 to 140 U/ml only when nitrogen content is increased from 0.3 to 0.7 % which comprises only 16.66 % increase in activity. The initial pH of 9.5 of growth medium in this study indicates the alkalophilic nature of the organism understudy. Alkalophilic microorganism produces alkaline pectinase, most suited for various industrial processes; such an alkaline pectinase was produced by Bacillus halodurans isolated by Mei et al. [53]. It was able to grow under highly alkaline conditions at pH 10. The optimal initial pH value for producing pectinases depends on the microorganism nature for example, a much higher initial optimal pH of 8.5 has been reported for pectinase production by Bacillus pumilus dcsr1 [20] and pH 8.0 has also been noted for pectinase production by Cocci bacteria [6].
Incubation Time and pH Interaction
The 3D surface plot for incubation time verses pH interaction clearly demonstrates that keeping the pH at low level while increasing the incubation time has negative impact on pectinase activity. On the other hand high pH and increased incubation period together contribute to high pectinase yield. Figure 6d, d′ shows that there is a steep rise from 120 to 200 U/ml in pectinase activity at high incubation period and high pH. In our results as shown in Table 2, maximum pectinase activity of 219 U/ml was achieved at high incubation period of 120 h and at high pH of 9.5 which validate the Fig. 6d, d′.
Inoculums Size and Temperature Interaction
Figure 7e, e′ shows the 3D surface plot and contour plot for temperature and inoculums size. Graph show moderate tendencies in activity increase. Low inoculums size and moderate temperature seems favorable for high yield of pectinase. From results shown in Table 2, it is obvious that maximum pectinase production is favored at low inoculumn size of 0.3 ml containing 0.3 mg bacterial dry cell mass and moderate temperature of 37 °C. Incubation temperature is also an important factor for enzyme production by any organism. The optimum incubation temperature in our case was 37 °C. Below and above this, the organism did not produce as much enzyme as compared to its optimum temperature. Our findings are in accordance with Rehman et al. [54] in which case the isolated bacterium was Bacillus licheniformis KIBGE IB-21. Embaby et al. [45] found maximum polygalacturonase production by Bacillus licheniformis SHG10 at 37.8 °C, respectively. Prakash et al. [36] obtained maximum activity by Bacillus subtilis at 37 °C.
Nitrogen Content and Temperature Interaction
Figure 8f, f′ illustrates the interaction between these two factors. Low temperature and low nitrogen content favors pectinase yield to some extent while low temperature and high nitrogen content inhibits pectinase yield. The graph show a marked decrease in pectinase production when the nitrogen content increases from 0.5 to 0.7 %.
Incubation Time and Temperature Interaction
Incubation time has been shown to have an impact on enzyme yield. Reports have shown that many bacteria and fungi produce pectinase maximally at incubation time of 72 h and above [41] which is in concordance with results of this study. Incubation time and temperature interaction as shown in Fig. 9g, g′ reflects a moderate impact of both factors on pectinase activity. The contour lines shown in (g′) are almost parallal indicating moderate impact of the factors. Increase in temperature and incubation time favors pectinase production but not much noticeable increase is observed. Although keeping both factors at low level inhibits pectinase production.
Nitrogen Content and Inoculum Size
Nitrogen content and inoculums size interaction as shown in 3D surface plot Fig. 10h, h′ indicates that high inoculums size and high nitrogen content lowers the activity of pectinase and reflects negative effect but as both factors are brought to their minimum level, there is slight increase in pectinase activity but the overall impact is not very productive in sense of pectinase activity. If inoculumn size is very small, it will lead to reduced enzyme production due to insufficient growth of bacteria while high inoculumn size will lead to oxygen and nutrients depletion in culture broth [55].
Incubation Time and Inoculums Size
Figure 11i, i′ shows that low inoculums size and high incubation time has a remarkable impact. The greater impact of both factors can be observed at inoculums’s rate of 0.3 ml and incubation time ranging between 96 and 120 h. Slight increase in pectinase activity can be observed if temperature is kept at moderate level of 37 °C and inoculums size is increased gradually as is obvious from Table 2, Run # 16and 35. In these two experiments all other conditions are same except inoculums size. In Run # 16 inoculum size is 0.5 ml while in Run # 35, inoculum size is 0.3 ml and due to this change Run # 16 provides 161 U/ml while Run # 35 provides 191 U/ml.
Incubation Time and Nitrogen Content
Figure 12j, j′ demonstrates the interaction between incubation time and nitrogen content. The saddle shaped 3D graph illustrates somewhat unusual results. It is shown by both plots that lowest and highest levels of both factors inhibit the activity. Low nitrogen content and high incubation period encourages the production of pectinase. Incubation time has been shown to have an impact on enzyme yield. Reports have shown that many bacteria and fungi produce pectinase maximally at incubation time of 72 h and above [41] which is in concordance with results of this study. Maximum yield of pectinase is obtained by our indigenous B. licheniformis in 120 h of incubation period using citrus peel as a substrate. A decrease in yield was observed when incubation time was increased above 120 h.
Comparison of Actual and Predicted Values
Comparing actual and predicted values provide a way to evaluate the RSM model. Figure 13 shows a graphical presentation of the two values. As is obvious from Fig. 13, both values are laying close along the median line which shows that difference between actual and predicted values is less which is also obvious from Table 2. The actual and predicted values were also analyzed quantitatively in order to know their relatedness. For this purpose Root Mean Square Error (RMSE) analysis of both values was calculated. RMSE is a statistical method that measures how much error there is between two data sets. RMSE usually compares a predicted value and an observed value. RMSE value of our model is 3.708279 (Table 4) which is also a proof of the validity of the model. The value of R2, adjusted R2 and predicted R2 is another way to determine that how closely the actual and predicted values are. The adjusted R2 and predicted R2 of our model are 0.9749 and 0.9585 respectively (Table 4). The difference between the two R2 values is <0.2 so the difference between the two values is insignificant. All these statistical calculations aimed to evaluate the model suitability proved that model is best in terms of prediction.
So our results as well as the work of other researchers fully support the use of agro-industrial wastes as key ingredient in medium composition for maximum yield of pectinase. The use of RSM is also an easy and quick way to optimize conditions for microorganism growth and high quantity of enzyme production by them. Pranaw et al. [56] found 66.31 % increase in activity only due to proper interactions among variables in RSM. Optimization of a procedure can best be achieved by Derringer function or desirability [57]. The desirability function range is between d = 0–1. Zero desirability indicates that a response is completely undesirable while d = 1 shows a fully desired response. The value of desirability closer to 1 indicates that the conditions are fully optimum for a procedure. The desirability values shown in Table 2 were calculated using software Design of Experiment (DOE) version 9 to find the optimum conditions of the experiment understudy. Maximum desirability of 0.954 in Table 2, indicates 95 % desirability of the conditions understudy. So in our case the optimum conditions with 95 % desirability are pH 9.5, Temperature 37 °C, Inoculumn size 0.3 ml, Nitrogent content 0.7 % and Incubation period of 120 h. The results in Table 2, shows that these conditions are capable of producing maximum quantity of enzyme as compared to other conditions showing less desirability.
Conclusion
Our aim was to investigate agricultural waste for pectinase production. From this study, it is obvious that agricultural waste and agricultural by-products are a valuable raw material for enzyme production by fermentation technology. All the wastes used were able to produce pectinase in an appreciable quantity although orange peel and wheat bran were the best. RSM is also a good and reliable method of optimization with the help of which we were able to increase our pectinase production from 39 U/ml (Fig. 1) to 219 U/ml (Table 2). Further increase in pectinase production is expected if optimization is carried out in solid state fermentation. Mutagenesis will also be able to induce further increase in pectinase yield so there is always a room for further betterment.
References
Jayani, R.S., Saxena, S., Gupta, R.: Microbial pectinolytic enzymes: a review. Process Biochem. 9, 2931–2944 (2005)
Sarrouh, B., Santos, T.M., Miyoshi, A., Dias, R., Azevedo, V.: Up-to-date insight on industrial enzymes applications and global market. J. Bioprocess. Biotech. 4, 1–10 (2012)
Dewan, S.S.: Global Markets for Enzymes in Industrial Applications, Report Overview BIO030G (2014)
Ghani, M., Ansari, A., Aman, A., Zohra, R.R., Siddiqui, N.N., Qader, S.A.U.: Isolation and characterization of different strains of Bacillus licheniformis for the production of commercially significant enzymes. Pak. J. Pharm. Sci. 26, 691–697 (2013)
Qureshi, A.S., Bhutto, M.A., Chisti, Y., Khushk, I., Dahot, M.U., Bano, S.: Production of pectinase by Bacillus subtilis EFRL 01 in a date syrup medium. Afr. J. Biotechnol. 11, 12563–12570 (2012)
Ajit, K., Rita, S.: Production of alkaline pectinase by bacteria (Cocci sps.) isolated from decomposing fruit materials. J. Phytol. 4, 01–05 (2012)
Martos, M.A., Zubreski, E.R., Garro, O.A., Hours, R.A.: Production of Pectinolytic enzymes by the yeast Wickerhanomyces anomalus isolated from citrus fruits peels. Biotechnol. Res. Int. 2013, 1–7 (2013)
Merín, M.G., Mendoza, L.M., Morata de Ambrosini, V.I.: Pectinolytic yeasts from viticultural and enological environments: novel finding of Filobasidium capsuligenum producing pectinases. J. Basic Microbiol. 54, 835–842 (2014)
Göğüş, N., HakgüderTaze, B., DEMİR, H., Tari, C., Ünlütürk, S., Lahore, M.F.: Evaluation of orange peel, an industrial waste, for the production of Aspergillus sojae polygalacturonase considering both morphology and rheology effects. Turk. J. Biol. 38, 537–548 (2014)
Yadav, K.K., Garg, N., Kumar, D., Kumar, S., Singh, A., Mutukumar, M.: Application of response surface methodology for optimization of polygalacturonase production by Aspergillus niger. J. Environ. Boil. 36, 255–259 (2015)
Martos, M.A., Zubreski, E.R., Combina, M., Garro, O.A., Hours, R.A.: Isolation of a yeast strain able to produce a polygalacturonase with maceration activity of cassava roots. Food Sci. Technol. 33, 332–338 (2013)
Mehrnoush, A., Mustafa, S., Yazid, A.M.M.: Characterization of pectinase from mango (Mangifera indica cv. Chokanan) peel. J. Food Agric. Environ. 10, 85–88 (2012)
Ramirez, H.L., Gómez Brizuela, L., Úbeda Iranzo, J., Arevalo-Villena, M., Briones Pérez, A.I.: Pectinase immobilization on a chitosan-coated chitin support. J. Food Process Eng. 39, 97–104 (2015)
Thangaratham, T., Manimegalai, G.: Optimization and production of pectinase using agro waste by solid state and submerged fermentation. Int. J. Curr. Microbiol. App. Sci. 3, 357–365 (2014)
Mojsov, K., Ziberoski, J., Bozinovic, Z., Petreska, M.: A comparison of effects of three commercial pectolytic enzyme preparations in red wine making. Int. J. Pure Appl. Sci. Technol. 1, 127–136 (2010)
Sieiro, C., García-Fraga, B., López-Seijas, J., Silva, A.F. Da., Villa, T.G.: Microbial pectic enzymes in the food and wine industry. In: B. Valdez (ed.) Food Industrial Processes - Methods and Equipment, pp. 201–218 (2012). doi:10.5772/33403
Ajayi, A.A., Osunkoya, F.A., Peter-Albert, C.F., Olasehinde, G.I.: Clarification of apple juice with laboratoryproduced-pectinase obtained from the deteriorationof apple (Malus domestica) fruits by Aspergillus niger. Intl. J. Adv. Biotechnol. Res. 5, 134–140 (2014)
Sharma, H.P., Patel, H., Sharma, S.: Enzymatic extraction and clarification of juice from various fruits—a review. Trends Post Harvest Technol. 2, 01–14 (2014)
Uçan, F., Akyildiz, A., Erdal, A.L.: Effect of different enzymes and concentrations in the production of clarified lemon juice. J. Food Process. 2014, 14 (2014). doi:10.1155/2014/215854
Sharma, D.C., Satyanarayana, T.: Biotechnological potential of agro residues for economical production of thermoalkali-stable pectinase by Bacillus pumilus dcsr1 by solid-state fermentation and its efficacy in the treatment of ramie fibres. Enzyme Res. 2012, 7 (2012). doi:10.1155/2012/281384
Zhang, C., Yao, J., Zhou, C., Mao, L., Zhang, G., Yanhe, M.: The alkaline pectate lyase PEL168 of Bacillus subtilis heterologously expressed in Pichia pastoris is more stable and efficient for Degumming ramie fiber. BMC Biotechnol. 13, 26 (2013)
Tanabe, H., Yoshihara, Y., Tamura, K., Kobayashi, Y., Terashita, T., Sakai, T.: Preteatment of pectic waste water from orange canning process by an alkalophilic Bacillus sp. J. Ferment. Technol. 65, 243–246 (1987)
Murthy, P.S., Naidu, M.M.: Improvement of robusta coffee fermentation with microbial enzymes. Eur. J. Appl. Sci. 3, 130–139 (2011)
Mojtaba, A., Fardin, K.: Optimization of enzymatic extraction of oil from Pistacia Khinjuk seeds by using central composite design. Food Sci. Technol. 1, 37–43 (2013)
Mortabit, D., Mourad Zyani, M., Koraichi, S.I.: Improving olive oil yield from Moroccan Picholine by bacterial enzymes extract. IJISET 1, 717–728 (2014)
Perez, E.E., Fernández, M.B., Nolasco, S.M., Crapiste, G.H.: Effect of pectinase on the oil solvent extraction from different genotypes of sunflower (Helianthus annuus L.). J. Food Eng. 117, 393–398 (2013)
Dzogbefia, V.P., Ofosu, G.A., Oldham, J.H.: Evaluation of locally produced Saccharomyces cerevisiae pectinase enzyme for industrial extraction of starch from cassava in Ghana. Sci. Res. Essays 3, 365–369 (2008)
Wilkins, M.R., Widmer, W.W., Grohmann, K.: Simultaneous Saccharafication and Fermentation of citrus peel waste by Sacchromyces cerevisiea to produce ethanol. Process Biochem. 42, 1614–1619 (2007)
Altaf, N., Khan, A.R.: Growth and development of low seeded Kinnow mandarin fruits in dense plantation. J. Agric. Sci. Technol. 11, 191–198 (2009)
Diaz, A.B., Ory, I.D., Caro, I., Belandino, A.: Production of hydrolytic enzymes from grape pomace and orange peels mixed susbstrate fermentation by Aspergillus awamori. Chem. Eng. Trans. 17, 1143–1148 (2009)
Kumar, P.G., Suneetha, V.: Natural, culinary fruit peels as a potential substrate for pectinolytic enzyme int. J. Drug Dev. Res. 6, 109–118 (2014)
Anvari, M., Khayati, G.: The effect of citrus pulp type on pectinase production in solid-state fermentation: process evaluation and optimization by Taguchi design of experimental (DOE) methodology. J. Biosci. Biotech. 3, 227–233 (2014)
Miller, G.L.: Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 (1959)
Jagiasi, S.R.: Production of bacterial pectinase from agro-industrial wastes. National conference on biodiversity: status and challenges in conversion. FAVEO (2013)
Mandhania, S., Jain, V., Malhotra, S.P.: Culture optimization for enhanced production of microbial pectin methylesterase under submerged conditions. Asian J. Biotech. 5, 12–22 (2010)
Prakash, S., Karthik, R., M, T.V., Sridhar, B., Bharath, P.G.: Optimization and production of pectinase from Bacillus subtilis (mtcc 441) by using orange peel as a substrate. Int. J. Recent Sci. Res. 5, 1177–1179 (2014)
Motwani, D.R., Meshram, V.G., Jambhulkar, V.S.: Partial purification of pectinase produced by Aspergillus niger grown on wheat bran. IJSER 4, 345–365 (2013)
Patil, R.C., Murugkar, T.P., Shaikh, S.A.: Extraction of pectinase from pectinolytic bacteria isolated from carrot waste. Int. J. Pharma Bio Sci. 3, 261–266 (2012)
Kumari, B.L., Sudhakar, P., Hemamalini, K., Satya sree, N., Vijetha, P.: Studies on pectinase production by Bacillus Subtilis using agro-industrial wastes. RJPBCS 5, 330–339 (2014)
Patil, S.R., Dayanand, A.: Exploration of regional agrowastes for the production of pectinase by Aspergillus niger. Food Technol. Biotechnol. 44, 289–292 (2006)
Oyeleke, S., Oyewole, O., Egwim, E., Dauda, B., Ibeh, E.: Cellulase and pectinase production potentials of Aspergillus Niger isolated from corn cob. Bayero J. Pure Appl. Sci. 5, 78–83 (2012)
Irshad, M., Anwar, Z., Mahmood, Z., Aqil, T., Mehmmod, S., Nawaz, H.: Bio-processing of agro-industrial waste orange peel for induced production of pectinase by Trichoderma viridi; its purification and characterization. Turk. J. Biochem. 39, 9–18 (2014)
Neagu, D.A., Destain, J., Thonart, P., Socaciu, C.: Effects of different carbon sources on pectinase production by Penicillium oxalicum. Bull. UASVM Agric. 69, 327–333 (2012)
Dinarvand, M., Rezaee, M., Masomian, M.: Effect of C/N ratio and media optimization through response surface methodology on simultaneous productions of intra- and extracellular inulinase and invertase from Aspergillus niger ATCC 20611. BioMed Res. Int. 2013(Article ID 508968), 13 (2013)
Embaby, A.M., Masoud, A.A., Marey, H.S., Shaban, N.Z., Ghonaim, T.M.: Raw agro-industrial orange peel waste as a low cost effective inducer for alkaline polygalacturonase production from Bacillus licheniformis SHG10. Springerplus. 3, 327 (2014)
Montgomery, D.C.: Design and Analysis of Experiments: Response Surface Method and Designs. Wiley, New York (2005)
Oehlert, G.W.: Design and Analysis of Experiments: Response Surface Design. Freeman and Company, New York (2000)
Chaturvedi, S., Kohli, K.U., Rajni, S., Khurana, S.M.P.: Statistical optimization of medium composition for Xylanase production by solid state fermentation using Agroresidues. Am. J. Microbiol. Res. 3, 85–92 (2015)
Dave, B.R., Parmar, P., Sudhir, A., Panchal, K., Subramanian, R.B.: Optimization of process parameters for cellulase production by Bacillus licheniformis MTCC 429 using RSM and molecular characterization of cellulase gene. J. Bioprocess. Biotech. 5, 212 (2015). doi:10.4172/2155-9821.1000212
Bhunia, B., Dutta, D., Chaudhuri, S.: Extracellular alkaline protease from Bacillus licheniformis NCIM-2042: improving enzyme activity assay and characterization. Eng. Life Sci. 11, 207–215 (2011)
Sangeetha, R., Geetha, A., Arulpandi, I.: Concomitant production of protease and lipase by Bacillus licheniformis VSG1: production, purification and characterization. Braz. J. Microbiol. 41, 179–185 (2010) ISSN 1517-8382
Kutner, M., Nachtsheim, C., Neter, J., Li, W.: Applied Linear Statistical Models. McGraw-Hill, New York (2005)
Mei, Y., Chena, Y., Zhai, R., Liu, Y.: Cloning, purification and biochemical properties of a thermostable pectinase from Bacillus halodurans M29. J. Mol. Catal. B Enzym. 94, 77–81 (2013)
Rehman, H.U., Qader, S.A.U., Aman, A.: Polygalacturonase: production of pectin depolymerising enzyme from Bacillus licheniformis KIBGE IB-21. Carbohydr. Polym. 90, 387–391 (2012)
Rahman, R.N.Z.A., Lee, P.G., Basri, M., Salleh, A.B.: Phisical factors affecting the production of organic solvent- tolarent protease by Pseudomonas aeruginosa strain K. Bioresour. Technol. 96, 429–436 (2005)
Pranaw, K., Singh, S., Dutta, D., Chaudhuri, S., Ganguly, S., Nain, L.: Statistical optimization of media components for production of fibrinolytic alkaline metalloproteases from Xenorhabdus indica KB-3. Biotechnol. Res. Int. 2014, 11 (2014). doi:10.1155/2014/293434
Bezerra, M.A., Santelli, R.E., Oliveira, E.P., Villar, L.S., Escaleira, L.A.: Response surface methodology (RSM) as a tool for optimization in analytical chemistry. A Rev. Talanta 76, 965–977 (2008)
Acknowledgments
I am highly thankful to Dr. Romana Tabassam, Deputy Chief Scientist in Industrial Biotechnology Division NIBGE, Faisalabad. The entire study was carried out in her supervision. Also many thanks to Higher Education Commission, Pakistan for grant of fellowship for smooth conductance of study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bibi, N., Ali, S. & Tabassum, R. Statistical Optimization of Pectinase Biosynthesis from Orange Peel by Bacillus licheniformis Using Submerged Fermentation. Waste Biomass Valor 7, 467–481 (2016). https://doi.org/10.1007/s12649-015-9470-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-015-9470-4